Abstract
Although recent findings suggest that the F-box genes SFB/SLF control pollen-part S specificity in the S-RNase-based gametophytic self-incompatibility (GSI) system, how these genes operate in the system is unknown, and functional variation of pollen S genes in different species has been reported. Here, we analyzed the S locus of two species of Maloideae: apple (Malus domestica) and Japanese pear (Pyrus pyrifolia). The sequencing of a 317-kb region of the apple S9 haplotype revealed two similar F-box genes. Homologous sequences were isolated from different haplotypes of apple and Japanese pear, and they were found to be polymorphic genes derived from the S locus. Since each S haplotype contains two or three related genes, the genes were named SFBB for S locus F-box brothers. The SFBB genes are specifically expressed in pollen, and variable regions of the SFBB genes are under positive selection. In a style-specific mutant S haplotype of Japanese pear, the SFBB genes are retained. Apart from their multiplicity, SFBB genes meet the expected characteristics of pollen S. The unique multiplicity of SFBB genes as the pollen S candidate is discussed in the context of mechanistic variation in the S-RNase-based GSI system.
THE S-RNase-based gametophytic self-incompatibility (GSI) system has been found in the families Solanaceae, Rosaceae, and Scrophulariaceae. Haplotypes of a single S locus determine the specificity of self and nonself discrimination; when an S haplotype in the haploid pollen matches one of two S haplotypes in the pistil, then the pollen is recognized as “self” and rejected by the pistil (de Nettancourt 2001). The S haplotype contains two closely linked S-specificity genes, pistil S and pollen S. While pistil S has been known to be the S-RNase gene, identity of the pollen S has long been unknown until recently (Kao and Tsukamoto 2004; McClure and Franklin-Tong 2006). Findings of SLF/SFB as the pollen S gene suggested that the F-box protein determines the pollen S specificity (Entani et al. 2003; Sijacic et al. 2004; Ushijima et al. 2003, 2004). Since the well-documented function of F-box protein is substrate recognition as a component of SCF complex, a kind of E3 ubiquitin ligase, it has been hypothesized that SLF/SFB recognizes nonself S-RNase in compatible pollen tubes and ubiquitinylates it for degradation by the 26S proteasome (Ushijima et al. 2003, 2004; Qiao et al. 2004; Hua and Kao 2006). However, recent immunolocalization and immunoblot analyses have shown that S-RNase is incorporated into vacuoles inside pollen tubes and that the amount of S-RNase is not significantly different between compatible and incompatible pollinations (Goldraij et al. 2006). Consequently, how SLF/SFB and S-RNase interact to trigger the self-incompatibility reaction is still largely unclear.
Although Solanaceae and Prunus species use a similar molecule as the pistil S determinant (S-RNase), clear differences have been reported for pollen S. First, pollen S in Prunus (SFB) shows much higher allelic diversity (66–82.5% amino acid identity; Ikeda et al. 2004) than pollen S (SLF) in Solanaceae (88.4–89.4% amino acid identity; Sijacic et al. 2004). Second, diploid pollen from the Prunus tetraploid is frequently capable of normal self-incompatibility function (Hauck et al. 2006), but heteroallelic pollen from Solanaceae always shows breakdown of self-incompatibility (SI) (competitive interaction) (de Nettancourt 2001). Finally, in Solanaceae, SLF is considered to be essential for pollen viability because all the pollen-part mutations were duplications of pollen S and no deletion type was recovered even after large-scale screening of X-ray-induced mutants (Golz et al. 2001). In contrast, deletion of SFB results in pollen-part self-compatibility in Prunus (Sonneveld et al. 2005). These differences in pollen S may reflect a mechanistic diversity of GSI systems among species.
Rosaceae comprises four subfamilies: Spiraeoideae, Rosoideae, Maloideae, and Amygdaloideae. In species of Maloideae and Amygdaloideae, the GSI mechanism has been studied at a molecular level and S-RNase's have been characterized extensively; however, the pollen S gene (SFB) has been identified only in Prunus, a species of Amlygdaloideae. The recent finding that Prunus SFB barely causes competitive interaction in heteroallelic pollen prompted Hauck et al. (2006) to suggest that pollen S in Prunus may be different from pollen S in Solanaceae. However, competitive interaction of pollen S has been documented in pear (Pyrus communis), a species of Maloideae (Crane and Lewis 1941; Lewis and Modlibowska 1942). Characterization of pollen S in Maloideae and comparison of it to its counterparts in Prunus and Solanaceae are likely to shed light on the mechanism and evolution of the S-RNase-based GSI system. However, attempts to isolate pollen S in Maloideae through a homology-based approach with Prunus SFB sequence information have been unsuccessful (Y. Suzuki and H. Sassa, unpublished results), most likely because of sequence diversity of pollen S between these subfamilies.
Here, we analyzed the apple S locus, a species of Maloideae, to identify pollen S. A complete sequence of the 317-kb apple S9 haplotype identified two closely related F-box genes, which we have named SFBB (S locus F-box brothers). Two SFBB genes also were isolated from apple S3 haplotype BAC clones, and three SFBB genes were isolated in each of the Japanese pear S4 and S5 haplotypes. SFBB genes in apple and Japanese pear show S haplotype-specific sequence polymorphism and pollen-specific gene expression. Analysis showed that S4sm, a mutant Japanese pear haplotype that lacks the S4-RNase gene and confers pistil-specific self-compatibility, lacks at least 110 kb that contains the S4-RNase gene but retains three SFBB4 genes. A sequence analysis also revealed that variable regions of SFBB genes are under positive selection. Apart from their multiplicity, the data support the idea that SFBB genes are the pollen S genes of apple and Japanese pear. The unique multiplicity of SFBB genes as the pollen S candidate is discussed in the context of functional variation in the S-RNase-based GSI system.
MATERIALS AND METHODS
Plant materials:
An apple (Malus x domestica) cultivar, Sekai-ichi (S3S9), and 16 cultivars of Japanese pear [P. pyrifolia (syn. serotina)]—Hayatama (S1S2), Doitsu (S1S2), Suisei (S1S4), Imamuraaki (S1S6), Chojuro (S2S3), Kikusui (S2S4), Nijisseiki (S2S4), Osa-Nijisseiki (S2S4sm), Chikusui (S3S4), Akemizu (S3S5), Hosui (S3S5), Shinsui (S4S5), Kosui (S4S5), Hogetsu (S1S7), Okusankichi (S5S7), and Chukanbohon Nou No.1 (S4smS4sm)—were used. Forty progenies obtained by crossing Chikusui (S3S4) and Akemizu (S4S5) and 40 plants derived from a cross between Akemizu (S3S5) and Shinsui (S4S5) were also used.
Construction of BAC and cosmid contigs:
A BAC library of the apple cultivar Florina (Vinatzer et al. 1998) was obtained from Texas A&M University and screened using the apple S9-RNase cDNA (Sc-RNase) (Sassa et al. 1996) as a probe. Overlapping clones were obtained by screening the library with probes from different positions in the initial BAC clones.
The cosmid library for the cultivar Nijisseiki (S2S4; Sassa et al. 2002) was screened using the S4-RNase cDNA (Sassa et al. 1997) as a probe. Overlapping clones were then obtained using the method described by Ushijima et al. (2001).
Shotgun sequencing:
The S9 haplotype-derived BAC clones (34G16, 45M19, and 90A15) were subjected to shotgun sequencing at Hitachi High-Tech Science Systems (Ibaraki, Japan) (Iwashita et al. 2003). For each BAC clone, a fivefold sequence coverage was assembled, and gaps were filled by polymerase chain reaction (PCR) and by direct sequencing of BACs. The assembled sequence was further verified by PCR.
Construction of phylogenetic trees of F-box proteins:
Amino acid sequences of F-box proteins were aligned using ClustalX (Thompson et al. 1997) and manually optimized. A neighbor-joining tree was constructed using the alignment (Saitou and Nei 1987). Protein distances among pairs of sequences were produced using the PAM Dayhoff matrix (Dayhoff et al. 1979) implemented by the PROTDIST program in PHYLIP (Felsenstein 2005). For each distance matrix, a bootstrap analysis was performed by randomly selecting amino acid positions for replacement to produce 1000 replicate protein distance matrices upon which the neighbor joining was performed.
Isolation of nucleic acids:
Genomic DNAs were isolated from leaves as described by Doyle and Doyle (1990) and Sassa and Hirano (1998). RNAs were isolated from leaves and floral organs as described by McClure et al. (1990).
PCR and RACE:
SFBBs of the apple S3 haplotype were amplified from BAC clones 66L6 and 72N11 using primer pairs FMdSL21 (ATGTCCCAGGTGCGTGAAAG) and RMdSL21 (CAATTCACTTGACTGGAACAATAC) and FSMF1 (TACRTGWGAAKAWTTCHYGTG) and RSMF1 (CTCAAGCHTTGTATCATGCATAC), respectively. Flanking sequences of the 66L6-derived gene, MdSFBB3-α, were further amplified using the DNA Walking SpeedUp kit (Seegene, Seoul, Korea) to determine the full-length sequence of the coding region.
Amplification of SFBB genes from the Japanese pear cosmid clones and from the Nijisseiki genomic DNA was conducted using primers FjpFB1 (CCAAGTCTCTGATGMGRTTCAAATG) and RjpFB1 (SRGTTAGKWGTTTTGTCCATGAAC), which were designed to amplify all SFBB sequences.
Total RNA from the pollen of Kosui was used for 3′RACE, using FMdSL21 as a specific primer. 5′RACE was conducted using specific primers PpSLFLr1 (AGAAGGATACAAGTGGAGGATG) and PpSLFLr2 (AATTGCTGAGGTGTTTGGCC) essentially as described by Ushijima et al. (2003). Full-length cDNAs of PpSFBBs were amplified by 3′RACE, using specific primers listed in supplemental Table 1 at http://www.genetics.org/supplemental/.
DNA and RNA blot analysis:
Five micrograms of genomic DNAs digested with HindIII were separated and blotted onto a nylon membrane. The membrane was probed with the digoxygenin-labeled cDNAs for genes expressed in pollen, washed, and visualized as described by Ushijima et al. (2001). An RNA blot analysis also was conducted as described by Sassa et al. (1997).
Cleaved amplified polymorphic sequence and RT–PCR/cleaved amplified polymorphic sequence:
Genomic DNAs of Japanese pear cultivars were used as templates for PCR amplification of SFBB genes. The PCR products were digested with restriction enzymes to detect specific cleaved amplified polymorphic sequence (CAPS) bands. The primers and enzymes are listed in supplemental Table 2 at http://www.genetics.org/supplemental/. A CAPS analysis of the S-RNase genes also was conducted using the method described by Takasaki et al. (2004).
RNAs from the leaf and the floral organs of apple “Sekai-ichi” and Japanese pear “Kosui” were treated with DNaseI (Nippongene). Their cDNAs were synthesized by SuperScript II (CLONTECH, Palo Alto, CA) with an oligo(dT) primer. The resultant cDNAs were used as templates for PCR amplification with gene-specific primers, and PCR products were treated with restriction enzymes to detect target-specific CAPS. A PCR was performed with ExTaq (TaKaRa), using a program of 30 cycles at 94° for 30 sec, 53° for 30 sec, and 72° for 45 sec and an initial denaturing of 94° for 2 min 30 sec and final extension of 72° for 7 min. PCR products were separated on a 1.5% agarose gel and stained with ethidium bromide.
Identification of the most variable sites in the amino acid sequences:
Amino acid sequences of 10 SFBB genes from apple and Japanese pear were aligned using the ClustalX program (Thompson et al. 1997) and manually adjusted. On the basis of the alignment, a normed variability index (NVI) was calculated for each site (Kheyr-Pour et al. 1990). Sites with an NVI > −0.25 were identified as the most variable sites.
Calculation of Ka- and Ks-values:
DNA sequences were aligned using GENETYX-MAC (version 13; Software Development, Tokyo). After gaps were removed, a codon-by-codon alignment was carried out manually. On the basis of the alignment, DNAsp (Rozas et al. 2003) was used for the calculation of Ka- and Ks-values.
RESULTS
Construction of BAC contigs for the apple S locus:
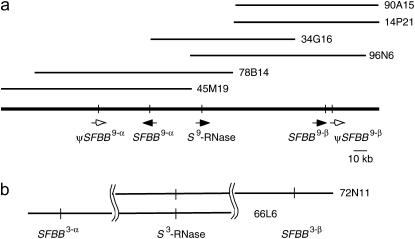
A BAC library from the apple cultivar Florina (Vinatzer et al. 1998) was screened using S9-RNase cDNA as a probe. Of the five clones obtained, three contained an S9-RNase gene and two included an S3-RNase gene. For the S9 haplotype-derived clones, overlapping BAC clones were further screened. End-sequence probes derived from the initial BAC clones produced smear patterns on apple genomic DNA blots, suggesting that they contained repetitive sequences and were not suitable for library screening. The BAC clones were then subjected to shotgun sequencing, and the draft data were used to select candidate probes to identify further BAC clones. The library was screened with probes that produced single bands on genomic DNA blots. Ultimately, a BAC contig consisting of seven overlapping clones corresponding to the S9 haplotype was constructed. A schematic representation of the BAC contig is shown in Figure 1a.
Figure 1.—
The S locus BAC contig of apple. (a) Apple S9 haplotype. Thin bars are in scale and show BAC clones. Solid arrows denote the transcriptional direction of genes. Open arrows show pseudogenes. (b) BAC clones for the apple S3 haplotype. Bars are not in scale.
Analysis of a 317-kb sequence of the S9 haplotype identified multiple, related F-box genes from the apple S locus:
Of the seven BAC clones in the S9-haplotype contig, three clones (90A15, 34G16, and 45M19) were completely sequenced. The entire 317-kb sequence contains a 169-kb region upstream of the S9-RNase gene and a 148-kb region downstream of the S9-RNase gene. The 317-kb sequence was annotated by the Rice Genome Automated Annotation System (RiceGAAS) (Sakata et al. 2002; http://ricegaas.dna.affrc.go.jp/index.html), which automatically analyzes large sequences by using several programs for prediction and analysis of protein-coding sequence, for example, Blast (Altschul et al. 1990), AutoPredLTR (Sakata et al. 2002), and GENSCAN (Burge and Karlin 1997).
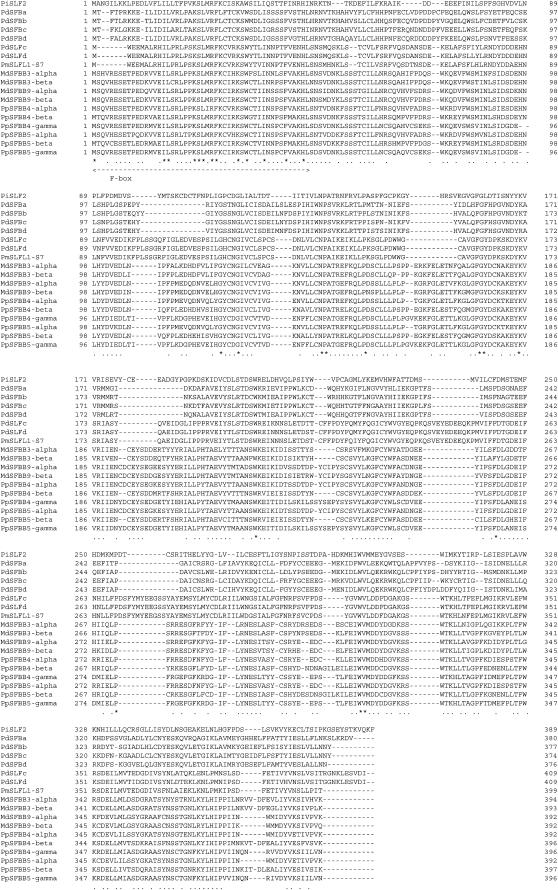
GENSCAN identified 71 open reading frames (ORFs) (Table 1). Of the 71 ORFs, 27 are homologous to retrotransposons, 36 show no significant homology to sequences in the databases, and 3 are similar to hypothetical proteins of rice, Medicago and Arabidopsis. Five ORFs—ORF20, ORF29, ORF43, ORF61, and ORF63—show homology to known genes. ORF43 was identified as the S9-RNase gene, and ORF63 was shown to be homologous to a putative aminotransferase of rice. ORF20, ORF29, and ORF61 are homologous to Prunus SLFL1, a monomorphic F-box gene found in the S locus (Entani et al. 2003; Ushijima et al. 2003). ORF29 and ORF61 were named MdSFBB9-α and MdSFBB9-β (S locus F-box brothers of M. domestica), respectively. MdSFBB9-α and MdSFBB9-β are located 42 kb upstream and 93 kb downstream of the S9-RNase gene, respectively (Figure 1a). Although ORF20 is homologous to SLFL1 and to the MdSFBB genes, the predicted amino acid sequence of 87 amino acid residues is much shorter than that of the SFBB genes (392 aa). Furthermore, while the downstream sequence of the stop codon for ORF20 is also homologous to MdSFBB genes, it contains several indels including a 1.4-kb insertion of unknown sequence; thus, ORF20 was considered to be a pseudogene and named ΨMdSFBB9-α. ΨMdSFBB9-α showed 84.0 and 83.3% nucleotide identity to MdSFBB9-α and MdSFBB9-β, respectively. Similarly, at a position ∼3 kb downstream of MdSFBB9-β, another pseudogene was identified, ΨMdSFBB9-β. ΨMdSFBB9-β contained several indels including a 980-base insertion of unknown sequence. ΨMdSFBB9-β showed a 75.5 and a 78.1% nucleotide identity to MdSFBB9-α and MdSFBB9-β, respectively. In addition, an ∼800-base sequence with similarity to SFBB was found ∼8.3 kb upstream of ΨMdSFBB9-α. With the exception of the SFBB genes, no other F-box genes were found in the 317-kb sequence of the S9-haplotype. MdSFBB genes are more homologous to Prunus SLFL1 (34.4–37.0% amino acid identity) than they are to SFB (21.3–28.2% amino acid identity; Table 2).
TABLE 1.
Predicted ORFs in the Apple S9 haplotype
| ORF | Homolog, accession no. (E-value) |
|---|---|
| ORF1 | Medicago truncatula putative retrotransposon, ABE87982 (2e−90) |
| ORF2 | Oryza sativa retrotransposon, XP_473330 (e−59) |
| ORF3 | Unknown |
| ORF4 | Unknown |
| ORF5 | Unknown |
| ORF6 | Unknown |
| ORF7 | O. sativa hypothetical protein, ABA95009 (3e−16) |
| ORF8 | Unknown |
| ORF9 | Unknown |
| ORF10 | Solanum demissum putative retroelement, AAT40504 (2e−43) |
| ORF11 | Unknown |
| ORF12 | Unknown |
| ORF13 | M. truncatula putative retroelement, ABE91625 (5e−57) |
| ORF14 | M. truncatula hypothetical protein, ABE93286 (3e−6) |
| ORF15 | O. sativa putative retroelement, ABA98201 (0) |
| ORF16 | O. sativa putative retroelement, ABA98732 (6e−27) |
| ORF17 | O. sativa putative retroelement, AAV24758 (4e−36) |
| ORF18 | Unknown |
| ORF19 | Arabidopsis thaliana putative retroelement, AAC33961 (9e−12) |
| ORF20 | Prunus mume S1-SLFL2, AB092625 (6e−36) |
| ORF21 | Unknown |
| ORF22 | A. thaliana unknown protein, BAD44360 (3e−8) |
| ORF23 | M. truncatula putative retroelement, ABE83303 (3e−153) |
| ORF24 | Unknown |
| ORF25 | Unknown |
| ORF26 | Phaseolus vulgaris putative retroelement, AAR13317 (8e−7) |
| ORF27 | O. sativa putative retroelement, ABA93344 (e−3) |
| ORF28 | Unknown |
| ORF29 | P. mume S7-SLFL1, AB092624 (2e−41) |
| ORF30 | Unknown |
| ORF31 | Unknown |
| ORF32 | Unknown |
| ORF33 | Unknown |
| ORF34 | Vitis vinifera putative retroelement, BAD18986 (2e−23) |
| ORF35 | Unknown |
| ORF36 | Unknown |
| ORF37 | Unknown |
| ORF38 | Unknown |
| ORF39 | Malus x domestica retrotransposon, AY603367 (3e−122) |
| ORF40 | Unknown |
| ORF41 | Unknown |
| ORF42 | A. thaliana putative retroelement, AAD15534 (4e−18) |
| ORF43 | M. x domestica S9-RNase, AY187627 (4e−148) |
| ORF44 | O. sativa putative retroelement, ABF98943 (3e−68) |
| ORF45 | Unknown |
| ORF46 | O. sativa putative retroelement, ABA93430 (3e−23) |
| ORF47 | Unknown |
| ORF48 | Unknown |
| ORF49 | Unknown |
| ORF50 | O. sativa putative retroelement, XP_474875 (4e−25) |
| ORF51 | Unknown |
| ORF52 | O. sativa putative retroelement, AAP52462 (5e−4) |
| ORF53 | O. sativa putative retroelement, XP_470707 (2e−58) |
| ORF54 | M. truncatula putative retroelement, ABE94393 (0) |
| ORF55 | Unknown |
| ORF56 | Unknown |
| ORF57 | M. truncatula putative retroelement, ABE87633 (6e−8) |
| ORF58 | Unknown |
| ORF59 | Unknown |
| ORF60 | M. truncatula putative retroelement, ABE90802 (4e−8) |
| ORF61 | P. mume S1-SLFL1, AB092623 (2e−46) |
| ORF62 | Unknown |
| ORF63 | O. sativa putative aminotransferase, XP_467987 (2e−7) |
| ORF64 | O. sativa putative retroelement, NP_919970 (5e−37) |
| ORF65 | O. sativa putative retroelement, NP_920511 (e−42) |
| ORF66 | O. sativa putative retroelement, XP_474437 (8e−148) |
| ORF67 | O. sativa putative retroelement, ABA95102 (5e−50) |
| ORF68 | Unknown |
| ORF69 | Unknown |
| ORF70 | Unknown |
| ORF71 | M. truncatula putative retroelement, ABE87982 (4e−88) |
TABLE 2.
Amino acid sequence identities (%) among the S locus-encoded F-box proteins
| MdSFBB9-β | PdSFBa | PdSFBb | PdSFBc | PdSFBd | PdSLFc | PdSLFd | PmS7-SLFL1 | PmS7-SLFL2 | PmS7-SLFL3 | PiSLF2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MdSFBB9-α | 87.5 | 22.6 | 28.2 | 25.3 | 22.5 | 34.4 | 34.7 | 34.7 | 32.6 | 34.4 | 28.9 |
| MdSFBB9-β | — | 22.4 | 23.1 | 21.3 | 21.5 | 36.8 | 37.0 | 36.1 | 33.0 | 33.2 | 29.3 |
| PdSFBa | — | 69.0 | 70.1 | 68.4 | 25.8 | 22.8 | 25.8 | 40.4 | 24.9 | 22.1 | |
| PdSFBb | — | 75.6 | 76.4 | 25.0 | 23.7 | 22.9 | 41.4 | 44.9 | 27.8 | ||
| PdSFBc | — | 75.8 | 25.0 | 23.7 | 22.9 | 41.4 | 44.9 | 27.8 | |||
| PdSFBd | — | 21.9 | 21.5 | 21.0 | 36.8 | 44.9 | 21.5 | ||||
| PdSLFc | — | 95.1 | 92.5 | 35.7 | 47.6 | 25.4 | |||||
| PdSLFd | — | 94.2 | 35.7 | 47.3 | 26.3 | ||||||
| PmS7-SLFL1 | — | 34.6 | 46.1 | 25.4 | |||||||
| PmS7-SLFL2 | — | 33.1 | 30.4 | ||||||||
| PmS7-SLFL3 | — | 24.2 |
Abbreviations for the F-box proteins: Md, apple (Malus domestica); Pd, almond (Prunus dulcis); Pm, Japanese apricot (Prunus mume); Pi, Petunia inflata. Sequences of almond, Japanese apricot, and P. inflata F-box proteins are from Ushijima et al. (2003), Entani et al. (2003), and Sijacic et al. (2004), respectively.
S-haplotype-specific sequence polymorphism of SFBB:
Apple SFBB homologs were obtained using a PCR from the S3-RNase gene containing two BAC clones, 66L6 and 72N11. The SFBB homolog sequences were related but not identical with each other, suggesting that they were derived from nonoverlapping regions of the two BAC clones. The two SFBB homologs were named MdSFBB3-α and MdSFBB3-β, respectively (Figure 1b).
An RT–PCR was used to clone pollen-expressed SFBB homologs from Japanese pear, using the primers derived from MdSFBB sequences. Six cDNAs were obtained from the Kosui (S4S5) pollen, a cultivar of Japanese pear: PpSFBB4-α, PpSFBB4-β, PpSFBB4-γ, PpSFBB5-α, PpSFBB5-β, and PpSFBB5-γ.
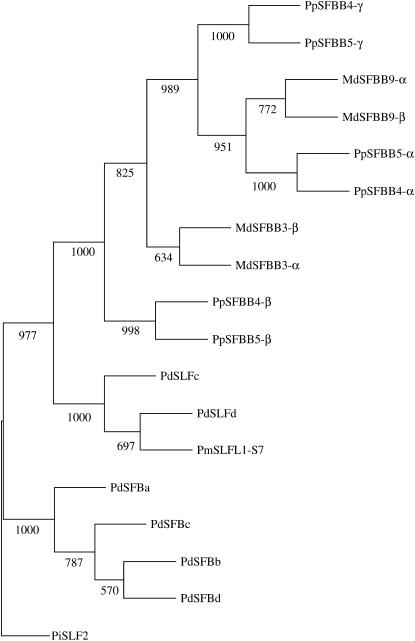
SFBB genes showed 58.4–99.0% deduced amino acid identity with each other (Table 3). While apple SFBB genes from the same haplotypes were more similar to other haplotypes, Japanese pear genes were more related to other haplotype genes of the same group (i.e., α-, β-, and γ-groups) (Table 3, Figures 2 and 3, see below).
TABLE 3.
Amino acid sequence identities (%) among the SFBBs
| MdSFBB3-β | MdSFBB9-α | MdSFBB9-β | PpSFBB4-α | PpSFBB4-β | PpSFBB4-γ | PpSFBB5-α | PpSFBB5-β | PpSFBB5-γ | |
|---|---|---|---|---|---|---|---|---|---|
| MdSFBB3-α | 82.2 | 70.5 | 69.0 | 67.3 | 71.7 | 63.0 | 67.3 | 69.3 | 62.3 |
| MdSFBB3-β | — | 73.7 | 73.4 | 71.6 | 72.5 | 66.0 | 71.1 | 71.8 | 66.1 |
| MdSFBB9-α | — | 87.5 | 83.9 | 67.6 | 70.6 | 83.7 | 65.6 | 70.1 | |
| MdSFBB9-β | — | 80.6 | 66.6 | 68.1 | 80.9 | 63.3 | 67.6 | ||
| PpSFBB4-α | — | 66.9 | 66.8 | 96.4 | 65.3 | 66.3 | |||
| PpSFBB4-β | — | 60.4 | 65.9 | 89.4 | 59.9 | ||||
| PpSFBB4-γ | — | 65.8 | 58.6 | 99.0 | |||||
| PpSFBB5-α | — | 64.6 | 65.3 | ||||||
| PpSFBB5-β | — | 58.4 |
Pp, Japanese pear (Pyrus pyrifolia). See Table 2 legend for other abbreviations.
Figure 2.—
Amino acid sequence alignment of the S locus F-box genes. Amino acid sequences of SFBB genes and other S locus F-box genes were aligned using ClustalX. Abbreviations of species are listed in Tables 2 and 3. Conserved sites and relatively conservative sites are marked with asterisks and dots, respectively. Double-headed arrow shows the F-box region.
Figure 3.—
Neighbor-joining tree of the S locus F-box genes with 1000 bootstraps.
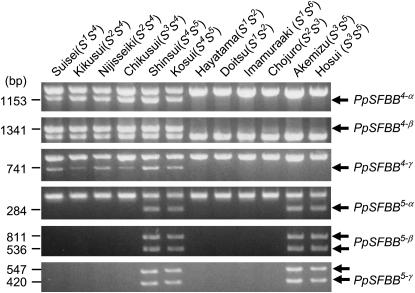
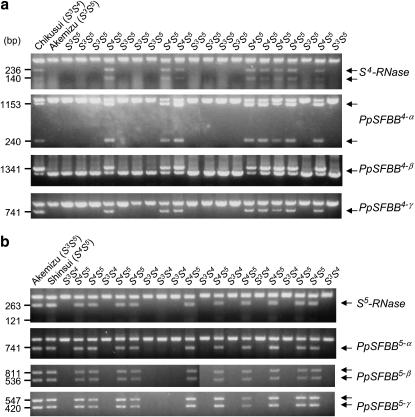
S-haplotype specificity was analyzed by correlating Japanese pear S genotype with PpSFBB gene polymorphism. Since the SFBB genes are similar to each other, a CAPS procedure was used to detect polymorphism. Amplification by group-specific primers was followed by digestion with a restriction enzyme to reveal gene-specific patterns. This CAPS analysis showed that PpSFBB genes are specific to their respective S haplotypes (Figure 4).
Figure 4.—
S-haplotype-specific sequence polymorphism of Japanese pear SFBB genes. Japanese pear cultivars with different S genotypes were subjected to CAPS analysis for SFBB genes.
A CAPS analysis also was used to examine the linkage between PpSFBB and S-RNase genes. A segregating population derived from a cross between Chikusui (S3S4) and Akemizu (S3S5) was analyzed for a linkage between S4-RNase and the PpSFBB4 genes. Figure 5 shows the representative results of this CAPS analysis. Three of the PpSFBB4 genes were detected specifically in the S4-containing progenies (18 of 40 plants analyzed), suggesting a linkage between S4-RNase and PpSFBB4. A similar analysis of 40 progenies derived from a cross between Akemizu (S3S5) and Shinsui (S4S5) also showed a linkage between the PpSFBB5 genes and S5-RNase.
Figure 5.—
Linkage analysis of SFBB genes and the S-RNase genes in Japanese pear. (a) Linkage between S4-RNase and PpSFBB4 genes. (b) Linkage between S5-RNase and PpSFBB5 genes. Parents (left two lanes) and their progenies were analyzed using CAPS.
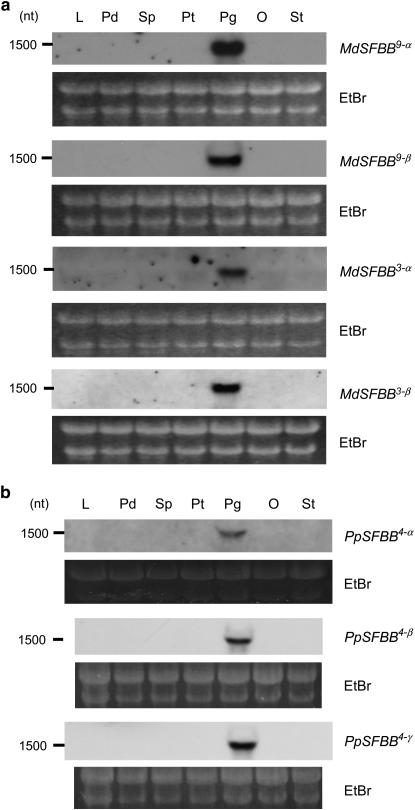
SFBB genes are specifically expressed in the pollen:
Organ-specific expression of the SFBB genes was analyzed using RNA blotting. Pollen-specific signals were detected for all SFBB genes (Figure 6). Since the SFBB genes show relatively high homology with each other in some pairs as is the case with SLF (Sijacic et al. 2004), these RNA blot results may suggest that each respective SFBB gene and/or its homologous gene(s) are specifically expressed in the pollen.
Figure 6.—
RNA gel blot analysis of SFBB genes. (a) Apple SFBB genes. (b) Japanese pear SFBB genes. Lf, leaf; Pd, pedicel; Sp, sepal; Pt, petal; Pg, pollen grain; Ov, ovary; St, style.
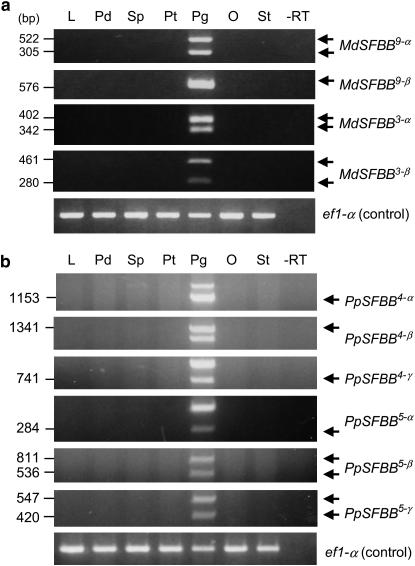
To determine whether each SFBB gene is specifically expressed in the pollen, an RT–PCR was performed in combination with a CAPS analysis (RT–PCR/CAPS). cDNAs derived from different organs were subjected to CAPS analysis to detect target, sequence-specific restriction fragments. Results showed that all the SFBB genes are actually and specifically expressed in pollen (Figure 7).
Figure 7.—
RT–PCR/CAPS analysis of SFBB expression in different organs. (a) Apple SFBB genes. (b) Japanese pear SFBB genes. −RT, pollen grain RNA negative control experiment performed without reverse transcriptase.
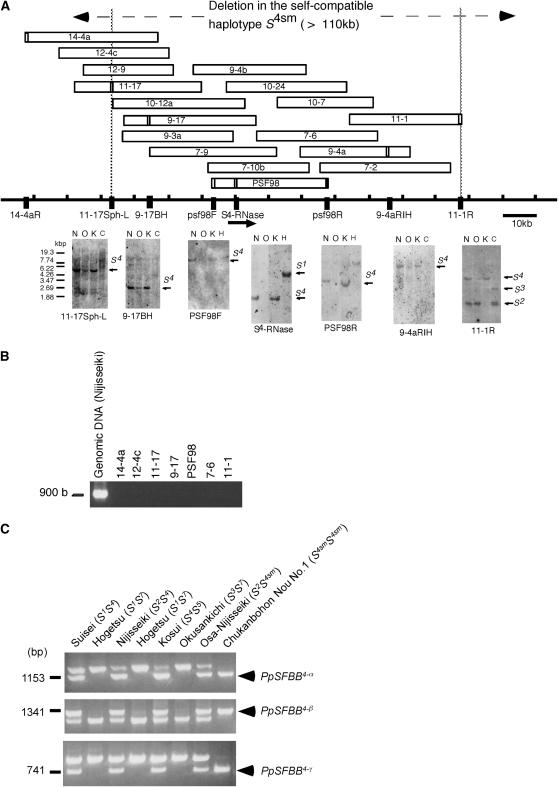
The S4sm haplotype, a style-specific self-compatible Japanese pear mutant, lacks the S4-RNase-containing region and retains SFBB4 genes:
We have previously analyzed a style-specific self-compatible Japanese pear mutant “Osa-Nijisseiki,” which has a defective S4 haplotype named S4sm, and shown that the S4sm haplotype lacks the S4-RNase gene-containing region for at least 4 kb (Sassa et al. 1997). Since pollen that has the S4sm haplotype is normally rejected by an S4 pistil, it is expected that the S4sm haplotype retains the pollen S4 gene and that this gene is located outside the deletion region.
To ascertain whether SFBB genes are retained in the S4sm haplotype, an ∼130-kb cosmid contig for the normal S4 haplotype was constructed and used to analyze the deletion region. Using probes derived from different positions on the contig, a genomic DNA blot analysis was conducted to determine if the corresponding region is deleted in the S4sm haplotype. Many of the cosmid end probes displayed smear patterns on a genomic DNA blot, probably because of the repetitive sequences that are rich in the S loci (Coleman and Kao 1992; Matton et al. 1995; Ushijima et al. 2001). Among the probes that displayed a single band on a DNA blot, the most upstream probe, 11-17Sph-L (located 37 kb upstream of S4-RNase), showed nearly no signal in Osa-Nijisseiki, suggesting that the corresponding region is deleted in the S4sm haplotype (Figure 8A). Similarly, the most downstream probe, 11-1R, detected nearly no signal in Osa-Nijisseiki. Faint signals found in Osa-Nijisseiki are derived from wild-type cells retained in the mutant, which was somaclonally derived from the original variety Nijisseiki (S2S4) and is chimeric for the S4 haplotype (Sassa et al. 1997). Therefore, a >110-kb region, located between 11-17Sph-L and 11-1R, is deleted in the S4sm haplotype.
Figure 8.—
Analysis of the style-specific, self-compatible haplotype S4sm. (A) A cosmid contig for the S4 haplotype and DNA gel blot analysis with cosmid-derived probes. Open boxes denote cosmid clones. N, Nijisseiki (S2S4); O, Osa-Nijisseiki (S2S4sm); K, Kosui (S4S5); C, Chojuro (S2S3); H, Hayatama (S1S2). (B) PCR amplification of SFBB genes from genomic DNA of Nijisseiki and cosmid clones for the S4 haplotype. (C) CAPS analysis of the S4smS4sm genotype, Chukanbohon Nou No.1, and other S genotypes.
Subsequently, we examined whether the SFBB4 genes are retained in the S4sm haplotype. SFBB genes could not be amplified by PCR from the cosmid clones of the S4 haplotype (Figure 8B). A subsequent CAPS analysis using an S4smS4sm homozygous genotype “Chukanbohon Nou No.1” did successfully amplify the SFBB4 genes from the S4smS4sm plant, indicating that the SFBB4 genes are retained in the S4sm haplotype (Figure 8C).
Nucleotide substitution patterns of SFBB genes:
For SI genes, new specificity is considered to have a reproductive advantage and tends to be maintained in a population. A sequence comparison has shown that SI genes have excess nonsynonymous substitutions, which supports the hypothesis that they are positively selected (Newbigin and Uyenoyama 2005). Consequently, we analyzed SFBB sequences to determine whether they show nucleotide substitution patterns similar to those of known SI genes.
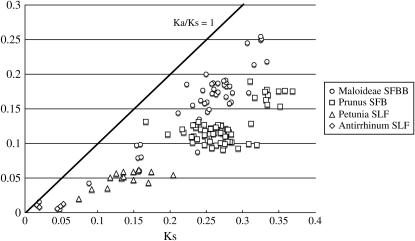
We used a codon-by-codon alignment of SFBB sequences to calculate the ratio of the nonsynonymous substitutions per nonsynonymous site (Ka) to the synonymous substitutions per synonymous site (Ks), or Ka/Ks. A similar analysis also was conducted for Prunus SFB, Petunia SLF, and Antirrhinum SLF. Figure 9 shows the results of the pairwise comparison of the Ka/Ks-values. The average Ka/Ks-value for SFBB (0.69) was higher than the Ka/Ks-values for Prunus SFB (0.45), Petunia SLF (0.34), and Antirrhinum SLF (0.27) (Table 4). These values indicate that SFB and SFBB are more diverged than SLF. As whole molecules, the Ka/Ks-values for all the genes were <1 and were lower than the Ka/Ks-values for S-RNase's: Maloideae, 0.83; Prunus, 0.54; and Solanaceae, 0.75 (Ma and Oliveira 2002). This may be due, partly, to the larger size of the F-box genes compared to the S-RNase genes and, partly, to the limited region(s) critical for recognition. For genes involved in recognition systems such as SI and disease resistance, it is known that the portions related to recognition are under positive selection (Ishimizu et al. 1998; Bergelson et al. 2001; Ikeda et al. 2004).
Figure 9.—
Pairwise comparisons of synonymous (Ks) and nonsynonymous (Ka) substitution frequencies in the SFBB and other S locus F-box genes. Circles, squares, triangles, and diamonds denote the data for SFBB of Maloideae, SFB of Prunus, SLF of Petunia, and SLF of Antirrhinum, respectively.
TABLE 4.
Ka/Ks-values of the S locus F-box genes
| Maloideae SFBB | Prunus SFB | Petunia SLF | Antirrhinum SLF | |
|---|---|---|---|---|
| Ks average (SD) | 0.2315 (0.0606) | 0.2648 (0.0413) | 0.1392 (0.0330) | 0.0337 (0.0168) |
| Ka average (SD) | 0.1586 (0.0573) | 0.1202 (0.0242) | 0.0482 (0.0121) | 0.0092 (0.0025) |
| Ka/Ks | 0.6853 | 0.4539 | 0.3463 | 0.2717 |
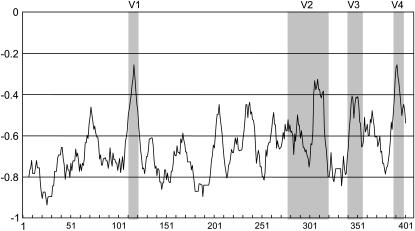
To detect diverged amino acid sites that may be important for recognition, an NVI (Kheyr-Pour et al. 1990) was calculated for the SFBB genes. Forty-eight variable sites were detected (supplemental Figure 1 at http://www.genetics.org/supplemental/). Four regions with particularly rich variable sites were named V1–V4 (Figure 10 and supplemental Figure 1). Ka/Ks-values were calculated for these four regions as well as for the F-box region. The Ka/Ks-values of the F-box, V1, V2, V3, and V4 regions were 0.22, 1.18, 1.33, 0.64, and 0.58, respectively (Table 5). These values suggest that V1 and V2 are under positive selection and that the F-box region is under purifying selection.
Figure 10.—
Window-averaged plot of normed variability index at each site in the alignment of the SFBB genes. Variable regions are shaded.
TABLE 5.
Ka/Ks-values of F-box and variable regions of SFBB genes
| F-box | V1 | V2 | V3 | V4 | |
|---|---|---|---|---|---|
| Ka (SD) | 0.070 (0.025) | 0.470 (0.293) | 0.246 (0.108) | 0.262 (0.121) | 0.763 (0.593) |
| Ks (SD) | 0.324 (0.149) | 0.397 (0.239) | 0.185 (0.073) | 0.409 (0.246) | 0.440 (0.266) |
| Ka/Ks | 0.218 | 1.181 | 1.331 | 0.640 | 0.576 |
| Pair no. of Ka > Ks (Total 44) | 1 | 21 | 30 | 12 | 9 |
DISCUSSION
Organization of the S locus of Maloideae:
Aiming to identify the pollen S gene in Maloideae, a subfamily of Rosaceae, we completely sequenced the 317-kb apple S9 haplotype. This represents—along with the 328-kb Petunia S2 haplotype (Wang et al. 2004)—one of the largest sequences for the S locus. In the 328-kb Petunia S2 haplotype sequence, 31 ORFs showed high similarity to retrotransposons (Wang et al. 2004). Comparable numbers were found in the 317-kb apple S9 haplotype: 27 of 71 predicted ORFs were homologous to retrotransposons. Although it has been suggested that the retrotransposon-rich organization of the Petunia S locus may reflect its centromeric location (Wang et al. 2004), there are no data showing the subcentromeric localization of the Maloideae S locus.
Several findings show that the Maloideae S locus is larger than the Prunus S locus. Analysis of the Japanese pear S4sm haplotype showed that the pollen S gene must be located outside the deletion region by at least 110 kb. Additionally, the distances of MdSFBB9-α and MdSFBB9-β from the S9-RNase gene are 42 and 93 kb, respectively. In contrast, the distances between Prunus S-RNase's and SFBs are 380 bases to 36 kb (Yamane et al. 2003; Ushijima et al. 2004). Differences in the size of the S locus between species also have been reported in Brassica, a species with sporophytic SI. The S locus region of Brassica oleracea is much larger than that of B. rapa (Fujimoto et al. 2006). Expansion of the S locus region in B. oleracea has been partly attributed to the insertion of retrotransposons, which suggests higher retrotransposon activity in B. oleracea than in B. rapa (Fujimoto et al. 2006). It should be noted that Maloideae is considered to be of polyploid origin (Evans and Campbell 2002) and polyploidization can activate retrotransposons (Madlung et al. 2005). The abundant retrotransposons found in the apple S locus may help to prevent recombination at the chromosomal region and to maintain the tight linkage between S-RNase and the pollen S allele.
Related, multiple, polymorphic, and pollen-specific F-box genes in the S locus of Maloideae:
Sequence analyses have revealed that the S loci of Prunus, Petunia, and Antirrhinum contain several F-box genes in addition to the pollen determinant SFB/SLF (Lai et al. 2002; Entani et al. 2003; Ushijima et al. 2003; Wang et al. 2004). In these species, however, each F-box gene is a single copy in a haplotype. In contrast, the S haplotypes of apple and Japanese pear contain two or three copies of the SFBB genes. During preparation of this article, Cheng et al. (2006) isolated S locus-linked and pollen-expressed F-box genes from apple by PCR and named them SLF of apple. The apple SLF was highly homologous to the SFBBs of apple and Japanese pear; however, Cheng et al. (2006) described a single SLF gene from each haplotype. SFBB genes, therefore, represent the first case of related and multiple F-box genes in the S locus. Initially, occurrence of multiple SFBB genes in a haplotype may appear inconsistent with the idea that they are the pollen determinant of GSI; the pollen-part factor F-box genes are single-copy genes in Prunus (SFB: Entani et al. 2003; Ushijima et al. 2003; Yamane et al. 2003) and in Petunia (SLF: Sijacic et al. 2004). However, apart from their multiplicity feature, SFBB genes are a good candidate for pollen S in Maloideae, as they show linkage to the S-RNase gene, S haplotype-specific sequence divergence, and pollen-specific expression. Unlike in Solanaceae, there is no report of subcentromeric localization of the S locus in Maloideae. Therefore, taking into consideration that Petunia SLF2 is located 161 kb from the S2-RNase gene (Wang et al. 2004), it seems unlikely that another genuine pollen S gene is located outside the 317-kb region of the apple S9 haplotype. Additionally, our analysis of the pistil-specific, self-compatible haplotype S4sm showed that PpSFBB4 genes are retained in the S4sm haplotype and are located outside the known deletion region. Taken together, these findings support the idea that the SFBB genes are the pollen S determinant in Maloideae.
An analysis of nucleotide substitution patterns of the SFBB genes and other pollen S genes showed that the SFBB genes have a higher average Ka/Ks-value than SFB of Prunus, SLF of Petunia, and SLF of Antirrhinum. Among the S-RNase genes, the Ka/Ks-value was also higher in Maloideae than in Prunus, suggesting that the S-RNase genes of Maloideae diverged more recently than those of Prunus (Ma and Oliveira 2002). The higher Ka/Ks-value of SFBB than of SFB may reflect the coevolution of the F-box and the S-RNase genes in Rosaceae. An amino acid sequence analysis of the SFBB detected four variable regions with high NVI values, and two of them (V1 and V2) were found to be under positive selection. Positive selection has been suggested in variable regions of S-RNase genes and SFB genes of Rosaceae, which supports the idea that these regions are critical for S specificity (Ishimizu et al. 1998; Ikeda et al. 2004). Positive selection detected in the variable regions of the SFBB genes is also consistent with the possible “self” recognition function of the protein. Although the Ka/Ks-values of the V3 and the V4 regions were <1, this may be due, partly, to high Ks-values in these regions and/or to gaps in the V4 region and may not exclude their potential importance in recognition.
The exceptional feature of SFBB as the pollen S candidate is its multiplicity. It is possible that only one SFBB gene in a haplotype is the pollen determinant. However, this seems unlikely, since pear shows competitive interaction (Crane and Lewis 1941; Lewis and Modlibowska 1942) and multiple SFBB genes with S-specific polymorphisms are expressed in pollen with normal GSI function. Expressed non-S SFBB genes may competitively interact with the pollen S SFBB to breakdown GSI in pollen. Another possibility is that all the expressed SFBB genes act together as the pollen determinant. Analyses of pollen-part, self-compatible mutations of Prunus have found that all mutations, both natural and X-ray-induced ones, were loss-of-function type (Ushijima et al. 2004; Sonneveld et al. 2005). However, loss-of-function mutations have not been reported in Maloideae, and pollen-part breakdown of GSI has been interpreted as a result of competitive interaction in tetraploid plants (Crane and Lewis 1941; Lewis and Modlibowska 1942). The occurrence of multiple pollen S genes also may explain the absence of deletion type of the pollen self-compatible mutation in Maloideae. However, sequence divergence of SFBB copies in a single haplotype may be unusual if the copies are only for backup function. Although duplication of a pollen S gene was reported for the Sb-haplotype of self-incompatible Arabidopsis lyrata, which exhibits sporophytic SI, sequences of the two copies of the SCRb genes were identical to each other (Kusaba et al. 2001). An interesting possibility is that SFBB proteins form a multimer in pollen as suggested by Luu et al. (2001). However, other possibilities that only one of the SFBBs in a haplotype or none of them are involved in pollen S specificity cannot be excluded at present. Functional characterization of SFBB in pollen and a comparative analysis of the apple S locus structure with those of other species will shed light on the mechanism, variation, and evolution of the the S-RNase-based GSI system.
Acknowledgments
We thank R. Tao of Kyoto University for helpful comments; T. Seki and K. Kawashima of Kanagawa Agricultural Technology Center and Y. Yamamoto, T. Hirai, and H. Katsura of Chiba Prefectural Agriculture Research Center for plant materials; K. Ohshima of Hitachi High-Tech Science Systems for shotgun sequencing; H. Zhang of Texas A&M University for the apple BAC library; and Melody Kroll for proofreading this manuscript. This work was supported by the Grants-in-Aid for Young Scientists (A, 16688001) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to H.S.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. AB270792 (genomic sequence of apple S9 haplotype), AB270793 (MdSFBB9-α), AB270794 (MdSFBB9-β), AB270795 (MdSFBB3-α), AB270796 (MdSFBB3-β), AB270797 (PpSFBB4-α), AB270798 (PpSFBB4-β), AB270799 (PpSFBB4-γ), AB270800 (PpSFBB5-α), AB270801 (PpSFBB5-β), and AB270802 (PpSFBB5-γ).
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bergelson, J., M. Kreitman, E. A. Stahl and D. Tian, 2001. Evolutionary dynamics of plant R-genes. Science 292: 2281–2285. [DOI] [PubMed] [Google Scholar]
- Burge, C., and S. Karlin, 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Cheng, J., Z. Han, X. Xu and T. Li, 2006. Isolation and identification of the pollen-expressed polymorphic F-box genes linked to the S-locus in apple (Malus x domestica). Sex Plant Reprod. 19: 175–183. [Google Scholar]
- Coleman, C. E., and T.-h. Kao, 1992. The flanking regions of two Petunia inflata S alleles are heterogeneous and repetitive sequences. Plant Mol. Biol. 18: 499–511. [DOI] [PubMed] [Google Scholar]
- Crane, M. B., and D. Lewis, 1941. Genetical studies in pears. III. Incompatibility and sterility. J. Genet. 43: 31–44. [Google Scholar]
- Dayhoff, M. O., R. M. Schwartz and B. C. Orcutt, 1979. A model of evolutionary change in proteins, pp. 345–352 in Atlas of Protein Sequence and Structure, Vol. 5 (Suppl. 3), edited by M. O. Dayhoff. National Biomedical Research Foundation, Washington, DC.
- de Nettancourt, D., 2001. Incompatibility and Incongruity in Wild and Cultivated Plants. Springer-Verlag, Berlin.
- Doyle, J. J., and J. L. Doyle, 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Entani, T., M. Iwano, H. Shiba, F.-S. Che, A. Isogai et al., 2003. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213. [DOI] [PubMed] [Google Scholar]
- Evans, R. C., and C. S. Campbell, 2002. The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. Am. J. Bot. 89: 1478–1484. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 2005. PHYLIP (Phylogeny Inference Package), Version 3.65. Distributed by the author (http://evolution.genetics.washington.edu/phylip.html). Department of Genome Sciences, University of Washington, Seattle.
- Fujimoto, R., K. Okazaki, E. Fukai, M. Kusaba and T. Nishio, 2006. Comparison of the genome structure of the self-incompatibility (S) locus in interspecific pairs of S haplotypes. Genetics 173: 1157–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij, A., K. Kondo, C. B. Lee, C. N. Hancock, M. Sivaguru et al., 2006. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439: 805–810. [DOI] [PubMed] [Google Scholar]
- Golz, J. F., V. Su, H.-Y. Oh, M. Kusaba and E. Newbigin, 2001. Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc. Natl. Acad. Sci. USA 98: 15372–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, N. R., H. Yamane, R. Tao and A. Iezzoni, 2006. Accumulation of nonfunctional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172: 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, A., and T.-h. Kao, 2006. Identification and characterization of components of a putative Petunia S-locus F-box–containing E3 ligase complex involved in S-RNase–based self-incompatibility. Plant Cell 18: 2531–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K., B. Igic, K. Ushijima, H. Yamane, N. R. Hauck et al., 2004. Primary structural features of the S haplotype-specific F-box proteins, SFB, in Prunus. Sex. Plant Reprod. 16: 235–243. [Google Scholar]
- Ishimizu, T., T. Endo, Y. Yamaguchi-Kabata, K. T. Nakamura, F. Sakiyama et al., 1998. Identification of regions in which positive selection may operate in S-RNase of Rosaceae: implication for S-allele-specific recognition sites in S-RNase. FEBS Lett. 440: 337–342. [DOI] [PubMed] [Google Scholar]
- Iwashita, S., N. Osada, T. Itoh, M. Sezaki, K. Oshima et al., 2003. A transposable element-mediated gene divergence that directly produces a novel type bovine Bcnt protein including the endonuclease domain of RTE-1. Mol. Biol. Evol. 20: 1556–1563. [DOI] [PubMed] [Google Scholar]
- Kao, T.-h., and T. Tsukamoto, 2004. Molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16: S72–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheyr-Pour, A., S. B. Bintrim, T. R. Ioerger, R. Remy, S. A. Hammond et al., 1990. Sequence diversity of pistil S-proteins associated with gametophytic self-incompatibility in Nicotiana alata. Sex. Plant Reprod. 3: 88–97. [Google Scholar]
- Kusaba, M., K. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643. [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., W. Ma, B. Han, L. Liang, Y. Zhang et al., 2002. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50: 29–42. [DOI] [PubMed] [Google Scholar]
- Lewis, D., and I. Modlibowska, 1942. Genetical studies in pears IV. Pollen-tube growth and incompatibility. J. Genet. 43: 211–222. [Google Scholar]
- Luu, D.-T., X. Qin, G. Laublin, Q. Yang, D. Morse et al., 2001. Rejection of S-heteroallelic pollen by a dual-specific S-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics 159: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, R.-C., and M. M. Oliveira, 2002. Evolutionary analysis of S-RNase genes from Rosaceae species. Mol. Genet. Genomics 267: 71–78. [DOI] [PubMed] [Google Scholar]
- Madlung, A., A. Tyagi, B. Watson, H. Jiang, T. Kagochi et al., 2005. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 41: 221–230. [DOI] [PubMed] [Google Scholar]
- Matton, D. P., S. L. Mau, S. Okamoto, A. E. Clarke and E. Newbigin, 1995. The S-locus of Nicotiana alata: genomic organization and sequence analysis of two S-RNase alleles. Plant Mol. Biol. 28: 847–858. [DOI] [PubMed] [Google Scholar]
- McClure, B. A., and V. Franklin-Tong, 2006. Gametophytic self-incompatibility: understanding the cellular mechanisms involved in “self” pollen tube inhibition. Planta 224: 233–245. [DOI] [PubMed] [Google Scholar]
- McClure, B. A., J. E. Gray, M. A. Anderson and A. E. Clarke, 1990. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347: 757–760. [Google Scholar]
- Newbigin, E., and M. K. Uyenoyama, 2005. The evolutionary dynamics of self-incompatibility systems. Trends Genet. 21: 500–505. [DOI] [PubMed] [Google Scholar]
- Qiao, H., H. Wang, L. Zhao, J. Zhou, J. Huang et al., 2004. The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16: 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sakata, K., Y. Nagamura, H. Numa, B. A. Antonio, H. Nagasaki et al., 2002. RiceGAAS: an automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa, H., and H. Hirano, 1998. Style-specific and developmentally regulated accumulation of a glycosylated thaumatin/PR5-like protein in Japanese pear (Pyrus serotina Rehd.). Planta 205: 514–521. [DOI] [PubMed] [Google Scholar]
- Sassa, H., T. Nishio, Y. Kowyama, H. Hirano, T. Koba et al., 1996. Self-incompatibility (S) alleles of the Rosaceae encode members of a distinct class of the T2/S-ribonuclease superfamily. Mol. Gen. Genet. 250: 547–557. [DOI] [PubMed] [Google Scholar]
- Sassa, H., H. Hirano, T. Nishino and T. Koba, 1997. Style-specific self-compatible mutation caused by deletion of the S-RNase gene in Japanese pear (Pyrus serotina). Plant J. 12: 223–227. [Google Scholar]
- Sassa, H., K. Ushijima and H. Hirano, 2002. A pistil-specific thaumatin/PR5-like protein gene in Japanese pear (Pyrus serotina): sequence and promoter activity of the 5′ region in transgenic tobacco. Plant Mol. Biol. 50: 371–377. [DOI] [PubMed] [Google Scholar]
- Sijacic, P., X. Wang, A. L. Skirpan, Y. Wang, P. E. Dowd et al., 2004. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Sonneveld, T., K. R. Tobutt, S. P. Vaughan and T. Robbins, 2005. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17: 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki, T., K. Okada, C. Castillo, Y. Moriya, T. Saito et al., 2004. Sequencing of the S9-RNase cDNA and PCR-RFLP system for discriminating S1- to S9-allele in Japanese pear. Euphytica 135: 157–167. [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., H. Sassa, M. Kusaba, R. Tao and M. Tamura, 2001. Characterization of the S-locus region of almond (Prunus dulcis): analysis of a somaclonal mutant and a cosmid contig for an S haplotype. Genetics 158: 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., H. Sassa, A. M. Dandekar, T. M. Gradziel, R. Tao et al., 2003. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., H. Yamane, A. Watari, E. Kakehi and K. Ikeda, 2004. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 39: 573–586. [DOI] [PubMed] [Google Scholar]
- Vinatzer, B. A., H.-B. Zhang and S. Sansavini, 1998. Construction and characterization of a bacterial artificial chromosome library of apple. Theor. Appl. Genet. 97: 1183–1190. [Google Scholar]
- Wang, Y., T. Tsukamoto, K.-w. Yi, X. Wang, S. Huang et al., 2004. Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S2-RNase. Plant Mol. Biol. 54: 727–742. [DOI] [PubMed] [Google Scholar]
- Yamane, H., K. Ikeda, K. Ushijima, H. Sassa and R. Tao, 2003. A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol. 44: 764–769. [DOI] [PubMed] [Google Scholar]