Abstract
Evolutionary biologists have identified several factors that could explain the widespread phenomena of sex and recombination. One hypothesis is that host–parasite interactions favor sex and recombination because they favor the production of rare genotypes. A problem with many of the early models of this so-called Red Queen hypothesis is that several factors are acting together: directional selection, fluctuating epistasis, and drift. It is thus difficult to identify what exactly is selecting for sex in these models. Is one factor more important than the others or is it the synergistic action of these different factors that really matters? Here we focus on the analysis of a simple model with a single mechanism that might select for sex: fluctuating epistasis. We first analyze the evolution of sex and recombination when the temporal fluctuations are driven by the abiotic environment. We then analyze the evolution of sex and recombination in a two-species coevolutionary model, where directional selection is absent (allele frequencies remain fixed) and temporal variation in epistasis is induced by coevolution with the antagonist species. In both cases we contrast situations with weak and strong selection and derive the evolutionarily stable (ES) recombination rate. The ES recombination rate is most sensitive to the period of the cycles, which in turn depends on the strength of epistasis. In particular, more virulent parasites cause more rapid cycles and consequently increase the ES recombination rate of the host. Although the ES strategy is maximized at an intermediate period, some recombination is favored even when fluctuations are very slow. By contrast, the amplitude of the cycles has no effect on the ES level of sex and recombination, unless sex and recombination are costly, in which case higher-amplitude cycles allow the evolution of higher rates of sex and recombination. In the coevolutionary model, the amount of recombination in the interacting species also has a large effect on the ES, with evolution favoring higher rates of sex and recombination than in the interacting species. In general, the ES recombination rate is less than or equal to the recombination rate that would maximize mean fitness. We also discuss the effect of migration when sex and recombination evolve in a metapopulation. We find that intermediate parasite migration rates maximize the degree of local adaptation of the parasite and lead to a higher ES recombination rate in the host.
THE vast majority of species reproduce sexually, at least occasionally. Such a widespread success of sex and recombination is problematic given the strong fitness costs associated with sexual reproduction (e.g., the twofold cost of sex, the cost of breaking a favorable combination of genes, the cost of finding and courting a mate, etc.). This problem has attracted considerable theoretical attention (Maynard-Smith 1978; Kondrashov 1993; Barton and Charlesworth 1998). Although some theories invoke proximate (or mechanistic) explanations (e.g., sex might be induced to promote DNA repair), we focus on evolutionary (or generative) hypotheses that focus on the effects of sex and recombination on genetic associations. The modifier theory approach, introduced by Nei (1967), has helped to clarify the conditions under which sex and recombination might evolve by considering the dynamics at genes that alter (“modify”) the mode of reproduction and the frequency of crossover events.
In a general model of recombination evolution, Barton (1995) showed that two conditions favor a modifier allele that increases recombination. First, recombination can be favored if it breaks apart less-fit combinations of genes and, consequently, increases the mean fitness of the descendants, a so-called short-term benefit of recombination (because a modifier inducing more recombination immediately increases in frequency). Second, recombination can be favored if it increases the variance in fitness of the descendants and, consequently, the efficacy of selection, a so-called long-term benefit of recombination (because the frequency of the modifier changes by hitchhiking with the most fit of the variants produced, which take several generations to spread). In either case, selection on the modifier of recombination is indirect, with frequency changes at the modifier locus occurring via associations with other loci that are directly under selection.
The main determinant of whether these short-term and long-term effects actually do benefit recombination is the type of genetic associations (linkage disequilibria) that exist among selected alleles. In general, evolutionary hypotheses can be classified according to the forces that generate these genetic associations (Kondrashov 1993). Here, we review four leading hypotheses:
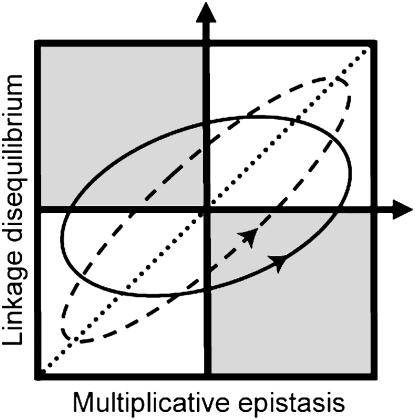
If selection is constant in time, linkage disequilibrium develops with the same sign as the multiplicative epistasis, which is a measure of the curvature of the fitness surface measured on a log scale (Felsenstein 1965; Eshel and Feldman 1970). Basically, selection builds up allelic combinations that work well together, which causes disequilibrium to have the same sign as epistasis and implies that genetic associations among alleles tend to increase fitness, on average. In this case, the main short-term effect of recombination is to decrease the average fitness of offspring. Nevertheless, evolution favors higher rates of recombination via a long-term benefit, as long as epistasis is negative and sufficiently weak that the short-term costs are not too severe (Feldman et al. 1980; Barton 1995; Otto and Feldman 1997). With negative epistasis, advantageous alleles at one locus become associated with disadvantageous alleles at a second locus (negative disequilibrium), which hinders a population's response to selection. Higher rates of recombination can be favored in this situation because it breaks this linkage disequilibrium and thus facilitates the response to natural selection.
If epistasis fluctuates over time, a lag between epistasis and linkage disequilibrium can develop, leading to a mismatch at some points in time between which combinations of alleles are most fit and are most common (Sturtevant and Mather 1938; Charlesworth 1976; Maynard Smith 1978; Barton 1995). In this situation, recombination can be favored because it breaks apart the currently maladapted allele combinations and increases the mean fitness of descendants (a short-term benefit). Because such mismatches must occur often to have much influence, however, this mechanism works only under restrictive parameter values (Charlesworth 1976; Barton 1995). In particular, Barton (1995) found that epistasis must fluctuate very rapidly for this mechanism to work: epistasis must change sign every 2–5 generations, implying cycles with a period of 4–10 generations (the so-called “Barton zone”; Peters and Lively 1999) to account for high rates of recombination.
When the population is finite, the interaction between genetic drift and selection yields negative linkage disequilibrium (Hill and Robertson 1966). In this case, increased recombination can evolve because recombination increases the variance in fitness of the descendants (a long-term benefit) through the production of high-fitness genotypes that are rare or absent due to genetic drift (Felsenstein and Yokoyama 1976; Otto and Barton 2001; Iles et al. 2003; Barton and Otto 2005; Keightley and Otto 2006; Martin et al. 2006). This process can select for recombination over a range of both positive and negative epistasis, but it requires that drift is neither too weak (no effect on linkage disequilibrium) nor too strong (polymorphisms are rapidly lost). Nevertheless, a drift-based advantage to recombination can occur even in very large populations as long as they are spatially structured (Martin et al. 2006) and/or selection acts on a large number of loci (Iles et al. 2003).
Selection that varies over space can promote the evolution of recombination in the absence of genetic drift. With migration among populations, spatial heterogeneity in selection can generate positive or negative linkage disequilibrium and select for or against recombination, depending on the sign of epistasis (Pylkov et al. 1998; Lenormand and Otto 2000). Modifier theory thus provides a general framework that allows us to formalize and compare different evolutionary hypotheses for the evolution of sex and recombination.
All of the theories reviewed above focus on a single species in which recombination is evolving. Where do species interactions fit within this framework? The Red Queen hypothesis posits that the biotic environment of a species is continually changing due to the coevolution of surrounding species, including parasites, pathogens, predators, competitors, etc. Within this changing biotic environment, sex and recombination might be favored as mechanisms that generate rare combinations of alleles to which antagonistic species are not adapted (akin to the advantage of recombination in an abiotically fluctuating environment). Unfortunately, the intrinsic complexity of coevolutionary dynamics impedes a general analytical treatment of the problem. In fact, most theoretical articles on the Red Queen hypothesis are based on the exploration of complex simulation models, making it difficult to determine exactly what selects for sex (Otto and Michalakis 1998). For example, in one of the first simulation models used to formalize the Red Queen hypothesis in a metapopulation (Ladle et al. 1993), all four of the factors listed above may have been responsible for the evolution of host recombination: (i) directional selection, (ii) fluctuating epistasis, (iii) interaction between drift and selection, and (iv) spatial covariance in selection. Is it the synergistic action of these multiple factors that explains the success of sex in these models (West et al. 1999) or could each factor work in isolation?
To answer this question one needs to analyze simpler models with fewer factors affecting the evolution of sex and recombination. Recent studies of the Red Queen hypothesis (Peters and Lively 1999; Otto and Nuismer 2004) have focused on two forces: directional selection and fluctuating epistasis. Otto and Nuismer (2004) developed a general model of species interactions and showed that under weak selection, the epistasis generated by most genetic models underlying species interactions (matching-genotypes model, gene-for-gene model, a quantitative trait model) is too strong relative to the strength of selection to favor the evolution of high rates of recombination. When epistasis is strong relative to selection, the main effect of sex and recombination is to break apart the good gene combinations that allowed parents to survive and reproduce in the face of the current suite of coevolving species. Consequently, the short-term cost of sex (the recombination load) is too severe relative to the long-term benefit (the production of rare genotypes). Nevertheless when selection is strong, simulations revealed that recombination can be favored (Peters and Lively 1999; Otto and Nuismer 2004). What force selects for recombination in these cases? Peters and Lively (1999) examined the coevolutionary dynamics of a matching-alleles model, where parasites must match a host's alleles to infect, and found that there is often a discrepancy between which combinations of alleles are currently present (disequilibria) and which are currently favored (epistasis). They thus concluded that fluctuating epistasis is the primary force driving the evolution of recombination when selection is strong, although directional selection also contributed (see also Lythgoe 2000). In summary, when selection is weak, recombination induces a short-term cost by breaking apart good combinations of alleles, which prevents the evolution of recombination despite the potential for long-term benefits. In contrast, when selection is strong, the short-term effect of recombination is to break apart currently maladapted gene combinations, which allows modifiers that increase recombination to spread.
In the above studies (Peters and Lively 1999; Lythgoe 2000; Otto and Nuismer 2004), the case where selection is strong relative to the rates of sex and recombination was examined only through simulation (Pomiankowski and Bridle 2004). In the present article, we examine simpler models that allow us to focus entirely on the analysis of fluctuating epistasis as a factor favoring the evolution of sex and recombination. Specifically, we use fitness functions that lead allele frequencies to equilibrate at 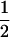 , after which point directional selection is absent. Although these models are not empirically justified, we consider them to represent extreme scenarios, where directional selection is absent and where we can get an analytical handle on the impact of fluctuations in epistasis, which are observed in more biologically realistic models. This approach was introduced by Nee (1989), who found that coevolutionary interactions provide an advantage to recombination that is independent of the period of the fluctuations. This contrasts with the results of Peters and Lively (1999, 2007), who found that host–parasite coevolution does not favor high rates of recombination much outside the Barton zone (i.e., fluctuating epistasis with a period between 4 and 10 generations).
, after which point directional selection is absent. Although these models are not empirically justified, we consider them to represent extreme scenarios, where directional selection is absent and where we can get an analytical handle on the impact of fluctuations in epistasis, which are observed in more biologically realistic models. This approach was introduced by Nee (1989), who found that coevolutionary interactions provide an advantage to recombination that is independent of the period of the fluctuations. This contrasts with the results of Peters and Lively (1999, 2007), who found that host–parasite coevolution does not favor high rates of recombination much outside the Barton zone (i.e., fluctuating epistasis with a period between 4 and 10 generations).
To elucidate the mechanisms at work, we adopt a two-step approach. We first analyze a one-species model where fluctuations in epistasis are governed by the abiotic environment (as in Sasaki and Iwasa 1987). We then study a second model where fluctuations in epistasis are driven by host–parasite coevolution (as in Nee 1989). Together these two models help clarify the extent to which fluctuating epistasis—with or without coevolution—favors the evolution of sex and recombination. In both cases, we consider whether evolutionarily stable rates of recombination coincide with the rate of recombination that maximizes mean fitness (Sasaki and Iwasa 1987; Nee 1989).
Finally, we use these two models to analyze the evolution of sex and recombination in a metapopulation, accounting for migration among patches. Furthermore, we discuss the link between the evolution of recombination and local adaptation (a measure of mean fitness in these models). Throughout, we consider only deterministic models with large local and global population sizes.
ONE-SPECIES MODEL
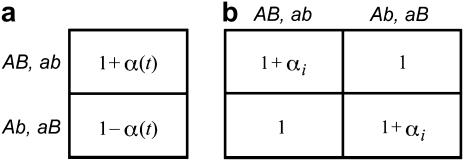
We first consider a single haploid and hermaphroditic organism with nonoverlapping generations living in an isolated and large population of constant size (no drift). Using a deterministic model, we follow haplotype frequencies through a life cycle consisting of a census, selection, and reproduction. We further assume random mating when sex occurs. Selection acts on two loci (A and B) with two alleles (A/a, B/b). The phenotype of an individual is set to 0 if it is either AB or ab and to 1 if it is aB or Ab. Phenotype 0 has fitness 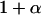 , while phenotype 1 has fitness
, while phenotype 1 has fitness 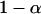 (Figure 1a). It is assumed that the quantity
(Figure 1a). It is assumed that the quantity 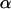 varies sinusoidally over time,
varies sinusoidally over time,
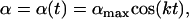 |
(1) |
where 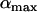 measures the amplitude of the fitness oscillations and
measures the amplitude of the fitness oscillations and 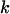 is the speed of these oscillations, which varies between 0 (infinitely slow oscillations) and
is the speed of these oscillations, which varies between 0 (infinitely slow oscillations) and 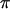 (
(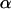 changes sign each generation). The corresponding period of the cycle is
changes sign each generation). The corresponding period of the cycle is 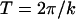 . This fitness regime was first introduced in the case of a single species by Sturtevant and Mather (1938) and was analyzed by Sasaki and Iwasa (1987).
. This fitness regime was first introduced in the case of a single species by Sturtevant and Mather (1938) and was analyzed by Sasaki and Iwasa (1987).
Figure 1.—
Coefficients of selection for (a) the one-species model and (b) the two-species coevolutionary model. The values in each case give the fitness of the genotypes of the focal species i (indicated to the left). In the one-species model, selection varies through time because α(t) fluctuates (extrinsically imposed). In contrast, in the coevolutionary model, the αi remain constant but selection fluctuates because of the temporal variation in the genotype frequencies of the coevolving species (b, top).
To investigate the evolution of sex and recombination, we allow the probability of sex and/or the rates of recombination to depend on a third modifier locus (M) with two alleles (M/m), with gene order MAB. Thus, there are eight possible haplotypes (MAB, MAb, MaB, Mab, mAB, mAb, maB, mab, labeled from 1 to 8), whose frequencies among adults are xj, for j = 1–8. Whenever convenient, pj will denote the frequency of allele j. The modifier locus is assumed to be neutral (but this assumption is relaxed in the discussion), and the fitness of genotype j is thus 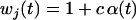 , where
, where 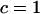 if
if 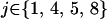 and
and 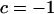 otherwise. After selection, the haplotypic frequencies are
otherwise. After selection, the haplotypic frequencies are 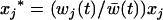 for haplotype j, where
for haplotype j, where 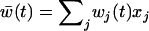 is the mean fitness of the population.
is the mean fitness of the population.
Following selection, the probability that two haploid individuals carrying modifier alleles k and l engage in sexual reproduction is σkl. Haploid individuals that do not mate reproduce asexually. Sexual mating is followed immediately by meiosis with recombination between loci A and B at rate rkl and between loci M and A at rate Rkl. We let ckl denote the probability of a double crossover. Appropriate choices of ckl thus allow interference among chiasmata and alternative gene orders to be considered. In deriving the recursions for a haploid population, the probabilities of undergoing sex and the rates of recombination always enter as products. Therefore, we simplify the equations using the compound parameters
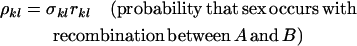 |
 |
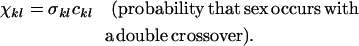 |
Because recombination affects the array of offspring produced only when it occurs between heterozygous loci, the compound parameters involving the M locus (ψkl and χkl) are relevant only in Mm heterozygotes, so we may drop the kl subscript. A major advantage of using these compound parameters is that in a haploid population, the same equations apply whether the modifier locus alters the probability of sex or the rates of recombination or both. The special case of a sexual form arising within an asexual population with no gene flow between them can also be modeled by setting ρmm > 0 and ρMm = ρMM = ψ = χ= 0, in which case only group selection acts on the frequency of sex (Felsenstein 1974). We used the recursion equations presented in Otto and Nuismer (2004) to follow the frequencies of each genotype after reproduction.
Change in modifier frequency:
To determine if higher rates of sex and recombination are favored we ask whether an allele m that increases the rate of sex and recombination, ρkl, rises in frequency. After selection and sexual reproduction the change in frequency of m exactly equals
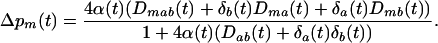 |
(2a) |
In the above equation, 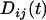 and
and 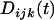 refer to the linkage disequilibria between two loci (i and j) and between three loci (i, j, and k) at time t (see appendix a). We simplify the notation in Equation 2a by defining new variables to measure the departure of the selected allele frequencies from
refer to the linkage disequilibria between two loci (i and j) and between three loci (i, j, and k) at time t (see appendix a). We simplify the notation in Equation 2a by defining new variables to measure the departure of the selected allele frequencies from  :
: 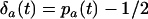 and
and 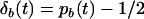 . If we assume that α and linkage disequilibria are small (of order ζ), Equation 2a becomes
. If we assume that α and linkage disequilibria are small (of order ζ), Equation 2a becomes
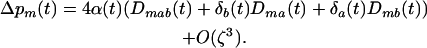 |
(2b) |
Equation 2 shows that the fate of the modifier depends on the associations between the modifier locus, M, and the loci A and B; that is, the frequency of the allele m evolves only through indirect selection (hitchhiking) with loci A and B. To proceed in the analysis, we thus need to understand the dynamics of these genetic associations.
A QLE analysis:
As a first attempt to predict the fate of the modifier, let us assume that the processes generating disequilibria (epistasis in this model) are weak relative to the processes reducing disequilibria (sex and recombination). In this case, the disequilibria between loci quickly reach their steady-state values predicted on the basis of current allele frequencies, the selection coefficients, and the rates of sex and recombination. This steady state is known as the “quasilinkage equilibrium” (QLE). Assuming a modifier that has only a small effect on sex and recombination, the dynamics of a modifier allele in a single population can be approximated at QLE by
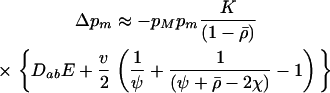 |
(3) |
Equation 3 was derived following the methods described in Barton (1995). The term K measures the efficacy of the modifier, which equals the average effect of the modifier, 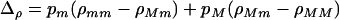 , divided by the probability that recombination breaks apart a three-locus haplotype
, divided by the probability that recombination breaks apart a three-locus haplotype 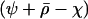 , thereby breaking down associations among the modifier and selected loci. The term
, thereby breaking down associations among the modifier and selected loci. The term 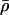 is the average rate of sex and recombination between loci A and B. The term E measures the amount of epistasis defined on a multiplicative scale:
is the average rate of sex and recombination between loci A and B. The term E measures the amount of epistasis defined on a multiplicative scale: 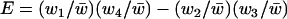 . Finally, v measures the contribution of linkage disequilibrium to the additive genetic variance in fitness and equals
. Finally, v measures the contribution of linkage disequilibrium to the additive genetic variance in fitness and equals 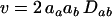 , where aj is the total selective force acting on allele j:
, where aj is the total selective force acting on allele j: 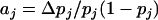 . When v is positive (negative), the linkage disequilibrium increases (decreases) the frequency of the currently most- and least-fit combinations of alleles, which accelerates (hinders) evolutionary change (Barton 1995).
. When v is positive (negative), the linkage disequilibrium increases (decreases) the frequency of the currently most- and least-fit combinations of alleles, which accelerates (hinders) evolutionary change (Barton 1995).
The direction of selection of the modifier depends on the sum of two terms (Barton 1995; Lenormand and Otto 2000). The first term (proportional to 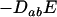 ) measures the short-term effect of recombination: recombination is beneficial when it breaks apart unfavorable genetic associations (i.e., when disequilibria have opposite sign to the current value of epistasis). The second term (proportional to
) measures the short-term effect of recombination: recombination is beneficial when it breaks apart unfavorable genetic associations (i.e., when disequilibria have opposite sign to the current value of epistasis). The second term (proportional to 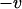 ) measures the long-term effect of recombination: recombination is beneficial when breaking down linkage disequilibria increases the additive genetic variance in fitness (this occurs when
) measures the long-term effect of recombination: recombination is beneficial when breaking down linkage disequilibria increases the additive genetic variance in fitness (this occurs when 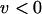 ).
).
Following Barton (1995), we can also determine the QLE value of the disequilibrium between loci A and B when epistasis fluctuates according to Equation 1 (appendix a),
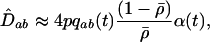 |
(4a) |
where
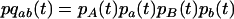 |
and
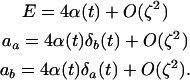 |
(4b) |
At this QLE, 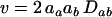 is of smaller order than
is of smaller order than 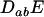 , and Equation 3 reduces to
, and Equation 3 reduces to
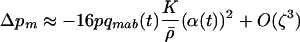 |
(5) |
with 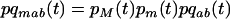 . If m is an allele increasing the frequency of sex and/or recombination, then
. If m is an allele increasing the frequency of sex and/or recombination, then 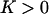 , and Equation 5 is always negative, predicting that sex and recombination should always be selected against. Essentially, sex and recombination are not favored because epistasis is too strong relative to the strength of directional selection, and the main effect of recombination is to break apart good gene combinations. The short-term cost of sex and recombination thus outweighs any potential long-term benefits.
, and Equation 5 is always negative, predicting that sex and recombination should always be selected against. Essentially, sex and recombination are not favored because epistasis is too strong relative to the strength of directional selection, and the main effect of recombination is to break apart good gene combinations. The short-term cost of sex and recombination thus outweighs any potential long-term benefits.
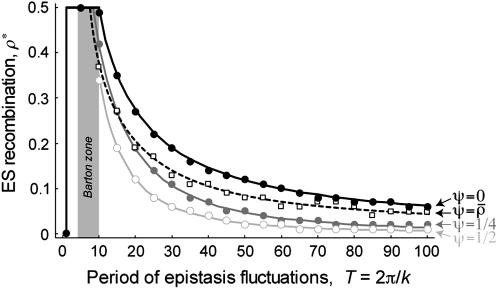
This prediction matches the results of simulations presented by Sasaki and Iwasa (1987) when the period is very large and recombination rates are initially high (see also our simulation results in Figure 2). When the period is short or recombination rates are low, however, some positive level of recombination can evolve. The discrepancy with the QLE analysis comes from the fact that fluctuations in epistasis that are rapid relative to the frequency of sex and recombination drag the linkage disequilibria away from their QLE values.
Figure 2.—
Evolutionarily stable recombination rates vs. the period of epistasis fluctuations in the one-species model. The circles are the ES values of 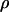 obtained from numerical simulations that allowed the recombination rate to evolve while
obtained from numerical simulations that allowed the recombination rate to evolve while 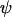 was fixed at
was fixed at 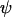 = 0 (solid circles),
= 0 (solid circles), 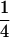 (shaded circles), or
(shaded circles), or 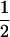 (open circles); the simulations coincide well with the analytical expectations based on Equation 16b (solid curves). The squares are the ES values of
(open circles); the simulations coincide well with the analytical expectations based on Equation 16b (solid curves). The squares are the ES values of  obtained from numerical simulations assuming that that the three loci are equidistant (i.e.,
obtained from numerical simulations assuming that that the three loci are equidistant (i.e., 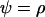 ); the simulations match the analytical expectation based on Equation 16a (dashed curve). While αmax = 0.01 was used, other values of αmax yielded similar results. The area in gray denotes the window of parameter values where epistasis fluctuates with a period between 4 and 10 generations (the so-called “Barton zone”; Peters and Lively 1999).
); the simulations match the analytical expectation based on Equation 16a (dashed curve). While αmax = 0.01 was used, other values of αmax yielded similar results. The area in gray denotes the window of parameter values where epistasis fluctuates with a period between 4 and 10 generations (the so-called “Barton zone”; Peters and Lively 1999).
Recursion analysis:
Following the analysis of Barton (1995, Appendix 4) we now take into account fluctuations in linkage disequilibria without assuming that linkage disequilibrium is always and instantaneously at its steady-state value (the QLE assumption). This recursion analysis is greatly simplified by the peculiar dynamical behavior of this simplified model. As pointed out by Sasaki and Iwasa (1987), the selection regime used rapidly yields a situation where allele frequencies converge toward 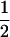 [i.e.,
[i.e., 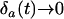 and
and 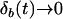 when
when 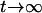 ]. Sasaki and Iwasa (1987) proved the convergence in a continuous-time model, and numerical simulations of our discrete-time model confirm this convergence as long as some recombination occurs. Once the allele frequencies have reached
]. Sasaki and Iwasa (1987) proved the convergence in a continuous-time model, and numerical simulations of our discrete-time model confirm this convergence as long as some recombination occurs. Once the allele frequencies have reached 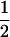 , the dynamics of the system are very simple, and only linkage disequilibria vary through time. Furthermore, the dynamics of the modifier locus are simplified, because
, the dynamics of the system are very simple, and only linkage disequilibria vary through time. Furthermore, the dynamics of the modifier locus are simplified, because 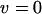 when directional selection is absent [
when directional selection is absent [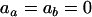 , see (4b)]. In other words, the evolution of a modifier of recombination is governed only by the short-term effect of recombination, and the change in the frequency of allele m on the M locus exactly equals
, see (4b)]. In other words, the evolution of a modifier of recombination is governed only by the short-term effect of recombination, and the change in the frequency of allele m on the M locus exactly equals
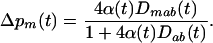 |
(6a) |
If we further assume that 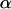 and linkage disequilibria are small (of order ζ) this yields (using 4b)
and linkage disequilibria are small (of order ζ) this yields (using 4b)
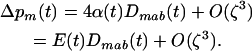 |
(6b) |
Thus the direction of selection on the modifier depends only on the product of epistasis and the three-locus disequilibrium. This three-locus disequilibrium depends on the two-locus disequilibrium between loci A and B. In appendix a, we assume that the effect of the modifier is weak (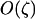 ) to derive an approximation for the three-locus disequilibrium,
) to derive an approximation for the three-locus disequilibrium,
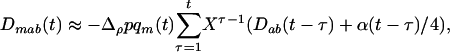 |
(7) |
where 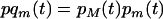 and
and 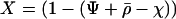 . Using (7) in (6b) we get
. Using (7) in (6b) we get
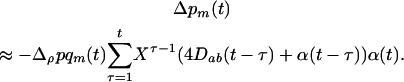 |
(8) |
Because 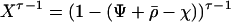 is a rapidly decreasing function of τ in the presence of sex and recombination, Barton (1995) argued that only the first few terms in the above sum have a major impact on the evolution of sex and recombination.
is a rapidly decreasing function of τ in the presence of sex and recombination, Barton (1995) argued that only the first few terms in the above sum have a major impact on the evolution of sex and recombination.
For sex and recombination to be favored, 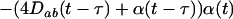 must be positive, which requires knowledge of the dynamics of the two-locus linkage disequilibrium. Again assuming that
must be positive, which requires knowledge of the dynamics of the two-locus linkage disequilibrium. Again assuming that 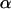 and linkage disequilibria are small (of order ζ), the two-locus disequilibrium is governed by the recursion equation
and linkage disequilibria are small (of order ζ), the two-locus disequilibrium is governed by the recursion equation
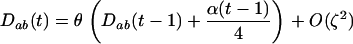 |
(9) |
(appendix a), where 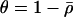 . The Z transform can be used to solve the recursion Equation 9, and assuming disequilibrium is initially absent, we obtain the general solution
. The Z transform can be used to solve the recursion Equation 9, and assuming disequilibrium is initially absent, we obtain the general solution
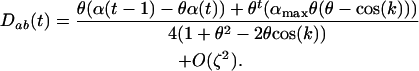 |
(10) |
When there is some sex and recombination (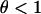 ), Equation 10 converges toward
), Equation 10 converges toward
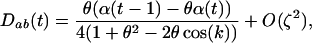 |
(11a) |
which can be evaluated using the fitness function (1),
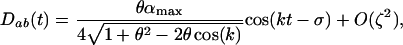 |
(11b) |
where
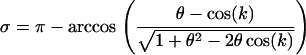 |
(11c) |
The term 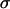 represents the lag between the fluctuations in epistasis and the two-locus linkage disequilibrium. This lag increases with the speed of the fluctuations, k, and it decreases with recombination.
represents the lag between the fluctuations in epistasis and the two-locus linkage disequilibrium. This lag increases with the speed of the fluctuations, k, and it decreases with recombination.
Using (11a) and (7) in (6b) yields the change in frequency of a modifier allele:
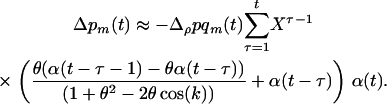 |
(12) |
Because 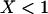 if there is some sex and recombination, the sum over τ can be evaluated explicitly,
if there is some sex and recombination, the sum over τ can be evaluated explicitly,
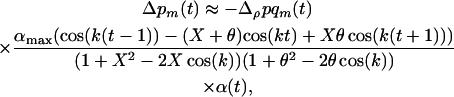 |
(13) |
where we have ignored transient dynamics due to the initial conditions. Averaging over a cycle (denoted 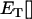 ), we get the per generation change in the frequency of the modifier allele:
), we get the per generation change in the frequency of the modifier allele:
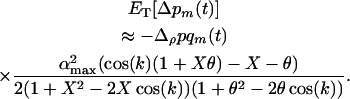 |
(14) |
The above expression can be used to predict the direction of selection on the modifier locus.
If epistasis fluctuates slowly (i.e.,  ), we get
), we get
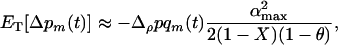 |
(15a) |
which is equivalent to the QLE prediction (5) averaged across a cycle. From the fact that (15a) is negative, we recover the conclusion obtained from the QLE analysis: sex and recombination are selected against when epistasis fluctuates slowly. Conversely, selection favors increased levels of sex and recombination if epistasis fluctuates rapidly (i.e., 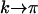 ):
):
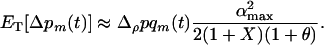 |
(15b) |
More generally, if a population engages in little sex and recombination relative to the speed of the cycles (i.e., 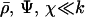 ), increased levels of sex and recombination are favored:
), increased levels of sex and recombination are favored:
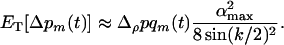 |
(15c) |
In summary, when the rate of genetic mixing is initially low or the period of the cycles is sufficiently short, higher rates of sex and recombination evolve. Otherwise, decreased levels of sex and recombination evolve.
Ultimately, the rates of sex and recombination will reach an evolutionarily stable (ES) level that is no longer susceptible to invasion by other modifier alleles; i.e., 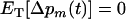 . We now determine the ES level of recombination in two cases: when the loci are equidistant and when the recombination rate between loci M and A is fixed {other cases can be investigated numerically by setting
. We now determine the ES level of recombination in two cases: when the loci are equidistant and when the recombination rate between loci M and A is fixed {other cases can be investigated numerically by setting 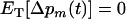 in Equation 14}. In both cases, we assume that the population is sexual with no interference in recombination among the three loci (i.e.,
in Equation 14}. In both cases, we assume that the population is sexual with no interference in recombination among the three loci (i.e., 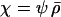 ).
).
When the loci are equidistant (i.e., 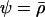 ) the ES level of sex and recombination equals
) the ES level of sex and recombination equals
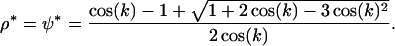 |
(16a) |
Alternatively, when the recombination rate between M and A is fixed at 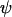 , the recombination rate between loci A and B evolves to a level that satisfies
, the recombination rate between loci A and B evolves to a level that satisfies
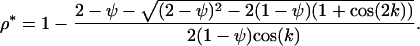 |
(16b) |
The evolutionary stability of these ES values was checked by verifying that 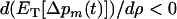 when
when 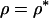 . Given the restriction that recombination rates must be
. Given the restriction that recombination rates must be 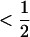 , we have the following expression for the ES recombination rate:
, we have the following expression for the ES recombination rate:
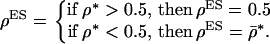 |
(17) |
When 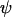 is held fixed, the ES value of recombination between loci A and B (16b) increases when there is tighter linkage between the modifier and the first selected locus (i.e., when
is held fixed, the ES value of recombination between loci A and B (16b) increases when there is tighter linkage between the modifier and the first selected locus (i.e., when 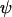 is smaller). This makes sense as the modifier can hitchhike for longer by association with favorable alleles if tightly linked to at least one of the selected loci. Furthermore, the ES value increases with the speed of the oscillations, k, and thus decreases with the period of the cycle. Indeed, the maximum level of recombination, 0.5, is the evolutionarily stable state for periods between 2 and 7 generations when the loci are equidistant [from (16a)] or when the modifier is loosely linked [from (16b)] and for periods between 2 and 9 generations when the modifier is tightly linked [from (16b)]. Thus, this analysis confirms the general claim that high recombination rates evolve in the Barton zone (with periods of 4–10 generations). Although we find that high rates of recombination can evolve with periods shorter than the Barton zone (including periods of 2 generations, i.e.,
is smaller). This makes sense as the modifier can hitchhike for longer by association with favorable alleles if tightly linked to at least one of the selected loci. Furthermore, the ES value increases with the speed of the oscillations, k, and thus decreases with the period of the cycle. Indeed, the maximum level of recombination, 0.5, is the evolutionarily stable state for periods between 2 and 7 generations when the loci are equidistant [from (16a)] or when the modifier is loosely linked [from (16b)] and for periods between 2 and 9 generations when the modifier is tightly linked [from (16b)]. Thus, this analysis confirms the general claim that high recombination rates evolve in the Barton zone (with periods of 4–10 generations). Although we find that high rates of recombination can evolve with periods shorter than the Barton zone (including periods of 2 generations, i.e., 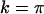 ), an examination of Barton's 1995 model indicates that this is possible in haploid models (as considered here), but not in diploid models as considered by Barton (1995).
), an examination of Barton's 1995 model indicates that this is possible in haploid models (as considered here), but not in diploid models as considered by Barton (1995).
As previously pointed out by several authors (Charlesworth 1976; Maynard-Smith 1978; Barton 1995), selection for high rates of recombination requires rapid fluctuations in the sign of epistasis. Interestingly, the amplitude of the oscillations (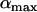 ) does not affect the direction of selection on a modifier or the ES value, although
) does not affect the direction of selection on a modifier or the ES value, although 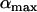 does affect the strength of selection and thus matters when there are direct costs associated with higher recombination (see discussion).
does affect the strength of selection and thus matters when there are direct costs associated with higher recombination (see discussion).
Mean fitness:
It is worth comparing the ES recombination rate to the recombination rate that would maximize the long-term geometric average of the mean fitness within the population, as studied by Sasaki and Iwasa (1987). In this one-species model with allele frequencies at 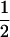 , the mean fitness is
, the mean fitness is
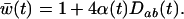 |
(18a) |
If selection is weak [i.e., the amplitude, 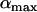 , is small and the disequilibrium is approximately (11b)], the geometric average of
, is small and the disequilibrium is approximately (11b)], the geometric average of 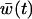 is well approximated by the arithmetic average over one cycle, which is
is well approximated by the arithmetic average over one cycle, which is
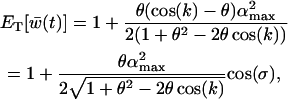 |
(18b) |
where again 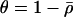 , and
, and 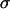 is the lag between fluctuations in epistasis and disequilibrium given by Equation 11c. Recombination affects the mean fitness by altering the amplitude of the fluctuations in linkage disequilibrium and the lag, σ. While increasing recombination decreases the amplitude by which linkage disequilibrium oscillates in response to fluctuations in epistasis (see Equation 11b), it also decreases the lag because disequilibrium responds more quickly to changes in epistasis. These two effects act on the arithmetic mean of fitness in opposite directions, and there is consequently a recombination rate that maximizes (18b):
is the lag between fluctuations in epistasis and disequilibrium given by Equation 11c. Recombination affects the mean fitness by altering the amplitude of the fluctuations in linkage disequilibrium and the lag, σ. While increasing recombination decreases the amplitude by which linkage disequilibrium oscillates in response to fluctuations in epistasis (see Equation 11b), it also decreases the lag because disequilibrium responds more quickly to changes in epistasis. These two effects act on the arithmetic mean of fitness in opposite directions, and there is consequently a recombination rate that maximizes (18b):
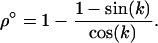 |
(19) |
According to (19), the optimal recombination rate increases with the speed of the cycles. For long periods (k small), Equation 19 is ∼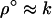 , as found by Sasaki and Iwasa (1987) using a similar model. For short periods (k large), Equation 19 more nearly equals
, as found by Sasaki and Iwasa (1987) using a similar model. For short periods (k large), Equation 19 more nearly equals 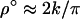 , as found by Sasaki and Iwasa (1987) using a square-wave model of selection with maximally strong selection.
, as found by Sasaki and Iwasa (1987) using a square-wave model of selection with maximally strong selection.
If group selection were governing the evolution of sex and recombination, the rate of sex and recombination would evolve toward 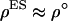 . Indeed, when the modifier is completely linked to the A locus (
. Indeed, when the modifier is completely linked to the A locus (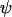 = 0), the optimal rate of sex and recombination (Equation 19) coincides with the ES level (Equation 16b). Consequently, the solid curve describing the ES level in Figure 2 when
= 0), the optimal rate of sex and recombination (Equation 19) coincides with the ES level (Equation 16b). Consequently, the solid curve describing the ES level in Figure 2 when 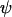 = 0 coincides with the optimal level of sex and recombination. In all other cases, the ES level of sex and recombination is lower than the value that would maximize mean fitness; this mismatch occurs because recombination between the modifier and the selected loci uncouples the fate of the modifier from its effects on the mean fitness of offspring.
= 0 coincides with the optimal level of sex and recombination. In all other cases, the ES level of sex and recombination is lower than the value that would maximize mean fitness; this mismatch occurs because recombination between the modifier and the selected loci uncouples the fate of the modifier from its effects on the mean fitness of offspring.
COEVOLUTIONARY MODEL
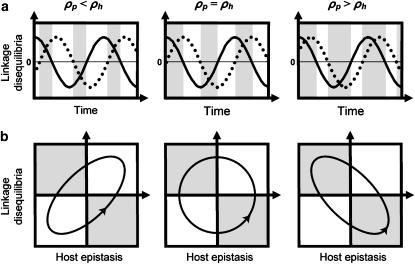
In the one-species model analyzed above, fluctuations in epistasis are imposed externally (see Equation 1). In contrast, we now focus on a situation where the temporal variability in epistasis emerges from coevolutionary interactions with an antagonist species. We follow haplotype frequencies in two haploid hermaphroditic species (the host and the parasite) through a life cycle consisting of a census, selection, and reproduction, with nonoverlapping generations. The two organisms form two large populations of constant size (no drift). Within each population we further assume random mating (when sex occurs). The species interaction is mediated via two loci (A and B) with two alleles (A/a, B/b) in each species. Every generation, each host encounters one parasite, chosen randomly from the parasite population. In each species, the phenotype of an individual is 0 if it is either AB or ab and is 1 if it is aB or Ab. When the host phenotype matches that of the one parasite that it encounters, the parasite gains a fitness advantage (αp > 0), while the host suffers a fitness cost (−1 ≤ αh < 0). The assumption regarding the specificity of the interaction (Figure 1b) was first introduced by Nee (1989) as a simple two-locus model that captures two essential features of the Red Queen hypothesis: fitness interactions are antagonistic and recombination influences the outcome of the species interaction. We thus refer to this coevolutionary model as the Nee model.
As in the one-species model, we allow the probability of sex and/or the rates of recombination to depend on a third modifier locus (M) with two alleles (M/m) and with gene order MAB. Thus, in each species, there are eight possible haplotypes (MAB, MAb, MaB, Mab, mAB, mAb, maB, mab, labeled from 1 to 8), whose frequencies among adults in species i are xi,j for j = 1–8. Where convenient, we denote the frequency of allele j by pi,j. We use the subscript i = h to refer to parameters and variables in the host species and i = p to refer to the parasite. The fitness of genotype j of species i is then 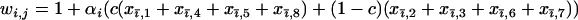 , where
, where 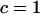 if
if 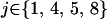 and
and 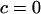 otherwise, and where
otherwise, and where 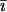 refers to the other species. After selection, the haplotypic frequencies are
refers to the other species. After selection, the haplotypic frequencies are 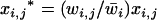 for haplotype j in species i, where
for haplotype j in species i, where 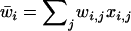 is the mean fitness of species i.
is the mean fitness of species i.
Following selection, the probability that two haploid individuals carrying modifier alleles k and l engage in sexual reproduction follows the rules defined in the single-species case. To simplify matters, we consider the evolution of sex and recombination in one species (the focal species) at a time, setting 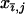 = 0 in the interacting species for j = 5–8. To determine if higher rates of sex and recombination can be favored we track changes in the frequency of allele m in the focal species i.
= 0 in the interacting species for j = 5–8. To determine if higher rates of sex and recombination can be favored we track changes in the frequency of allele m in the focal species i.
QLE analysis:
When the fluctuations in epistasis are slow relative to the rates of recombination (i.e., when selection is weak, see Equation 27 below), a QLE analysis can be used to predict the fate of the modifier allele from Equation 3. In this model, the speed of these fluctuations is not a parameter but depends on the coevolutionary dynamics and, in particular, on the strength of selection on the host and the parasite (see Equation 27 below). Under the assumption that 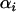 and linkage disequilibria are small (of order
and linkage disequilibria are small (of order 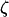 ), we have
), we have
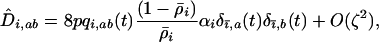 |
(20a) |
with 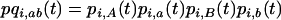 . The selection coefficients induced by the other species become
. The selection coefficients induced by the other species become
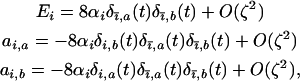 |
(20b) |
where 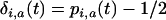 and
and 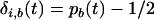 . Consequently, as in the one-species model, the product
. Consequently, as in the one-species model, the product 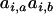 can be neglected relative to the strength of epistasis, and (3) reduces to
can be neglected relative to the strength of epistasis, and (3) reduces to
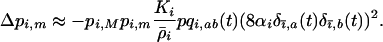 |
(21) |
Thus, if the modifier allele m increases recombination in species i, 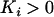 and (21) is always negative. Here, as in the one-species model and as in most models analyzed by Otto and Nuismer (2004), sex and recombination are not favored because epistasis is too strong relative to the strength of directional selection.
and (21) is always negative. Here, as in the one-species model and as in most models analyzed by Otto and Nuismer (2004), sex and recombination are not favored because epistasis is too strong relative to the strength of directional selection.
Like the single-species model analyzed above, this coevolutionary model is peculiar in that allele frequencies converge toward 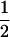 as long as there is some recombination. After this convergence, allele frequencies remain fixed, and only the fluctuations in linkage disequilibrium are needed to describe the dynamics of the selected loci. Equation 20 no longer applies once the allele frequencies have reached
as long as there is some recombination. After this convergence, allele frequencies remain fixed, and only the fluctuations in linkage disequilibrium are needed to describe the dynamics of the selected loci. Equation 20 no longer applies once the allele frequencies have reached 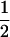 , however, as the
, however, as the 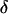 terms equal zero. It is thus necessary to consider smaller-order terms in the QLE approximation to predict the dynamics of the modifier. Assuming that sex and recombination are sufficiently frequent relative to the strength of selection in species i that the focal species (but not necessarily the interacting species) has reached QLE, and assuming that linkage disequilibria in both species are small (of order
terms equal zero. It is thus necessary to consider smaller-order terms in the QLE approximation to predict the dynamics of the modifier. Assuming that sex and recombination are sufficiently frequent relative to the strength of selection in species i that the focal species (but not necessarily the interacting species) has reached QLE, and assuming that linkage disequilibria in both species are small (of order 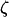 ), we have
), we have
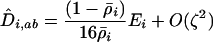 |
(22a) |
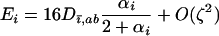 |
(22b) |
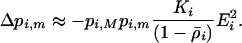 |
(22c) |
If the interacting species is also at QLE, Equations 22 are satisfied for both species only if  , indicating that disequilibria will not be maintained in this model when rates of sex and recombination are sufficiently high in both species.
, indicating that disequilibria will not be maintained in this model when rates of sex and recombination are sufficiently high in both species.
When the rate of sex and recombination is high relative to selection in the focal species but not in the interacting species, the QLE analysis predicts that rates of sex and recombination should decrease over evolutionary time in the focal species (i.e., Equation 22c is negative for modifier alleles with positive 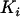 ). As the rate of genetic mixing decreases, however, the assumption that the focal species will be at QLE becomes tenuous. Indeed, simulations reveal that sex and recombination do not evolve to zero but to some positive level, which increases with the strength of selection (see Figure 3). To explore cases where selection is strong relative to the rate of sex and recombination, we follow Barton (1995) and study the dynamics of linkage disequilibria, just as we did in the one-species model.
). As the rate of genetic mixing decreases, however, the assumption that the focal species will be at QLE becomes tenuous. Indeed, simulations reveal that sex and recombination do not evolve to zero but to some positive level, which increases with the strength of selection (see Figure 3). To explore cases where selection is strong relative to the rate of sex and recombination, we follow Barton (1995) and study the dynamics of linkage disequilibria, just as we did in the one-species model.
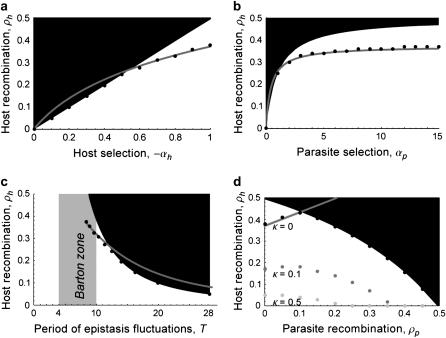
Figure 3.—
Evolutionarily stable host recombination rate in the two-species coevolutionary model as a function of (a) the strength of selection on the host, (b) the strength of selection on the parasite, (c) the period of the coevolutionary cycles, and (d) the parasite recombination rate. The ES values obtained from simulations (circles) are compared to Equation 34 (shaded curve). When recombination is so frequent that R < 1 (see Equation 36), the coevolutionary cycles and, hence, selection on recombination vanish (solid area). Except for the parameters illustrated on the horizontal axes, default parameters of 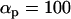 ,
, 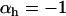 ,
, 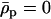 ,
, 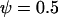 , and
, and 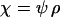 were used (in c,
were used (in c, 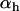 was varied as in a, from which the period was calculated). In d, we also incorporate different intrinsic costs of recombination (see Equation 44): κ = 0 (solid circles), κ = 0.1 (dark-shaded circles), and κ = 0.5 (light-shaded circles ).
was varied as in a, from which the period was calculated). In d, we also incorporate different intrinsic costs of recombination (see Equation 44): κ = 0 (solid circles), κ = 0.1 (dark-shaded circles), and κ = 0.5 (light-shaded circles ).
Recursion analysis:
When the allele frequencies have converged toward  at both loci in both species, the dynamics of linkage disequilibrium between selected loci can be approximated by the linear recursion equations
at both loci in both species, the dynamics of linkage disequilibrium between selected loci can be approximated by the linear recursion equations
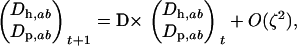 |
(23) |
where
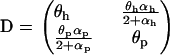 |
(24) |
and 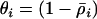 . Although similar to the QLE analysis in requiring that the disequilibria are small, this recursion analysis does not assume that the disequilibria instantaneously reach the steady-state values predicted by the current level of epistasis, as does the QLE analysis. Thus, Equation 23 can be applied even when selection is large relative to the rates of sex and recombination.
. Although similar to the QLE analysis in requiring that the disequilibria are small, this recursion analysis does not assume that the disequilibria instantaneously reach the steady-state values predicted by the current level of epistasis, as does the QLE analysis. Thus, Equation 23 can be applied even when selection is large relative to the rates of sex and recombination.
Given the linear recursions (23), the dynamics of the disequilibria in both species can be solved using standard algebraic techniques. It can be readily seen that the point where 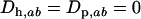 is an equilibrium of (23). A local stability analysis of this equilibrium reveals that it is unstable with complex eigenvalues as long as
is an equilibrium of (23). A local stability analysis of this equilibrium reveals that it is unstable with complex eigenvalues as long as
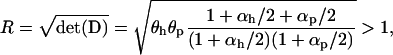 |
(25) |
where 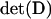 is the determinant of matrix D, and R is the magnitude of the complex eigenvalue of D. When (25) is satisfied the disequilibria of both species cycle sinusoidally over time, with an amplitude that increases over time. In other words, the coevolutionary cycles can persist over the long term only if selection is sufficiently strong relative to the rates of recombination of both species; otherwise the
is the determinant of matrix D, and R is the magnitude of the complex eigenvalue of D. When (25) is satisfied the disequilibria of both species cycle sinusoidally over time, with an amplitude that increases over time. In other words, the coevolutionary cycles can persist over the long term only if selection is sufficiently strong relative to the rates of recombination of both species; otherwise the 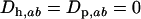 equilibrium is locally stable. We focus on the case where
equilibrium is locally stable. We focus on the case where 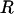 is near one (recombination rates low relative to selection), so that linkage disequilibria persist but remain small for extended periods of time.
is near one (recombination rates low relative to selection), so that linkage disequilibria persist but remain small for extended periods of time.
Measuring time as the first point at which the host disequilibrium passes zero, the general solution to Equation 23 is
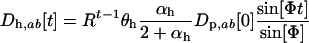 |
(26a) |
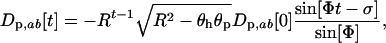 |
(26b) |
where Φ is the speed of the coevolutionary cycles (i.e., 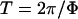 is the period),
is the period),
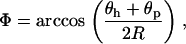 |
(27) |
and 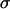 is the phase shift between the parasite and the host cycles,
is the phase shift between the parasite and the host cycles,
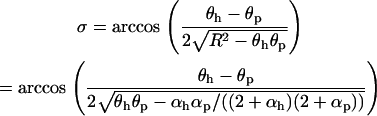 |
(28) |
with the parasite always lagging behind the host. Whenever (25) is satisfied, the radical terms in (26) and (28) are real and positive.
Because R is an increasing function of 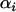 , Equation 27 indicates that the cycles become faster as selection in either species becomes stronger. The effect of recombination on the speed of the cycles is more complex, however. The cycles become faster as the recombination rates of the two species approach one another. Increasing both recombination rates (while holding their difference constant), however, slows down the cycles.
, Equation 27 indicates that the cycles become faster as selection in either species becomes stronger. The effect of recombination on the speed of the cycles is more complex, however. The cycles become faster as the recombination rates of the two species approach one another. Increasing both recombination rates (while holding their difference constant), however, slows down the cycles.
According to Equation 28, the parasite cycle lags 90° behind the host cycle when the average recombination rates between loci A and B are the same in both species (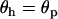 ). If, however, the host undergoes more recombination than the parasite then the phase shift increases. We can think of this case as the host “winning” the Red Queen race (the host is less susceptible to parasitic attack when averaged across a cycle). Conversely, if the host undergoes less recombination then the phase shift decreases, and the parasite is winning the Red Queen race. The speed of the cycle (and hence the selection coefficients) also influences the lag. Typically (i.e., for periods greater than four generations), increasing the speed of the cycle brings the lag closer to 90°.
). If, however, the host undergoes more recombination than the parasite then the phase shift increases. We can think of this case as the host “winning” the Red Queen race (the host is less susceptible to parasitic attack when averaged across a cycle). Conversely, if the host undergoes less recombination then the phase shift decreases, and the parasite is winning the Red Queen race. The speed of the cycle (and hence the selection coefficients) also influences the lag. Typically (i.e., for periods greater than four generations), increasing the speed of the cycle brings the lag closer to 90°.
After selection and sexual reproduction (under the assumption that allele frequencies remain constant at 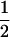 in both species), the change in frequency of allele m in species i is
in both species), the change in frequency of allele m in species i is
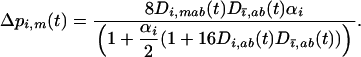 |
(29a) |
Under the assumption that linkage disequilibria are small (of order ζ) this yields
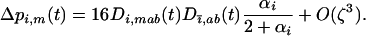 |
(29b) |
Using the fact that the epistasis experienced by species i at time t is
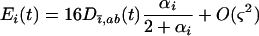 |
(30) |
we can rewrite (29b) as
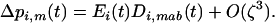 |
(31) |
We thus recover the result of the one-species model (compare Equations 31 and 6b).
To further analyze Equation 31 we need to understand the dynamics of the genetic associations. The analysis is simplified when we assume that the effect of the modifier of species i is of small order (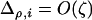 ), in which case the recursion for the three-locus disequilibrium may be approximated by
), in which case the recursion for the three-locus disequilibrium may be approximated by
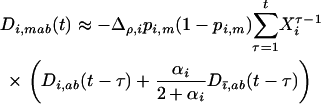 |
(32) |
(see appendix a, Three-locus disequilibrium), where 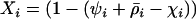 . Plugging (32) into (29b) yields
. Plugging (32) into (29b) yields
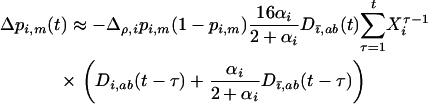 |
(33) |
Using the fact that 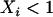 when there is some sex and recombination, the sum over τ can be evaluated explicitly using Equations 26. Then, as in the one-species model, we can average the change in modifier allele frequency over one coevolutionary cycle, ignoring transient dynamics due to the initial conditions. When calculating this average, we also average over all possible starting points in the host–parasite cycle. Using Equations 25, 27, and 28 to simplify the result, we find that a modifier allele m that increases the frequency of sex and/or recombination in species i spreads whenever the following is positive,
when there is some sex and recombination, the sum over τ can be evaluated explicitly using Equations 26. Then, as in the one-species model, we can average the change in modifier allele frequency over one coevolutionary cycle, ignoring transient dynamics due to the initial conditions. When calculating this average, we also average over all possible starting points in the host–parasite cycle. Using Equations 25, 27, and 28 to simplify the result, we find that a modifier allele m that increases the frequency of sex and/or recombination in species i spreads whenever the following is positive,
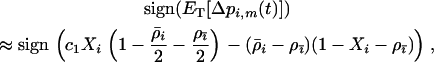 |
(34) |
where c1 is the positive constant (recall that −1 ≤ αh < 0 and 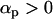 ):
):
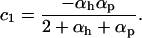 |
The first term in Equation 34 is always positive, favoring higher rates of sex and recombination, especially when selection is strong (large c1), the modifier is tightly linked (large 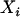 ), or host and parasite recombination rates are similar (
), or host and parasite recombination rates are similar (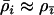 ). If the focal species has a much higher rate of sex and recombination than the species with which it interacts (
). If the focal species has a much higher rate of sex and recombination than the species with which it interacts (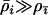 ), however, decreased levels of sex and recombination will evolve (second term in Equation 34). For a modifier that is completely linked (
), however, decreased levels of sex and recombination will evolve (second term in Equation 34). For a modifier that is completely linked (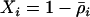 ), the first term dominates the second in Equation 34, and higher rates of sex and recombination are always favored (assuming that R ≥ 1). These results differ from that of the QLE analysis, where lower rates of recombination are always favored. This discrepancy is due to the QLE assumption that recombination rates are large relative to selection, not small as assumed in (34).
), the first term dominates the second in Equation 34, and higher rates of sex and recombination are always favored (assuming that R ≥ 1). These results differ from that of the QLE analysis, where lower rates of recombination are always favored. This discrepancy is due to the QLE assumption that recombination rates are large relative to selection, not small as assumed in (34).
To determine the ES level of recombination, we set Equation 34 to zero and solved for 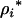 under the assumption that
under the assumption that 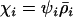 , giving
, giving
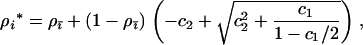 |
(35) |
where again c1 and c2 (as well as 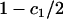 ) are strictly positive constants:
) are strictly positive constants:
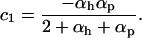 |
and
 |
Equation 35 indicates that the evolutionarily stable recombination rate for the focal species, 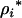 , is always larger than the recombination rate in the interacting species,
, is always larger than the recombination rate in the interacting species, 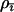 . Furthermore, it can be shown using Equation 35 that stronger selection (which induces faster cycles) increases the ES recombination rate in both the host and the parasite. Similarly, tighter linkage between the modifier and the selected loci (lower
. Furthermore, it can be shown using Equation 35 that stronger selection (which induces faster cycles) increases the ES recombination rate in both the host and the parasite. Similarly, tighter linkage between the modifier and the selected loci (lower 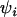 ) favors higher ES recombination rates. As in the one-species model, the amplitude does not, by itself, affect the direction of selection on the modifier; the amplitude of the cycles can be altered independently of the speed and the phase shift by changing
) favors higher ES recombination rates. As in the one-species model, the amplitude does not, by itself, affect the direction of selection on the modifier; the amplitude of the cycles can be altered independently of the speed and the phase shift by changing 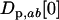 in Equation 26, yet the direction in which the modifier evolves (Equation 34) is independent of
in Equation 26, yet the direction in which the modifier evolves (Equation 34) is independent of 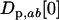 .
.
Figure 3 explores the relationship between the ES recombination rate in the host vs. the strength of selection (Figure 3, a–c) and the level of recombination in the parasite (Figure 3d). The evolution of host recombination was simulated by introducing a series of modifier alleles and tracking their spread or loss until further modifier alleles failed to spread. As expected, higher rates of recombination evolved in the host when selection was stronger (Figure 3, a and b). This was due to the faster coevolutionary cycles associated with stronger selection (Figure 3c), as was observed in the one-species model (compare to Figure 2). Equation 35 provides an excellent approximation of the evolutionary endpoint of these simulations when the strength of selection is high (Figure 3, a and b). When selection is weak, however, recombination evolution halts before it reaches the expected level (Figure 3, a and b, left). This occurs because selection on recombination vanishes when the cycles disappear (Figure 3, solid areas). Using Equation 25, the cycles disappear whenever the recombination rate rises above a threshold level given by
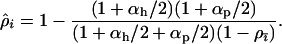 |
(36) |
This threshold also explains the nonmonotonic effect of parasite recombination on the evolution of host recombination (Figure 3d). When parasite recombination is rare, higher recombination in the parasite favors higher recombination in the host, as this helps the host “keep up” in the coevolutionary arms race (as predicted by Equations 34 and 35). When parasite recombination is common, however, coevolutionary cycles dissipate once the host reaches the critical level of recombination (Equation 36). As the rate of recombination in the parasites increases, this critical value becomes lower (Figure 3d), explaining why the evolutionarily stable rate of host recombination falls once recombination in the parasite becomes sufficiently common.
When selection is strong, the core assumption of our analysis, that linkage disequilibria remain small need not be valid. Nevertheless, Figure 3 indicates that Equation 35 continues to provide an excellent approximation for the evolutionarily stable rate of recombination even when selection in the host and/or parasite becomes very strong.
Mean fitness:
Nee (1989) used this coevolutionary model to determine the recombination rate maximizing the geometric mean fitness. When allele frequencies have converged upon 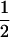 , the mean fitness of species i is
, the mean fitness of species i is
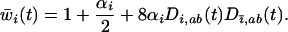 |
(37) |
We may use (37) to derive the geometric mean fitness over one coevolutionary cycle. To make progress, however, we approximate the geometric mean by the arithmetic mean over one coevolutionary cycle (as in the one-species model and Nee 1989). The arithmetic mean provides a good approximation for the geometric mean as long as the amplitude of the oscillations remains small. If we further assume that the system remains near a coevolutionary limit cycle (i.e., R = 1) this yields (using equation 26)
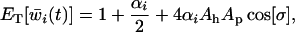 |
(38a) |
where 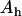 and
and 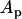 are the amplitudes of the linkage disequilibrium oscillations in the host and parasite (i.e., the magnitude of the factors multiplying the sine terms in Equation 26). We obtained a very similar expression for the arithmetic mean fitness in the one-species model [compare (18b) and (38a)].
are the amplitudes of the linkage disequilibrium oscillations in the host and parasite (i.e., the magnitude of the factors multiplying the sine terms in Equation 26). We obtained a very similar expression for the arithmetic mean fitness in the one-species model [compare (18b) and (38a)].
Nee (1989) pointed out that the impact of recombination on the long-term mean fitness depends only on the “geometry of the coevolution” (i.e., the amplitudes of and the lag between the fluctuations in the host and the parasite),
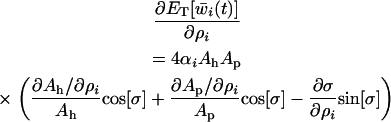 |
(38b) |
When hosts and parasites are nearly 90° out of phase (i.e., when recombination is similar in the two species, see Equation 28), 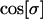 is approximately zero. In this case, whether recombination increases or decreases mean fitness depends only on its effect on the lag (the last term within parentheses in Equation 38b). The lag increases (the parasite falls further behind) with higher rates of recombination in the host (
is approximately zero. In this case, whether recombination increases or decreases mean fitness depends only on its effect on the lag (the last term within parentheses in Equation 38b). The lag increases (the parasite falls further behind) with higher rates of recombination in the host (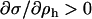 from Equation 28), while the lag decreases (the parasite more closely matches the host) with higher rates of recombination in the parasite (
from Equation 28), while the lag decreases (the parasite more closely matches the host) with higher rates of recombination in the parasite (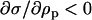 ). Thus, the sign of
). Thus, the sign of 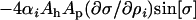 is always positive, implying that increasing recombination increases long-term mean fitness in either species (Nee 1989).
is always positive, implying that increasing recombination increases long-term mean fitness in either species (Nee 1989).
Further insight can again be gained by assuming that recombination rates between the selected loci are low. To linear order in 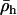 and
and 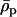 (replacing R,
(replacing R, 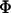 , and
, and 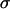 with Equations 25, 27, and 28), the mean fitness is
with Equations 25, 27, and 28), the mean fitness is
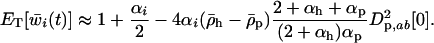 |
(38c) |
Recalling that 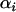 is defined to be negative in the host and positive in the parasite, Equation 38c indicates that increasing the frequency of sex and/or recombination would always increase the mean fitness of a species in the Nee model.
is defined to be negative in the host and positive in the parasite, Equation 38c indicates that increasing the frequency of sex and/or recombination would always increase the mean fitness of a species in the Nee model.
As with the single-species model, we find that examining the evolution of recombination using a mean fitness approach generates different predictions than using a modifier approach. As long as cycles are maintained (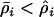 ), the mean fitness approach predicts that increased recombination is always favorable (Equation 38c, confirmed by a numerical exploration), while the modifier approach predicts that increased recombination evolves only up until the evolutionary stable level given by
), the mean fitness approach predicts that increased recombination is always favorable (Equation 38c, confirmed by a numerical exploration), while the modifier approach predicts that increased recombination evolves only up until the evolutionary stable level given by 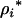 (Equation 35). Once again, this discrepancy occurs because recombination between the modifier and the selected loci uncouples the fate of a modifier allele from its long-term effects on mean fitness. Only when
(Equation 35). Once again, this discrepancy occurs because recombination between the modifier and the selected loci uncouples the fate of a modifier allele from its long-term effects on mean fitness. Only when 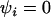 does the modifier approach predict that higher rates of sex and recombination should always evolve (Equation 34), as predicted by an analysis of mean fitness.
does the modifier approach predict that higher rates of sex and recombination should always evolve (Equation 34), as predicted by an analysis of mean fitness.
METAPOPULATION MODELS
We now briefly describe results extending the above analysis to situations where the habitat consists of a number n of local populations connected by gene flow. We assume that population sizes are large (locally and globally), so that genetic drift is negligible. We also assume that dispersal among populations follows the rules of an island model (no isolation by distance).
One-species model:
For the sake of simplicity, we assume that the amplitude and the speed of abiotic oscillations in selection remain constant over space, but we allow the phase of the cycle to vary, with all possible phases being equally common. Selection in a given population  (
(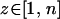 ) at time t is then given by
) at time t is then given by
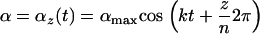 |
(39) |
The parameter 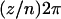 measures the phase shift of population
measures the phase shift of population  and ensures that selection among populations is asynchronous. (When selection is synchronized among populations the metapopulation behaves as a single population and, consequently, migration has no effect.) According to (39), the average selection over the whole metapopulation (denoted
and ensures that selection among populations is asynchronous. (When selection is synchronized among populations the metapopulation behaves as a single population and, consequently, migration has no effect.) According to (39), the average selection over the whole metapopulation (denoted 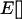 ) equals zero:
) equals zero: 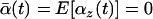 .
.
Migration among populations is assumed to take place just after selection and reproduction with a probability d. The haplotypic frequencies in a given population, z, after migration are thus given by
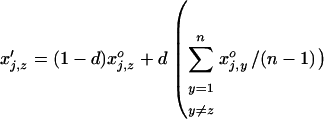 |
(40) |
For simplicity, we assume in the following that the number of populations is extremely large (i.e., 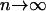 ). In this case, we can use the approximation
). In this case, we can use the approximation 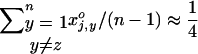 , because the negative frequency dependence tends to maintain all genotypes at the two loci at the same global frequency (i.e., when measured at the scale of the metapopulation).
, because the negative frequency dependence tends to maintain all genotypes at the two loci at the same global frequency (i.e., when measured at the scale of the metapopulation).
The recursion analysis can be readily adapted to study the effect of migration rates. This yields a new expression for the change in modifier frequency in population z (i.e., 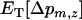 see appendix b). The evolutionarily stable recombination rate is defined as the strategy that cannot be invaded at the scale of the metapopulation. Because a modifier will spread throughout the metapopulation if and only if it spreads in the migrant pool, the ES value must satisfy
see appendix b). The evolutionarily stable recombination rate is defined as the strategy that cannot be invaded at the scale of the metapopulation. Because a modifier will spread throughout the metapopulation if and only if it spreads in the migrant pool, the ES value must satisfy 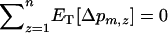 . In particular, in the absence of interference in recombination among the three loci (i.e.,
. In particular, in the absence of interference in recombination among the three loci (i.e., 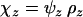 ), the ES recombination rate between loci A and B (holding the recombination rate between M and A fixed at
), the ES recombination rate between loci A and B (holding the recombination rate between M and A fixed at 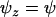 ) is
) is
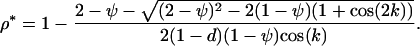 |
(41) |
This differs from Equation 16b only by the (1 − d) term in the denominator. Thus, the effect of gene flow is to decrease the ES recombination rate. Increasing migration, like increasing the recombination rate 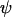 between the modifier and selected loci, decouples the dynamics at the modifier locus from the potential benefits of being associated with particular alleles at the selected loci. This decoupling decreases the strength of indirect selection at the modifier locus through the decrease of the three-locus linkage disequilibrium (appendix b).
between the modifier and selected loci, decouples the dynamics at the modifier locus from the potential benefits of being associated with particular alleles at the selected loci. This decoupling decreases the strength of indirect selection at the modifier locus through the decrease of the three-locus linkage disequilibrium (appendix b).
Because selection is spatially heterogeneous one might expect populations to be locally adapted. One measure of local adaptation at time t is the difference between the mean performance of the population in the original habitat [i.e., the mean fitness of the population, 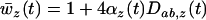 , see Equation 18a] and the mean performance of the population in a randomly chosen habitat in the metapopulation [i.e., the mean fitness averaged over the different habitats:
, see Equation 18a] and the mean performance of the population in a randomly chosen habitat in the metapopulation [i.e., the mean fitness averaged over the different habitats: 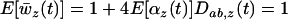 ]. This measure of local adaptation thus equals
]. This measure of local adaptation thus equals 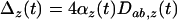 , which oscillates over time. It will be positive only when linkage disequilibrium and epistasis in the local patch (
, which oscillates over time. It will be positive only when linkage disequilibrium and epistasis in the local patch (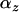 ) have the same sign. When local adaptation is averaged over the different populations this yields
) have the same sign. When local adaptation is averaged over the different populations this yields 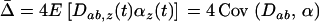 . Because we assume that all possible phases are equally common among the different populations, we would get the same result if we compared the mean fitness of a population with the mean fitness obtained by taking the same population, holding the genotype frequencies constant, and averaging over all time points in the cycle:
. Because we assume that all possible phases are equally common among the different populations, we would get the same result if we compared the mean fitness of a population with the mean fitness obtained by taking the same population, holding the genotype frequencies constant, and averaging over all time points in the cycle: 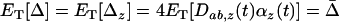 .
.
Several factors affect the mean level of local adaptation. In particular 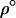 (Equation 19) is the recombination rate that maximizes mean local adaptation because maximizing arithmetic mean fitness also maximizes local adaptation. Migration also affects the long-term average mean fitness in a population [approximated by
(Equation 19) is the recombination rate that maximizes mean local adaptation because maximizing arithmetic mean fitness also maximizes local adaptation. Migration also affects the long-term average mean fitness in a population [approximated by 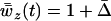 , given by Equation 18b with
, given by Equation 18b with  ]. When the speed of the oscillations is low, migration always decreases
]. When the speed of the oscillations is low, migration always decreases 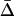 , as in classical models (Slatkin 1987; Lenormand 2002). When the oscillations are fast, however, migration can increase
, as in classical models (Slatkin 1987; Lenormand 2002). When the oscillations are fast, however, migration can increase 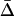 . The migration rate maximizing mean fitness and local adaptation is
. The migration rate maximizing mean fitness and local adaptation is
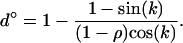 |
(42) |
The mean fitness is maximized when there is some migration because gene flow allows the population to better track fluctuations in epistasis.
Coevolutionary model:
In the coevolutionary model we again assume that the strengths of selection (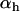 and
and 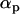 ) do not vary through space. Host and parasite genotype frequencies, however, may vary from one population to the next, and this generates heterogeneous selective pressures and local adaptation. Migration among populations is assumed to take place independently for the host and the parasite (with probabilities dh and dp, respectively). The haplotypic frequencies after migration are thus given by
) do not vary through space. Host and parasite genotype frequencies, however, may vary from one population to the next, and this generates heterogeneous selective pressures and local adaptation. Migration among populations is assumed to take place independently for the host and the parasite (with probabilities dh and dp, respectively). The haplotypic frequencies after migration are thus given by
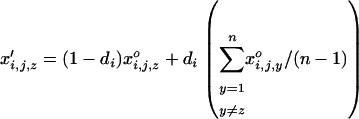 |
(43) |
In this deterministic metapopulation model, migration rapidly leads to the synchronization of coevolutionary cycles among populations. After synchronization, migration has no effect on coevolutionary dynamics, and the metapopulation behaves as a single population.
One way to desynchronize the dynamics among populations is to allow genetic drift within each population (Gandon 2002; Sasaki et al. 2002). In Figure 4 we present the results of numerical simulations that examine the effect of both host and parasite migration rates on the evolution of recombination (see details of the simulations in the Figure 4 legend). These simulations show that intermediate levels of parasite migration increase the mean fitness (and the level of local adaptation) of parasites (Figure 4a). Furthermore, these simulations show that an allele increasing host recombination is more strongly favored for intermediate parasite migration rates as long as the host migration rate is not much higher than the parasite migration rate (Figure 4b).
Figure 4.—
Effect of host and parasite migration rates on (a) parasite local adaptation and (b) the evolution of a modifier of recombination in the host. These results are obtained from numerical simulations of a stochastic version of the Nee coevolutionary model in a metapopulation. We considered 50 host and parasite populations of equal size (1000 individuals in each population), connected by host and parasite migration. In a we plot the level of local adaptation: in the solid area the parasite is locally adapted (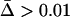 ); in the shaded area there is no local adaptation (
); in the shaded area there is no local adaptation (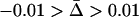 ); in the open area the parasite is locally maladapted (
); in the open area the parasite is locally maladapted (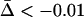 ). In b we plot the frequency of a modifier allele after 500 generations (on the basis of the mean of 100 runs using the same parameter values). The modifier allele m is codominant and induces free recombination in homozygotes, while the resident allele M induces no recombination. The contours give the final frequency of m, given an initial frequency of 0.5. Observe that the area where recombination is favored in the host corresponds to parameter values where the parasite is relatively more locally adapted (a). Other parameter values:
). In b we plot the frequency of a modifier allele after 500 generations (on the basis of the mean of 100 runs using the same parameter values). The modifier allele m is codominant and induces free recombination in homozygotes, while the resident allele M induces no recombination. The contours give the final frequency of m, given an initial frequency of 0.5. Observe that the area where recombination is favored in the host corresponds to parameter values where the parasite is relatively more locally adapted (a). Other parameter values: 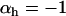 ,
, 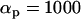 ,
, 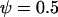 ,
, 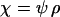 , and
, and  .
.
Perhaps not surprisingly, the parameter combinations promoting parasite adaptation are also the ones most conducive to the evolution of host recombination. This is consistent with the results of the coevolutionary model in a single population, where parasite recombination allowed parasites to be better adapted to their hosts and promoted the evolution of higher rates of host recombination (Equation 35). These stochastic simulations introduce two factors ignored in the deterministic models considered above, however. First, the simulations allow genetic drift, which can have a strong effect on the evolution of recombination (see Introduction). Second, genetic drift prevents the convergence of allele frequencies to  and, consequently, allows directional selection to act in the simulations. Thus, the simulations include all four factors known to influence the evolution of recombination: (i) directional selection, (ii) fluctuating epistasis, (iii) interaction between drift and selection, and (iv) spatial covariance in selection, making it difficult to pinpoint their relative importance.
and, consequently, allows directional selection to act in the simulations. Thus, the simulations include all four factors known to influence the evolution of recombination: (i) directional selection, (ii) fluctuating epistasis, (iii) interaction between drift and selection, and (iv) spatial covariance in selection, making it difficult to pinpoint their relative importance.
DISCUSSION
The speed, shape, and size of cycles matter:
Using one-species and two-species models that focus solely on fluctuating epistasis in the absence of directional selection, we have derived analytical predictions for the evolution of a modifier of sex and recombination under a pure Sturtevant–Mather process (Sturtevant and Mather 1938). These models are highly symmetric in that the “extreme” genotypes (ab and AB) always have equal fitness, as do the “intermediate” genotypes (Ab and aB). This type of selection leads allele frequencies to converge upon 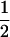 , after which point directional selection disappears. While this selection regime is arguably not very realistic, we believe that these models have a heuristic value and provide interesting insights regarding the effect of fluctuating epistasis and host–parasite coevolution. In particular, these models can be solved analytically across a broad range of recombination rates, whereas other analytical results use QLE approximations that require high rates of recombination relative to selection (Barton 1995; Otto and Nuismer 2004) or are based on models where selection is constant over time. Our analyses reveal that the evolution of recombination depends on three aspects of the fluctuations in epistasis, whether epistasis is extrinsically induced or results from coevolution.
, after which point directional selection disappears. While this selection regime is arguably not very realistic, we believe that these models have a heuristic value and provide interesting insights regarding the effect of fluctuating epistasis and host–parasite coevolution. In particular, these models can be solved analytically across a broad range of recombination rates, whereas other analytical results use QLE approximations that require high rates of recombination relative to selection (Barton 1995; Otto and Nuismer 2004) or are based on models where selection is constant over time. Our analyses reveal that the evolution of recombination depends on three aspects of the fluctuations in epistasis, whether epistasis is extrinsically induced or results from coevolution.
First, faster fluctuations in epistasis favor the evolution of higher recombination rates. We derive explicit expressions for the evolutionarily stable recombination rate in both the one-species model (Equation 16) and the two-species model (Equation 35). In both cases, we find that higher rates of recombination are favored for cycles of low-to-intermediate periods, as pointed out earlier by Charlesworth (1976) and Barton (1995). Fluctuating epistasis is often presented as an unlikely mechanism explaining the evolution of recombination because it requires such fast oscillations. It is important to emphasize, however, that some recombination is always favored by fluctuating epistasis (Figures 2 and 3), even outside the window of parameter values described in earlier studies (the so-called Barton zone of periods between 4 and 10 generations). For example, when the modifier and selected loci are equidistant (i.e., 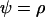 ), cycles with a period of 40 generations can select for recombination rates as high as 0.1. This result does not conflict with the general analysis of Barton (1995), because Barton focused on the periods necessary to evolve high rates of recombination (near
), cycles with a period of 40 generations can select for recombination rates as high as 0.1. This result does not conflict with the general analysis of Barton (1995), because Barton focused on the periods necessary to evolve high rates of recombination (near 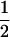 ). Indeed, we also find that the evolutionarily stable recombination rate is near
). Indeed, we also find that the evolutionarily stable recombination rate is near 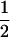 only in or near the Barton zone in both the one-species model (Figure 2) and the host-parasite model (Figure 3c). Our analysis also allows us to demonstrate that stronger selection on either the host or the parasite generates faster coevolutionary cycles, bringing the species into the zone where sex and recombination are favored (Figure 3a). This confirms previous claims based on simulation results (Peters and Lively 1999, 2007; Otto and Nuismer 2004).
only in or near the Barton zone in both the one-species model (Figure 2) and the host-parasite model (Figure 3c). Our analysis also allows us to demonstrate that stronger selection on either the host or the parasite generates faster coevolutionary cycles, bringing the species into the zone where sex and recombination are favored (Figure 3a). This confirms previous claims based on simulation results (Peters and Lively 1999, 2007; Otto and Nuismer 2004).
Second, we find that higher levels of recombination are favored when the linkage disequilibrium and epistasis within a species are more out of phase (Figures 5 and 6). Linkage disequilibrium results from past selection and always lags behind the fluctuations in epistasis. This lag yields a short-term benefit of recombination, because it is almost always beneficial to break down linkage disequilibrium that is opposite in sign to the form of epistasis (see Box 1 in Otto and Lenormand 2002). Interestingly, in the one-species model, this lag is tightly governed by the speed of the fluctuations in epistasis (see Equation 11c). The slower the cycles the smaller the lag, and it is thus difficult to distinguish between the effect of the speed and the effect of the lag on the evolution of recombination (Figure 5). As a consequence, analyses of the one-species model have focused on the speed of the cycles rather than on the lag between epistasis and linkage disequilibrium (Sasaki and Iwasa 1987; Barton 1995). In the coevolutionary model, however, the lag does not depend just on the speed of the coevolutionary cycles (see Equation 28). Indeed, the lag depends strongly on the difference between host and parasite recombination rates (see Equation 28 and Figure 6). Parasite recombination allows the parasite population to track the temporal fluctuations of host genotypes more closely. The fact that parasites are better adapted to their hosts than vice versa causes a larger phase shift between linkage disequilibrium and epistasis in the host population (Equations 22, 26, and 28), which favors higher recombination in the host (Equation 34).
Figure 5.—
Schematic description of the fluctuations between multiplicative epistasis and linkage disequilibrium between selected loci A and B in the one-species model. There is an advantage (short-term benefit) of a modifier that increases recombination in the shaded area (where epistasis and linkage disequilibrium have opposite signs). In the absence of epistasis fluctuations, linkage disequilibrium is expected to be proportional to epistasis (dotted line, Lenormand and Otto 2000). When epistasis fluctuates slowly, linkage disequilibrium lags behind epistasis (counterclockwise movement), and this yields the elongated ellipses (dashed). When epistasis fluctuates rapidly, linkage disequilibrium lags far behind epistasis and this yields “fatter” ellipses (solid). The slope of the main axis of these ellipses also decreases with the speed of the cycle. The faster the fluctuations, the longer the period of time spent in the shaded area where a modifier increasing recombination is favored.
Figure 6.—
Schematic description of (a) the fluctuations in host and parasite linkage disequilibria over time and (b) the multiplicative epistasis induced in the host vs. the host linkage disequilibrium between selected loci A and B, in the coevolutionary model. In a we show how the ratio between host and parasite recombination rates affects the phase shift between host disequilibrium (solid curves) and parasite disequilibrium (dashed curves). Higher parasite recombination rates allow the parasite to track host genotype frequencies more closely. In b we show how the ratio between host and parasite recombination rates affects the geometry of the cycles between linkage disequilibrium and epistasis in the host. The fact that host linkage disequilibrium and epistasis more often have opposite signs (shaded regions) as parasite recombination increases indicates that there is more often a short-term benefit to breaking down disequilibrium in the host. This explains why higher parasite recombination selects for higher host recombination (assuming that the cycles are maintained, 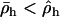 ).
).
Our article also resolves an apparent discrepancy between claims that the speed of the cycles is critical to the evolution of recombination (Charlesworth 1976; Barton 1995) and claims that it is irrelevant when coevolution is explicitly modeled (Nee 1989). While the claims of Charlesworth (1976) and Barton (1995) are based on modifier models, Nee (1989) focused on the mean fitness of a population. As shown in Equation 38c, the mean fitness of a species is always improved by increasing its recombination rate. Nevertheless, modifier alleles that increase the rate of recombination need not be able to spread within the species. This is because modifiers can recombine away from the gene combinations that they produce; this distributes the long-term mean fitness advantage of increased recombination among all of the alleles at a modifier locus, not just the ones that increase recombination. Furthermore, the period of the cycle has an important impact on the spread of modifier alleles. If the period is very long, disequilibria generated by epistasis typically remain favorable for long periods of time, and there is a short-term cost to breaking apart these favorable genetic associations. Using a modifier approach, we found that the evolutionarily stable recombination rate does decline with the period of the cycles in Nee's model (i.e., increases with the strength of selection in Equation 35, see Figure 3c), as found in one-species models (Charlesworth 1976; Barton 1995, Figure 2). Thus both the speed and the phase difference between host and parasite cycles strongly affect the evolution of recombination.
The “size” of these cycles (i.e., the amplitude of host and parasite fluctuations) appears to have less effect on the direction of evolution. Indeed, we found that the ES recombination rate is independent of the terms that set the amplitude of epistasis fluctuations: 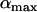 in the one-species model (Equation 17) and
in the one-species model (Equation 17) and 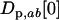 in the coevolutionary model (Equation 35). This inference relies on the assumption that there is no direct cost of sex and/or recombination. If one assumes that the m modifier allele inducing a higher rate of sex and/or recombination carries a small cost (
in the coevolutionary model (Equation 35). This inference relies on the assumption that there is no direct cost of sex and/or recombination. If one assumes that the m modifier allele inducing a higher rate of sex and/or recombination carries a small cost (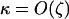 ), which is paid regardless of whether recombination actually occurs, the average change in frequency of m over one cycle in the one-species model changes from (14) to
), which is paid regardless of whether recombination actually occurs, the average change in frequency of m over one cycle in the one-species model changes from (14) to
 |
(44) |
Clearly, the benefits of sex and recombination (the first term in brackets) can compensate for the costs (the second term) only when the amplitude of the cycles, 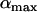 , is sufficiently large. Indeed, with costs, the ES rate of sex and recombination rises with higher-amplitude oscillations (
, is sufficiently large. Indeed, with costs, the ES rate of sex and recombination rises with higher-amplitude oscillations (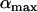 ). Thus, when sex and recombination are costly, the outcome of evolution depends not only on the speed and the phase relationship of the cycles between epistasis and linkage disequilibria but also on the size of these cycles. Similar conclusions are reached with the coevolutionary model. For example, costs of recombination were incorporated in the simulations reported in Figure 3d, always leading to a lower ES rate of recombination. Furthermore, because higher rates of recombination in the parasite tend to decrease the amplitude of the fluctuations, the advantages of recombination become less able to pay for the costs. This explains why the ES rate of host recombination declines as a function of the rate of recombination in the parasite (right-hand side of Figure 3d), even though cycles are still maintained.
). Thus, when sex and recombination are costly, the outcome of evolution depends not only on the speed and the phase relationship of the cycles between epistasis and linkage disequilibria but also on the size of these cycles. Similar conclusions are reached with the coevolutionary model. For example, costs of recombination were incorporated in the simulations reported in Figure 3d, always leading to a lower ES rate of recombination. Furthermore, because higher rates of recombination in the parasite tend to decrease the amplitude of the fluctuations, the advantages of recombination become less able to pay for the costs. This explains why the ES rate of host recombination declines as a function of the rate of recombination in the parasite (right-hand side of Figure 3d), even though cycles are still maintained.
Other coevolutionary models:
In most coevolutionary models, both fluctuating epistasis and directional selection are present and influence the evolution of recombination (Peters and Lively 1999, 2007; Otto and Nuismer 2004). When selection is weak relative to recombination, host–parasite models select against sex and recombination because directional selection dominates (Otto and Nuismer 2004). When selection is strong, however, fluctuating epistasis plays a more important role (Peters and Lively 1999, 2007; Lythgoe 2000), and the models analyzed in this article provide us with a better understanding of how much sex and recombination could evolve in the extreme case where fluctuating epistasis dominates. In fact, it is possible to conduct a similar analysis to that above using the matching-genotypes model considered by Peters and Lively (1999) if we fix the allele frequencies at 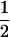 for the selected loci in hosts and parasites. In contrast with the Nee model, the allele frequencies do not converge to
for the selected loci in hosts and parasites. In contrast with the Nee model, the allele frequencies do not converge to 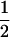 in the matching-genotypes model. Holding the allele frequencies at
in the matching-genotypes model. Holding the allele frequencies at 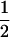 , however, allows us to isolate the importance of fluctuating epistasis. Doing so, we obtained qualitatively similar conclusions regarding the effect of the speed and shape of coevolutionary cycles (not shown). Thus, we speculate that results similar to those described in this article will hold in models where directional selection is present, but weak relative to fluctuating epistasis. The interaction between fluctuating epistasis and directional selection may, however, yield interesting evolutionary dynamics. In a matching-genotypes model, Peters and Lively (2007) found a bimodal relationship between mean recombination rate and parasite virulence for a linked modifier. We confirmed that this bimodality is also true of the ES level of recombination, by simulating the appearance of multiple linked modifiers until an ES was reached (not shown). As pointed out by Peters and Lively (2007), this could be due to the balance of the effects of directional selection (effective when selection is not too strong) and fluctuating epistasis (effective when selection is strong).
, however, allows us to isolate the importance of fluctuating epistasis. Doing so, we obtained qualitatively similar conclusions regarding the effect of the speed and shape of coevolutionary cycles (not shown). Thus, we speculate that results similar to those described in this article will hold in models where directional selection is present, but weak relative to fluctuating epistasis. The interaction between fluctuating epistasis and directional selection may, however, yield interesting evolutionary dynamics. In a matching-genotypes model, Peters and Lively (2007) found a bimodal relationship between mean recombination rate and parasite virulence for a linked modifier. We confirmed that this bimodality is also true of the ES level of recombination, by simulating the appearance of multiple linked modifiers until an ES was reached (not shown). As pointed out by Peters and Lively (2007), this could be due to the balance of the effects of directional selection (effective when selection is not too strong) and fluctuating epistasis (effective when selection is strong).
Metapopulation models:
Previous results, from both simulation (Ladle et al. 1993; Keeling and Rand 1995; Sasaki et al. 2002) and analytical studies (Agrawal 2006), demonstrate that the direction in which sex and recombination evolves depends strongly on whether or not random mixing between hosts and parasites is assumed. Our metapopulation models explicitly allow for nonrandom mixing because of limited migration. We show that migration of the focal species (say the host in the coevolutionary model) selects for lower sex and recombination in that species. Migration effectively breaks the association between the modifier locus and selected loci. Because recombination evolves only by hitchhiking, decreasing this association impedes the evolution of recombination. In simulations of the coevolutionary model, we have shown that migration of the interacting species (say, the parasite) can, however, promote the evolution of recombination in the host. This is consistent with results obtained from previous simulation studies (Ladle et al. 1993; Sasaki et al. 2002). Parasite migration yields parasite local adaptation, with a shorter lag between parasite and host dynamics (Gandon et al. 1996; Gandon 2002). In response, higher rates of sex and recombination (increasing the lag) can evolve in the host. Although Figure 4 is broadly consistent with this interpretation, it was obtained from a complex simulation model involving evolutionary forces not included in our models (i.e., genetic drift and directional selection). Further work is needed to investigate the relationship between fluctuating epistasis, local adaptation, and the evolution of recombination.
Recombination, mutation, migration, and canalization:
We studied the effect of fluctuating epistasis on the evolution of recombination rates but other traits may also evolve as an adaptation to a variable environment. Nee (1989) pointed out the similarity between the selective pressure acting on the evolution of recombination and on the evolution of mutation rates in his coevolutionary model. Haraguchi and Sasaki (1996) showed with a different host–parasite model that coevolution may indeed favor modifiers of mutation rates in both the host and the parasite. The present analysis could be extended to study the evolution of mutation rates to see how various quantities, such as the speed, the shape, and the size of fluctuations, influence the evolution of mutation rates.
The metapopulation models reveal that migration among populations acts like recombination in breaking down the genetic associations in the species that migrates. In the models we considered, migration tends to buffer the effect of selection on local genotypic frequencies because it pushes the population toward a trajectory where all genotypes have the same frequency. This buffering effect of migration is analogous to the effect of canalization. Kawecki (2000) showed that a modifier locus that acts to canalize fluctuations in fitness can be favored when selection varies sufficiently fast. The evolution of migration rates and canalization may thus represent yet other strategies to adapt to variable environments.
Conclusion:
Here we analyze simple models where the only factor selecting for recombination and sex is fluctuating epistasis. Our analyses reveal that the evolution of recombination is governed by the speed, the shape, and the size of the fluctuations between epistasis and linkage disequilibrium. It also demonstrates that models with or without coevolution have very similar effects on the evolution of recombination. Despite the artificial nature of this highly simplified model, our results have heuristic value in improving our understanding of those cases where sex and recombination are most likely to be favored by cyclic dynamics (when fluctuations in epistasis are substantial relative to directional selection). Future work should test the robustness of our predictions by incorporating fluctuations in allele frequencies (and thereby directional selection) in addition to fluctuating epistasis. Ultimately, the analysis of these and more complex coevolutionary models will allow us to identify more precisely the conditions under which the Red Queen hypothesis for the evolution of sex and recombination is most effective.
Acknowledgments
We thank F. Rousset and C. Lively for comments on an early version of the manuscript and A. Peters for insightful discussions on Nee's model of host–parasite dynamics. S.G. is funded by the Centre National de la Recherche Scientifique (France) and S.P.O. by the National Science and Engineering Research Council (Canada).
APPENDIX A: LINKAGE DISEQUILIBRIA—DEFINITIONS AND RECURSION EQUATIONS
Linkage disequilibria measure genetic associations among two or more loci within a haploid genome. Within the general framework proposed by Kirkpatrick et al. (2002), one may describe the allelic state of an individual sampled at a biallelic locus i using the variable 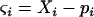 , where
, where 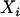 may take a value of 1 or 0 (depending on the presence or absence of the focal allele) and
may take a value of 1 or 0 (depending on the presence or absence of the focal allele) and 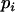 is the expected frequency of the focal allele in the population. In this article we assume that m, a, and b are the focal alleles at loci M, A, and B, respectively. One may then define various measures of associations among a set U of different loci. For example, if we are interested in the linkage disequilibria between three loci (
is the expected frequency of the focal allele in the population. In this article we assume that m, a, and b are the focal alleles at loci M, A, and B, respectively. One may then define various measures of associations among a set U of different loci. For example, if we are interested in the linkage disequilibria between three loci (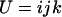 ), then
), then
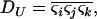 |
where the bar over the product 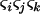 denotes the expectation over the distribution of genotype frequencies in the population. The above definition is used to derive the linkage disequilibria that appear in Equation 4. In particular, the two-locus disequilibrium between loci A and B is
denotes the expectation over the distribution of genotype frequencies in the population. The above definition is used to derive the linkage disequilibria that appear in Equation 4. In particular, the two-locus disequilibrium between loci A and B is 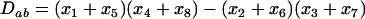 , which corresponds to the standard measure of two-locus linkage disequilibrium.
, which corresponds to the standard measure of two-locus linkage disequilibrium.
One-species model:
Two-locus disequilibrium:
The recursion for the linkage disequilibrium between A and B is
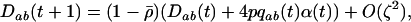 |
where 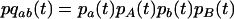 .
.
The quasilinkage equilibrium is the value  (Equation 6) that satisfies
(Equation 6) that satisfies  .
.
Three-locus disequilibrium:
When allele frequencies at loci A and B have converged upon 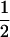 , the effect of the modifier is small (
, the effect of the modifier is small (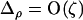 ), and the disequilibria are small (
), and the disequilibria are small (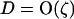 ), the recursion for the linkage disequilibrium among the three loci M, A, and B is
), the recursion for the linkage disequilibrium among the three loci M, A, and B is
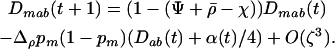 |
Technically, the term 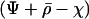 should read
should read 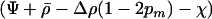 , but we assume that the effect of the modifier on the recombination rates,
, but we assume that the effect of the modifier on the recombination rates, 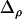 , is small relative to the recombination rates themselves. Following Barton (1995), this recursion may be iterated to yield
, is small relative to the recombination rates themselves. Following Barton (1995), this recursion may be iterated to yield
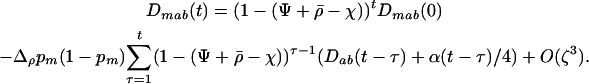 |
With recombination, the influence of the initial conditions measured by the first term in the above expression decays, leaving Equation 7.
Two-species model:
Two-locus disequilibrium:
The recursion for the linkage disequilibrium between A and B in species i is
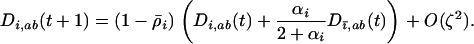 |
The quasilinkage equilibrium is the value  (Equation 26) that satisfies
(Equation 26) that satisfies  .
.
Three-locus disequilibrium:
The three-locus disequilibrium in species i is
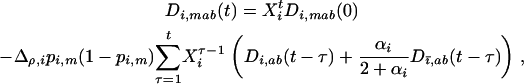 |
where 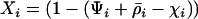 . With recombination and migration, the influence of the initial conditions measured by the first term in the above expression decays, leaving (32).
. With recombination and migration, the influence of the initial conditions measured by the first term in the above expression decays, leaving (32).
APPENDIX B: METAPOPULATION MODELS
In the one-species metapopulation model the dynamics of the linkage disequilibrium between loci A and B is given by (10) after replacing 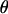 with
with 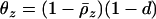 . The recursion analysis yields the average change in frequency of the modifier allele m (with an effect
. The recursion analysis yields the average change in frequency of the modifier allele m (with an effect 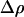 on recombination) over a cycle of the fluctuations of selection,
on recombination) over a cycle of the fluctuations of selection,
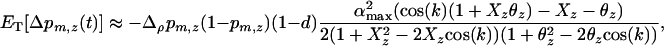 |
where 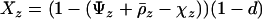 .
.
References
- Agrawal, A. F., 2006. Similarity selection and the evolution of sex: revisiting the Red Queen. PLoS Biol. 4: 1364–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. H., 1995. A general model for the evolution of recombination. Genet. Res. 65: 123–144. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and B. Charlesworth, 1998. Why sex and recombination? Science 281: 1986–1990. [PubMed] [Google Scholar]
- Barton, N. H., and S. P. Otto, 2005. Evolution of recombination due to random drift. Genetics 169: 2353–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., 1976. Recombination modification in a fluctuating environment. Genetics 83: 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel, I., and M. W. Feldman, 1970. On the evolutionary effect of recombination. Theor. Popul. Biol. 1: 88–100. [DOI] [PubMed] [Google Scholar]
- Feldman, M. W., F. B. Christiansen and L. D. Brooks, 1980. Evolution of recombination in a constant environment. Proc. Natl Acad. Sci. USA 77: 4838–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1965. The effect of linkage on directional selection. Genetics 52: 349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J., and S. Yokoyama, 1976. Evolutionary advantage of recombination. 2. Individual selection for recombination. Genetics 83: 845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandon, S., 2002. Local adaptation and the geometry of host-parasite coevolution. Ecol. Lett. 5: 246–256. [Google Scholar]
- Gandon, S., Y. Capowiez, Y. Dubois, Y. Michalakis and I. Olivieri, 1996. Local adaptation and gene-for-gene coevolution in a metapopulation model. Proc. R. Soc. Lond. Ser. B 263: 1003–1009. [Google Scholar]
- Haraguchi, Y., and A. Sasaki, 1996. Host-parasite arms race in mutation modifications: Indefinite escalation despite a heavy load? J. Theor. Biol. 183: 121–137. [DOI] [PubMed] [Google Scholar]
- Hill, W. G., and A. Robertson, 1966. The effect of linkage on the limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Iles, M. M., K. Walters and C. Cannings, 2003. Recombination can evolve in large finite populations given selection on sufficient loci. Genetics 165: 2249–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki, T. J., 2000. The evolution of genetic canalization under fluctuating selection. Evolution 54: 1–12. [DOI] [PubMed] [Google Scholar]
- Keeling, M. J., and D. A. Rand, 1995. A spatial mechanism for the evolution and maintenance of sexual reproduction. Oikos 74: 414–424. [Google Scholar]
- Keightley, P. D., and S. P. Otto, 2006. Selective interference among deleterious mutations favours sex and recombination in finite populations regardless of the nature of epistasis. Nature 443: 89–92. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, M., T. Johnson and N. H. Barton, 2002. General models of multilocus evolution. Genetics 161: 1727–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1993. Classification of hypotheses on the advantage of amphimixis. J. Hered. 84: 372–387. [DOI] [PubMed] [Google Scholar]
- Ladle, R. J., R. A. Johnstone and O. P. Judson, 1993. Coevolutionary dynamics of sex in a metapopulation: escaping the Red Queen. Proc. R. Soc. Lond. Ser. B 253: 155–160. [Google Scholar]
- Lenormand, T., 2002. Gene flow and the limits to natural selection. Trends Ecol. Evol. 17: 183–189. [Google Scholar]
- Lenormand, T., and S. P. Otto, 2000. The evolution of recombination in a heterogeneous environment. Genetics 156: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe, K. A., 2000. The coevolution of parasites with host-acquired immunity and the evolution of sex. Evolution 54: 1142–1156. [DOI] [PubMed] [Google Scholar]
- Martin, G., S. P. Otto and T. Lenormand, 2006. Selection for recombination in structured populations. Genetics 172: 593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith, J., 1978. The Evolution of Sex. Cambridge University Press, Cambridge/London/New York.
- Nee, S., 1989. Antagonistic co-evolution and the evolution of genotypic randomization. J. Theor. Biol. 140: 499–518. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1967. Modification of linkage intensity by natural selection. Genetics 57: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, S. P., and N. H. Barton, 2001. Selection for recombination in small populations. Evolution 55: 1921–1931. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and M. W. Feldman, 1997. Deleterious mutations, variable epistatic interactions, and the evolution of recombination. Theor. Popul. Biol. 51: 134–147. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and T. Lenormand, 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3: 252–261. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and Y. Michalakis, 1998. The evolution of recombination in changing environments. Trends Ecol. Evol. 13: 145–151. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and S. Nuismer, 2004. Species interactions and the evolution of sex. Science 304: 1018–1020. [DOI] [PubMed] [Google Scholar]
- Peters, A. D., and C. M. Lively, 1999. The Red Queen and fluctuating epistasis: a population genetic analysis of antagonistic coevolution. Am. Nat. 154: 393–405. [DOI] [PubMed] [Google Scholar]
- Peters, A. D., and C. M. Lively, 2007. Short- and long-term benefits and detriments to recombination under antagonistic coevolution. J. Evol. Biol. (in press). [DOI] [PubMed]
- Pomiankowski, A., and J. R. Bridle, 2004. Evolutionary genetics: no sex please, we're at QLE (quasi linkage equilibrium). Heredity 93: 407. [DOI] [PubMed] [Google Scholar]
- Pylkov, K. V., L. A. Zhivotovsky and M. W. Feldman, 1998. Migration versus mutation in the evolution of recombination under multilocus selection. Genet. Res. 71: 247–256. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., and Y. Iwasa, 1987. Optimal recombination rate in fluctuating environments. Genetics 115: 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A., W. D. Hamilton and F. Ubeda, 2002. Clone mixture and a pacemaker: new facets of Red-Queen theory and ecology. Proc. R. Soc. Lond. Ser. B 269: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin, M., 1987. Gene flow and the geographic structure of natural populations. Science 236: 787–792. [DOI] [PubMed] [Google Scholar]
- Sturtevant, A. H., and K. Mather, 1938. The interrelations of inversions, heterosis and recombination. Am. Nat. 72: 447–452. [Google Scholar]
- West, S. A., C. M. Lively and A. F. Read, 1999. A pluralist approach to sex and recombination. J. Evol. Biol. 12: 1003–1012. [Google Scholar]