Abstract
During animal development, transcription factor activities are modulated by several means, including subcellular localization. The Hox cofactor Extradenticle (Exd) has a dynamic subcellular localization, such that Exd is cytoplasmic by default, but is nuclear when complexed with another homeodomain protein, Homothorax (Hth). These observations raise the question of whether dimerization with Hth simply induces Exd's nuclear localization or, alternatively, if Hth is also necessary for Exd activity. To address this question, we analyzed the nuclear transport signals in Exd, including a divergent nuclear export signal (NES) and two nuclear localization signals (NLSs). We show that, although these signals are weak compared to canonical signals, they balance each other in Exd. We also provide evidence that Exd contains an NLS mask that contributes to its cytoplasmic localization. With these signals characterized, we generated forms of Exd that are nuclear localized in the absence of Hth. Surprisingly, although these Exd forms are functional, they do not phenocopy Hth overexpression. These findings suggest that Hth is required for Exd activity, not simply for inducing its nuclear localization.
THE regulation of transcription factor activity plays an important role in many biological processes. For example, post-translational modification, expression levels, and protein stability are all mechanisms known to influence transcription factor activity (Whitmarsh and Davis 2000; Gill 2003). Another potent mechanism, the control of nuclear localization, is also commonly used to determine where and when a particular transcription factor can regulate its target genes (Xu and Massagué 2004).
In Drosophila melanogaster, the homeodomain protein Extradenticle (Exd) is regulated at the level of its subcellular localization (Mann and Abu-Shaar 1996; Aspland and White 1997). To date, Exd's nuclear localization correlates perfectly with the expression of another homeodomain protein, Homothorax (Hth) (Rieckhof et al. 1997; Kurant et al. 1998). In cells where hth is transcribed, both factors are nuclear. In cells where hth is not transcribed, Exd is cytoplasmic. Conversely, the presence of Exd also regulates Hth: in the absence of Exd, Hth is unstable and degraded (Abu-Shaar and Mann 1998; Kurant et al. 1998). Thus, the nuclear localization, and consequently the activities, of both Hth and Exd require the presence of both proteins in the same cell. Accordingly, both exd and hth carry out a wide range of indistinguishable functions during Drosophila development (Mann and Morata 2000). These functions include working as Hox cofactors, patterning the proximal–distal axes of the fly appendages, and performing critical functions in both eye and antennal development.
The mechanism governing the codependency of Hth and Exd involves a direct protein–protein interaction between these two homeodomain proteins that is mediated by evolutionarily conserved N-terminal domains present in both proteins. For Hth, this N-terminal domain is called the Homothorax-Meis (HM) domain, so named due to its high degree of amino acid identity with Hth's vertebrate homologs, encoded by the Meis genes (Ryoo et al. 1999). Interestingly, there exist isoforms of Hth that contain an HM domain, but no homeodomain (Glazov et al. 2005; Noro et al. 2006). These isoforms are sufficient to mediate the nuclear localization of Exd and can also carry out many of hth's genetically defined functions in vivo. Exd contains two highly conserved N-terminal domains, referred to as PBC-A and PBC-B (Bürglin and Ruvkun 1992), both of which contribute to the direct interaction with the HM domain of Hth (Abu-Shaar et al. 1999).
For proteins >40 kDa, such as Exd (at ∼45 kDa), translocation through the nuclear pore is facilitated by nuclear transport receptors known as karyopherins (Mosammaparast and Pemberton 2004). To recognize cargo, karyopherins bind short amino acid motifs in the primary sequence of protein cargo. Nuclear localization signals (NLSs) are formed by stretches of basic amino acids, which are bound by the importin-α protein for recognition by the importin-β karyopherin (Kaffman and O'Shea 1999). On the other hand, the CRM1 nuclear export karyopherin binds to nuclear export signals (NESs), which have classically been defined as a series of leucines with a particular spacing pattern (Kutay and Güttinger 2005). The activity of these nuclear transport signals can be regulated through protein modifications that enhance or decrease receptor–cargo interactions. Alternatively, nuclear transport signals can be masked to prevent receptor–cargo binding (Kaffman and O'Shea 1999).
Several studies have aimed to understand the regulation of the subcellular localization of Exd, initially focusing on nuclear localization. Between Hth and Exd, there are three predicted NLSs in the heterodimer. The predicted NLS in Hth is in its homeodomain but, because the HM domain is sufficient for nuclear localization of an Exd/HM complex, this NLS is dispensable (Ryoo et al. 1999). Consistently, sequence analyses revealed two putative NLSs within the homeodomain of Exd and its homologs (Abu-Shaar et al. 1999; Berthelsen et al. 1999; Saleh et al. 2000). The first, NLS1, is located within the N-terminal arm, while NLS2 is in helix 3. Notably, both of these NLSs are within regions of the homeodomain that mediate DNA binding.
The mechanisms governing the cytoplasmic localization of Exd and its homologs have been investigated using several approaches. For instance, protein interaction studies indicate that nonmuscle myosin retains Pbx1 in the cytoplasm (Huang et al. 2003). Extensive studies, however, have analyzed the role of nuclear export in the cytoplasmic localization of Exd. Pharmacological disruption of the nuclear export pathway consistently causes the nuclear accumulation of Exd or Pbx, indicating the presence of a conserved, functional NES in these proteins (Abu-Shaar et al. 1999; Berthelsen et al. 1999). However, despite the high degree of sequence identity between these two proteins, the location of this NES has been proposed to be in the PBC-A domain of Pbx (Berthelsen et al. 1999) vs. the PBC-B domain of Exd (Abu-Shaar et al. 1999). One interesting way to reconcile this discrepancy was provided by Saleh et al. (2000), who suggested that the PBC-A domain of Pbx inhibits nuclear localization indirectly, by binding intramolecularly to its own homeodomain, thus masking two NLSs present in this region of the protein. According to this idea, deletion of PBC-A would lead to nuclear localization by revealing these NLSs. In another study, phosphorylation of serine residues in the PBC-B domain of Pbx1 was suggested as regulating its nuclear localization (Kilstrup-Nielsen et al. 2003). This study also proposed the existence of two NESs in the PBC-A domain of Pbx1. One potential complication with these studies, however, is that many of the experiments were carried out in cultured cells that express an uncharacterized number of Hth family members. In addition, these putative NESs were analyzed by large deletions, which also have the potential of interfering with the function of the NLS mask proposed by Saleh et al. (2000).
Exploiting the genetic techniques available in D. melanogaster, where there is only a single hth-like gene, Abu-Shaar et al. (1999) expressed a series of N-terminal truncations of Exd to identify regions that govern its subcellular localization. The levels of Exd were kept low to avoid saturating the nuclear export system. An N-terminally truncated protein without a PBC-A domain was predominantly, although not exclusively, in the cytoplasm in the absence of Hth. However, upon further deletion into the PBC-B domain, the protein was found to be nuclear, suggesting the presence of an NES in this region of the protein. In this study, we extend these findings to more precisely identify and characterize the nuclear transport signals in Exd by studying the effect of point mutations in these signals. Our results show how the balance between two homeodomain NLSs and an NES present in the PBC-B domain controls Exd's localization. Additional experiments are consistent with the suggestion of Saleh et al. (2000) that Exd has an NLS mask that effectively blocks NLS activity in the absence of Hth. Finally, by manipulating these signals, we show, surprisingly, that nuclear localization of Exd is not sufficient for biological activity in the absence of Hth. This work thus provides a high-resolution view of the signals within Exd that control both its localization and its subsequent ability to regulate target genes.
MATERIALS AND METHODS
Construction of pK2 plasmids:
For the creation of transgenic UAS lines, exd cDNAs were cloned into the pK2 vector. pK2 is a derivative of the p131 vector (Abu-Shaar et al. 1999), itself a derivative of the pUAS-T vector. To make pK2, a PCR-amplified GFP coding region was inserted in frame with the myc tag of p131 to make the myc–GFP tag. All pieces of exd were PCR amplified from cDNA for insertion in frame with the myc–GFP tag of pK2. Point mutations were engineered using a PCR-based strategy. For exogenous nuclear transport signals, oligonucleotides encoding the NESMAPKK and NLSSV40 signals were inserted into the Asp718 restriction site upstream of the myc tag in pK2. To create the ΔAla construct, HindIII restriction sites were engineered in the primers for nucleotides 467 and 530. After PCR amplification, a three-way ligation was performed to insert the fragments into pK2. All constructs were sequenced to confirm that there were no PCR-induced mutations. For analysis of the relationships between the nuclear transport signals along the proximal–distal axis, we made single, double, and triple mutations of the NES and NLSs in Exd178–300: A209HKKA213SSA216QA218 (ExdNES), RA240A241A242R (ExdNLS1), and A291A292IRYKK (ExdNLS2).
Fly stocks:
yw exd1 FRT19A/FM7 was a generous gift of Claude Desplan. yw hs-flp FRT19A; UAS-LacZ tub-Gal4/CyO was a kind gift of Andreu Casali. yw hs-flp; tub-Gal4/TM3 was a helpful gift of Michael Crickmore. w hs-flp Gal80 FRT19A/FM7 was obtained from the Bloomington Stock Center. ptc-Gal4 and Dll-Gal4 were obtained from the Mann lab stock collection.
Immunohistochemistry:
Discs were dissected in 1× PBS. To fix the tissue for microscopy, discs were first fixed for 20 min at RT in 4% formaldehyde (Polysciences) in 1× PBS, followed by a second fixation period of 20 min at room temperature (RT) in 4% formaldehyde in PBT (1× PBS, 0.1% Triton X-100). The discs were incubated with primary antibodies for 2 hr at RT, washed, incubated with secondary antibodies for 1 hr 30 min at RT, washed overnight at 4°, and mounted in 70% glycerol with the DABCO additive to preserve fluorescence (1,4-diazabicyclo[2.2.2]octane; Sigma, St. Louis). The following primary antibodies were used at the indicated dilution (source): mouse anti-Dac 1:20 and mouse antilamin 1:10 (Hybridoma Bank), rabbit anti-myc 1:100 (Molecular Probes, Eugene, OR), rabbit anti-β-gal 1:2000 (Cappell), guinea pig anti-Hth 1:1000, and rabbit anti-Exd 1:1000.
Construction of cell culture plasmids:
exd and exdnes were PCR amplified from the pK2 plasmids described above and inserted into the p131 vector to be myc tagged (Abu-Shaar et al. 1999). The fragments were subcloned into the pRMHA3 vector for inducible expression.
Cell culture:
Schneider line 2 (S2) cells were maintained in Sf-900 II media (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) with penicillin (50 mg/ml) and streptomycin (50 mg/ml) (Sigma). Transient transfections were performed with Cellfectin (Invitrogen, San Diego) using eight-well slides for immunocytochemistry.
Immunocytochemistry:
For immunocytochemistry, 70 μm CuSO4 was added 6 hr prior to fixation. Leptomycin B (10 nm final concentration) (Sigma) or a mock treatment was added to the media 3 hr prior to fixation. Cells were first fixed for 20 min at RT in 4% formaldehyde (Polysciences) in 1× PBS, followed by a second fixation period of 20 min at RT in 4% formaldehyde in PBT (1× PBS, 0.1% Triton X-100). The cells were treated with RNase (QIAGEN, Chatsworth, CA) for 30 min in PBT. For immunocytochemistry, the cells were then incubated with primary antibodies for 2 hr at RT with rotation, washed, incubated with secondary antibodies for 1 hr 30 min at RT, and washed overnight at 4°. Cells were stained with propidium iodide at a final concentration of 1 μg/ml (Molecular Probes) to visualize nucleic acids. After a quick rinse, the cells were mounted in 50% glycerol in PBS. The primary antibodies were used at the indicated concentration (source): mouse antilamin 1:10 (Hybridoma Bank) and rabbit anti-myc 1:100 (Molecular Probes).
RESULTS
Identification of a NES in the PBC-B domain of Exd:
Previous studies in Drosophila suggested the presence of a sequence located between residues 178 and 220 that was required for Exd's cytoplasmic localization (Abu-Shaar et al. 1999). One explanation for this observation is that this sequence contains an NES, a short motif originally characterized as a series of leucines arranged in a typical spacing pattern: L-x2–3-L-x2–3-L-x-L (where “x” is any residue). As more NESs have been identified, most large hydrophobic amino acids have been found in the key positions, including isoleucine, valine, methionine, and phenylalanine (la Cour et al. 2004). With this expanded definition, we analyzed the primary sequence of Exd for potential NES-like sequences. The sequence I209hkkFssIqM218 best fits this consensus, particularly in the spacing of the hydrophobic residues, which is not true of other hydrophobic-rich sequences in Exd. This sequence has been conserved over evolution, with only conservative changes of the key residues within the Pbx family (Figure 1B). Further, this sequence is within the previously defined region of PBC-B that is required for cytoplasmic localization in Drosophila imaginal discs (Figure 1A) (Abu-Shaar et al. 1999).
Figure 1.—
Identification of the NES in Exd. (A) Schematic of Exd protein depicting the location of the nuclear transport signals within the conserved domains of Exd: PBC-A, an alanine-rich region (Ala), PBC-B, and the homeodomain (HD). Exd interacts with Hth through the PBC-A domain. (B) By sequence analysis, a putative NES was found in the conserved PBC-B domain in Exd orthologs. In ExdNES, each of the key amino acids was mutated to an Ala. (C) S2 cells transfected with plasmids to express myc-tagged Exd or myc–ExdNES in the presence and absence of LMB. Nuclei are marked with an antibody against nuclear lamin (blue) and the nucleic acid stain propidium iodide (red). (D) Immunofluorescence imaging of leg imaginal discs expressing myc–GFP–Exd+ and myc–GFP–ExdNES under the control of the ptc-Gal4 driver, with blow-up views of proximal cells (P) and distal cells (D). In this and following figures, proximal nuclei are marked with an antibody against Hth (blue), distal (Hth-non-expressing) nuclei are marked with an antibody against Dachshund (Dac, red), and myc staining is in green. In the trochanter region of the disc, Hth and Dac are coexpressed (purple). In the myc-only panels, examples of nuclei that show predominantly nuclear myc (red arrows), predominantly cytoplasmic myc (green arrows), and a mixture of cytoplasmic and nuclear myc (yellow arrows) are indicated.
We first tested a role for this putative NES in the D. melanogaster cell line S2, which provides a useful environment because hth is not expressed in these cells (Abu-Shaar et al. 1999). Using an inducible expression system, we also took care to generate low amounts of these proteins to avoid saturating the export machinery. myc-Tagged Exd+ localized to the cytoplasm of these cells (Figure 1C). The CRM1 export receptor, which facilitates the nuclear translocation of proteins with leucine-rich NESs, can be specifically inhibited by leptomycin B (LMB) (Fornerod et al. 1997). Upon treatment with LMB, myc–Exd+ accumulated in the nucleus, indicating that CRM1 recognizes Exd and transports it to the cytoplasm (Figure 1C). As noted previously (Abu-Shaar et al. 1999), nuclear localization is not complete, suggesting the existence of a CRM1-independent mechanism for keeping Exd in the cytoplasm (see below).
To test the activity of the putative NES, each of the key amino acids was mutated to alanine to create myc–ExdNES (see materials and methods). myc–ExdNES localized predominantly to the nucleus of these cells (Figure 1C), indicating that these four amino acids are essential for the cytoplasmic localization of the protein in the absence of Hth. As with the LMB treatment of myc–Exd+, myc–ExdNES was also found at low levels in the cytoplasm. If the putative NES and CRM1 are acting at the same step, the localization of myc–ExdNES should not be affected by LMB. Consistent with this idea, LMB did not alter the subcellular localization of myc–ExdNES (Figure 1C). This result suggests that the control by CRM1 of the localization of Exd is mediated by the sequence I209hkkFssIqM218, which forms a functional NES.
To extend these results to a more in vivo setting, we used the leg imaginal disc, which is composed of both hth-expressing proximal cells, where Exd is nuclear, and hth-nonexpressing distal cells, where Exd is cytoplasmic (González-Crespo and Morata 1995; Rauskolb et al. 1995; Mann and Abu-Shaar 1996; Aspland and White 1997). To monitor subcellular localization, wild-type and mutant Exd coding sequences were expressed as myc–GFP-tagged fusion proteins using the Gal4-UAS system (Brand and Perrimon 1993). For these experiments, we used the ptc-Gal4 driver, which is expressed along the anterior–posterior compartment boundary of the leg imaginal disc and is therefore active in both proximal hth-expressing and distal hth-nonexpressing cells. For all transformants, care was taken to use lines that express less than or equal to the levels of endogenous Exd. Expressing only myc–GFP (∼37 kDa) using this driver resulted in a similar staining intensity in both the nucleus and the cytoplasm in all cells, suggesting that this tag is not recognized by the subcellular localization machinery (Figure 1D).
When myc–GFP–Exd+ is expressed under these conditions, the protein was cytoplasmic in distal cells and predominantly nuclear in proximal cells (Figure 1D). In contrast, myc–GFP–ExdNES had distinct nuclear staining along the entire proximal–distal axis of the imaginal disc, as shown by costaining for a distal transcription factor, Dachshund (Dac) (Figure 1D). As in S2 cells, the nuclear localization of myc–GFP–ExdNES was not complete, again suggesting a second mechanism for retaining Exd in the cytoplasm. This localization pattern was also similar to expression of an Exd form in which the region with export activity identified by Abu-Shaar et al. (1999) was deleted in an otherwise full-length protein (data not shown). Taken together, these results suggest that the I209hkkFssIqM218 sequence is a bona fide NES that plays an important role in the cytoplasmic localization of Exd in the absence of Hth.
A fragment of Exd reveals a competition between nuclear transport signals:
As described above, mutation or deletion of the NES in the PBC-B domain of Exd resulted in significant, but incomplete, nuclear localization, suggesting that there may be a second mechanism of cytoplasmic localization. To analyze the role of the NES in a simpler setting, we characterized the localization of a fragment of Exd from residue 178 to 300. This fragment includes a portion of the PBC-B domain, containing the NES defined above, but does not contain the Hth-binding domain or the NLS mask as defined by Saleh et al. (2000), both of which are in the PBC-A domain. Therefore, Exd178–300 is expected to behave similarly in both proximal and distal cells. Further, this fragment contains the homeodomain, which was found by Abu-Shaar et al. (1999) to contain the nuclear localization activity of Exd. Indeed, there are two predicted NLSs within this region: R239RKRR (NLS1) and K291RIRYKK (NLS2). These NLSs were mutated either individually or together with the NES mutation (see materials and methods).
When expressed using the ptc-Gal4 driver, myc–GFP–Exd178–300+ had a mixed subcellular localization in both distal and proximal cells (Figure 2A). This localization suggests that, in the absence of the rest of the protein, the NLSs and the NES present in Exd178–300+ are competing with each other, resulting in a mixed nuclear and cytoplasmic distribution. Consistent with this idea, mutations in the nuclear transport signals dramatically shift this equilibrium. Mutation of the NES (myc–GFP–Exd178–300NES) resulted in an exclusively nuclear protein, demonstrating that the NES is the sole source of cytoplasmic localization activity in this fragment and that, in its absence, the NLSs efficiently direct the fragment to the nucleus (Figure 2E). Mutation of NLS1 (myc–GFP–Exd178–300NLS1) resulted in a protein that is distributed evenly throughout the cell, similarly to myc–GFP–Exd178–300+ (Figure 2B). Mutation of NLS2 (myc–GFP–Exd178–300NLS2), however, shifted the equilibrium in favor of cytoplasmic localization (Figure 2C). Mutation of both NLS1 and NLS2 (myc–GFP–Exd178–300NLS1,NLS2) shifted this equilibrium even further, resulting in a protein that was exclusively cytoplasmic (Figure 2D). These experiments indicate that NLS2 is stronger than NLS1, although both are active NLSs. Further, in the absence of all NLS activity, the wild-type NES is sufficient to direct the protein to the cytoplasm.
Figure 2.—
Exd178–300 contains an NES that competes with two NLSs. Single, double, and triple mutants of the NES and NLS signals were engineered in myc–GFP–Exd178–300 and the proteins were expressed in the ptc domain of leg imaginal discs. (A) myc–GFP–Exd178–300. (B) myc–GFP–Exd178–300NLS1. (C) myc–GFP–Exd178–300NLS2. (D) myc–GFP–Exd178–300NLS1,NLS2. (E) myc–GFP–Exd178–300NES. (F) myc–GFP–Exd178–300NES,NLS1. (G) myc–GFP–Exd178–300NES,NLS2. (H) myc–GFP–Exd178–300NES,NLS1,NLS2.
The NLS mutations were also examined in the context of a mutated NES to analyze the nuclear localization potential of the NLSs without competition from the NES. Like myc–GFP–Exd178–300NES, myc–GFP–Exd178–300NES,NLS1 was exclusively nuclear, again suggesting that NLS1 is not required in the presence of NLS2 (Figure 2F). Mutation of NLS2, however, was more severe. myc–GFP–Exd178–300NES,NLS2 had a mixed distribution throughout the cell, as did myc–GFP–Exd178–300NES,NLS1,NLS2 (Figure 2, G and H). These data provide further evidence that NLS2 is the stronger NLS within the protein. When NLS2 is the only signal in the fragment, the protein is exclusively nuclear, demonstrating that NLS2 is sufficient to establish nuclear localization. NLS1 is much weaker and has little discernible ability to influence the localization of the fragment at this resolution.
The nuclear transport signals in Exd are weak compared to classical nuclear transport signals:
The above experiments indicate that the NLSs have different strengths and that together they effectively compete with Exd's NES. To further assess the strengths of these signals, two smaller fragments were tested that separate the NLS-containing homeodomain from the NES-containing PBC-B domain. The subcellular localization of these fragments (fused to myc–GFP) were determined in isolation and when fused to canonical nuclear transport signals. As expected, the homeodomain alone (myc–GFP–Exd237–300) was exclusively nuclear (Figure 3A). The canonical NES from MAPKK is efficient at localizing cargo to the cytoplasm (Fukuda et al. 1996). To determine if Exd's NLSs are able to compete with NESMAPKK, we fused it to the N terminus of myc–GFP–Exd237–300 to make NESMAPKK–myc–GFP–Exd237–300. This protein was detected exclusively in the cytoplasm (Figure 3B), indicating that the NLSs of Exd are not able to promote nuclear localization in the presence of this strong NES. Conversely, myc–GFP–Exd178–236 contains NESExd, which directed the protein to the cytoplasm (Figure 3C). When the SV40 large T-antigen NLS was added, however, NLSSV40–myc–GFP–Exd178–236 was found exclusively in the nucleus (Figure 3D). Thus, NESExd is not strong enough to counter the activity of a strong NLS.
Figure 3.—
The nuclear transport signals in Exd are weak in comparison to classical signals. All proteins were expressed with ptc-Gal4 and analyzed in leg discs. (A) The homeodomain, Exd237–300, includes the two NLSs and is constitutively nuclear. (B) NESMAPKK-Exd237–300 has the NES of MAPKK fused at the N terminus of Exd's homeodomain and is constitutively cytoplasmic. (C) A fragment of the PBC-B domain, Exd178–236, includes the NES and is constitutively cytoplasmic. (D) NLSSV40-Exd178–236 has the NLS of the large T-antigen of SV40 fused to the N terminus of Exd178–236.
NESExd is not required for Exd function in the proximal leg:
In full-length Exd, the NES promotes its cytoplasmic localization in distal, non-Hth-expressing cells. In proximal cells, the NES may not be required because Hth overcomes this signal to promote its nuclear localization. Alternatively, Exd may have to shuttle in and out of the nucleus to function properly in proximal cells. To see if the NES is necessary for Exd activity in proximal cells, the mosaic analysis with a repressible cell marker (MARCM) technique (Lee and Luo 1999) was used to rescue exd− cells with myc–GFP–Exd+ and myc–GFP–ExdNES. For comparison, exd− clones expressing LacZ were generated in parallel. These clones replicated previous analyses of exd mutant tissue (González-Crespo and Morata 1995). Exd is required for Hth stability and, therefore, as expected, these clones did not stain for Hth (Abu-Shaar and Mann 1998) (Figure 4A). exd− clones caused the fusion of leg segments to the body wall and changed the fate of the tissue to a more distal identity, as evidenced by the presence of bracted bristles, which normally form only in distal leg segments (Figure 4B). In contrast, exd− clones had no phenotype in the distal leg (Figure 4C).
Figure 4.—
NES is dispensable for Exd function in proximal cells. Using the MARCM technique, exd− clones express LacZ (A–C), myc–GFP–Exd+ (D–F), or myc–GFP–ExdNES (G–I). (A, D, and G) Immunofluorescence was used to assay the ability of these proteins to rescue Hth stability. Clones are marked by expression of either LacZ (green) or the myc-tagged transgene product (green). The levels of both myc–GFP–Exd+ and myc–GFP–ExdNES are similar to endogenous Exd (red). Hth (blue) staining is lost in exd− tub>LacZ clones, but is detected and nuclear in exd−, tub>myc–GFP–Exd+ and exd−, tub>myc–GFP–ExdNES clones. (B) Small proximal y− exd−, tub>LacZ clones cause segmental fusions and a loss of proximal fate as distinguished by bracted bristles (inset, black arrowheads indicate y+ bristles and white arrowheads indicate y− bristles). (C) y− exd− cells do not have defects in distal patterning. (E) Proximal clones of exd−, tub>myc–GFP–Exd+ cells do not fuse to the body, nor do they lose proximal fates (inset). (F) exd−, tub>myc–GFP–Exd+ clones do not disrupt distal patterning. (H) exd−, tub>myc–GFP–ExdNES clones do not cause segmental fusions or the loss of proximal fates (inset). (I) exd−, tub>myc–GFP–Exdnes do not disrupt distal patterning.
All of these phenotypes were rescued by the expression of myc–GFP–Exd+ or by the expression of myc–GFP–ExdNES. Both proteins restored the stability of Hth (Figure 4, D and G) and a normal leg pattern (Figure 4, E and H). Clones in distal leg tissue also did not interfere with distal leg development (Figure 4, F and I). These results demonstrate (1) that the myc–GFP tag does not interfere with Hth–Exd activity and (2) that a wild-type NES is not required for Exd function in proximal cells.
Evidence for an NLS mask:
We next turned to two earlier observations suggesting that there is a second mechanism, in addition to the NES, that promotes Exd's cytoplasmic localization. First, when the NES was mutated in the context of an otherwise full-length protein, some residual cytoplasmic protein was still observed. Second, when the NES was mutated in the context of Exd178–300, no residual cytoplasmic protein was observed. These results suggest that sequences present in full-length Exd, but not in Exd178–300, contribute to Exd's cytoplasmic localization in an NES-independent manner. Interestingly, experiments by Saleh et al. (2000) demonstrated that the N terminus of Exd can bind its own homeodomain in “GST-pulldown” experiments. Taken together, these findings suggest that the N-terminal domain of Exd may promote the cytoplasmic localization of the protein by binding to and blocking the activity of the NLSs in the homeodomain (Saleh et al. 2000).
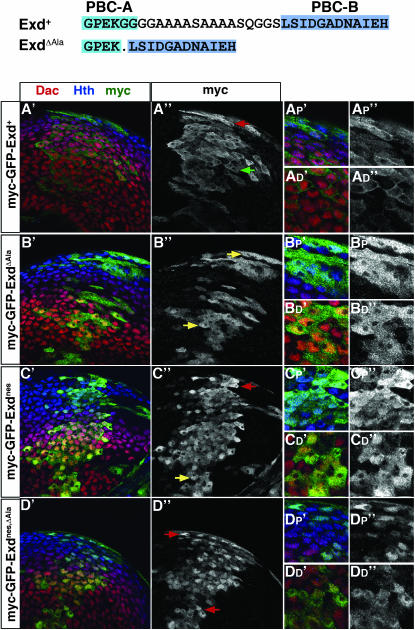
One striking feature of Exd's N terminus is a stretch of alanines that separates the PBC-A and PBC-B domains (Figure 5). The length of this polyalanine sequence varies from 8 to 12 residues among Exd homologs and is occasionally interspersed with glycine or serine residues. Structurally, stretches of alanines have a high propensity to form α-helices (Spek et al. 1999), which could be important for maintaining a conformation for nuclear or cytoplasmic localization of Exd, by helping to position the NLS mask. To determine if this region is important for regulating the subcellular localization of Exd, the polyalanine domain was deleted, thereby fusing the PBC-A and PBC-B domains. This deletion protein, ExdΔAla, was fused to myc–GFP and expressed in leg discs using ptc-Gal4. In contrast to myc–GFP–Exd+ (Figure 5A), myc–GFP–ExdΔAla was partially nuclear in distal cells, despite a wild-type NES (Figure 5B). We suggest that the deletion of the polyalanine stretch interferes with the ability of the PBC-A domain to interact with and mask the NLSs in the homeodomain.
Figure 5.—
Deletion of the polyalanine region between PBC-A and PBC-B promotes NLS activity. (Top) Schematic of the polyalanine region in Exd+ showing the deletion in ExdΔAla. myc–GFP–tagged derivatives were expressed with the ptc-Gal4 driver in leg discs. (A) myc–GFP–Exd+. (B) myc–GFP–ExdΔAla. (C) myc–GFP–ExdNES. (D) myc–GFP–ExdNES,ΔAla.
Instead of interfering with the NLS mask, we considered the possibility that the ΔAla deletion somehow interferes with the activity of the NES, resulting in nuclear localization. However, if the NES was nonfunctional in this protein, we would expect that mutating it should have no effect on the localization of myc–GFP–ExdΔAla. To test this, we examined the localization of myc–GFP–ExdNES,ΔAla and found that it was more nuclear than either myc–GFP–ExdΔAla or myc–GFP–ExdNES (Figure 5, B–D). Thus, these results are consistent with the idea that the polyalanine deletion interferes with a NES-independent mechanism for cytoplasmic localization, such as an NLS mask. We note, however, that myc–GFP–ExdNES,ΔAla is not as exclusively nuclear as the homeodomain alone, suggesting that the NLS mask may not be fully disabled by this deletion, or that there are additional mechanisms to keep this protein out of the nucleus.
Nuclear localization of Exd does not phenocopy ectopic expression of Hth:
Expression of Hth in the distal cells of the leg imaginal disc induces the nuclear localization of Exd and represses the response to Wg and Dpp, thus blocking proximal–distal axis formation (Abu-Shaar and Mann 1998; Ryoo et al. 1999; Azpiazu and Morata 2002). These previous results were unable to determine if Hth was required only for the nuclear localization of Exd, or if Hth was also required for gene regulation. Using the Dll-Gal4 driver (Figure 6A), we assayed the ability of nuclear Exd, in the absence of Hth, to disrupt distal patterning. Immunostains with the anti-Exd antibody demonstrated similar levels of nuclear Exd for all constructs. Using this driver, the expression of myc–GFP alone had no effect on Exd levels or localization (Figure 6B) or leg patterning (Figure 6C). GFP–Hth, however, induced the nuclear localization of Exd in distal cells (Figure 6D) and disrupted distal leg patterning, leading to a truncated leg (Figure 6E). Expression of the HM domain of Hth also induced nuclear localization of Exd (Figure 6F) and disrupted distal leg patterning (Figure 6G), confirming that this phenotype does not require the homeodomain of Hth (Ryoo et al. 1999; Jaw et al. 2000).
Figure 6.—
Nuclear Exd is not equivalent to Hth. UAS transgenes were expressed in distal cells under the control of the Distalless-Gal4 (Dll-Gal4) driver. (A) The Dll-Gal4 expression domain is shown with expression of myc–GFP (A, green). Exd (B, white) is nuclear in proximal cells, marked with Hth in A (blue). Note that the Hth+ cells in the distal domain are peripoedal cells in the focal plane of A and B. The following were expressed using the Dll-Gal4 driver: myc–GFP (A–C), GFP–Hth (D and E), GFP–HthHM (F and G), myc–GFP–Exd+ (H and I), myc–GFP–ExdNES (J and K), myc–GFP–ExdΔAla (L and M), and myc–GFP–ExdNES,ΔAla (N and O). Antibody stains demonstrate similar levels of nuclear Exd (white in B, D, F, H, J, and N) with Hth–Exd dimers, ExdNES, ExdΔAla, or ExdNES,ΔAla. Adult female T1 legs lose distal fates when nuclear Hth–Exd or HthHM–Exd dimers are expressed (E and G), but are not affected by undimerized nuclear Exd (K, M, and O).
When expressed with Dll-Gal4, myc–GFP–Exd+ is cytoplasmic in distal leg disc cells (Figure 6H) and has no effect on distal leg patterning (Figure 6I). Although mutation of the NES allows nuclear accumulation of the protein (Figure 6J), expression of myc–GFP–ExdNES with Dll-Gal4 also does not disrupt distal patterning (Figure 6K), suggesting that nuclear localization of Exd is not sufficient to disrupt distal cell fates. The above experiments, however, suggest that the homeodomain may still be masked in ExdNES and that deletion of the polyalanine residues at least partially disables this mask. When expressed in the Dll domain, however, neither myc–GFP–ExdΔAla nor myc–GFP–ExdNES,ΔAla had any effect on distal leg fate (Figure 6, L–O). Similarly, unlike Hth, these proteins also did not produce any phenotypes when ectopically expressed in the antenna or wing (data not shown). Thus, forcing the nuclear localization of Exd in distal cells of the leg or in other tissues is not able to phenocopy the expression of Hth. These results suggest that the HM domain of Hth contributes to the activity of Exd beyond the induction of nuclear localization.
DISCUSSION
Characterization of the nuclear transport signals in Exd:
In this study, we have identified a CRM1-dependent nuclear export signal of Exd. Mutation of this signal is sufficient to induce the nuclear accumulation of an otherwise wild-type, full-length protein. ExdNES is not responsive to leptomycin B treatment, indicating that amino acids I209hkkFssIqM218 compose the sole NES in Exd. This NES has been highly conserved over evolution and is found in all known Exd homologs. While CRM1-dependent NESs are typically composed of leucines, the export receptor can also recognize other large hydrophobic amino acids, including the isoleucines, methionine, and phenylalanine found in the NES of Exd (la Cour et al. 2004). The Exd export signal, however, appears to be weak. While sufficient to compete with the NLSs in the Exd homeodomain, the NES cannot compete with the strong NLS of the SV40 large-T antigen. The weakness of this NES may be due to its unusual lack of leucines, which are more typically found in previously characterized NESs.
An inefficient NES may be important for regulating the subcellular localization of the Hth–Exd dimer. In distal cells, the NES is crucial for the cytoplasmic localization of Exd. In proximal cells, however, the Hth–Exd dimer is exclusively nuclear. The fragment Exd178–300, which does not bind Hth, is evenly distributed between the nucleus and cytoplasm of all cells along the proximal–distal axis, demonstrating that the NES is not regulated differently in proximal cells than in distal cells. Therefore, association with Hth deactivates the NESExd, perhaps by physically masking this sequence and thus preventing its association with CRM1.
Exd has two NLSs in the homeodomain: a weaker NLS1 in the N-terminal arm and a stronger NLS2 in helix 3. These results contrast with those by Saleh et al. (2000), who observed similar strengths for the NLSs in the Pbx1 homeodomain. On the basis of sequence, we had predicted that NLS1 would be stronger than NLS2 (Abu-Shaar et al. 1999). Importin-α makes extensive peptide backbone contacts with NLS sequences, and as such, there is an important structural requirement for NLS function (Fontes et al. 2000, 2003). NLS1 and NLS2 may have differing abilities for making the structure necessary for strong importin-α binding. Indeed, upstream of NLS2, there is a potential bipartite NLS minor site, which may assist in forming a preferred conformation for importin-α binding. Compared to the nuclear localization activity of the NLSSV40, the NLSExd's are weak. The combined activity of the NLSs can compete with, but not overcome, the activity of the NESExd.
Evidence for an NLS mask in the N terminus of Exd:
In addition to a weak NES, Exd and Pbx also have a second mechanism for promoting cytoplasmic localization that has been referred to as an NLS mask (Saleh et al. 2000). Deleting the N terminus (PBC-A) of Exd removes this mask, allowing nuclear localization (Neuteboom and Murre 1997; Peltenburg and Murre 1997; Abu-Shaar et al. 1999; Calvo et al. 1999; Saleh et al. 2000). Such an idea is also consistent with previous results showing that full-length Exd or Pbx bind DNA poorly as a monomer while N-terminally truncated forms of Exd or Pbx bind DNA well (Lu and Kamps 1996; Neuteboom and Murre 1997; Peltenburg and Murre 1997; Green et al. 1998; Calvo et al. 1999; Saleh et al. 2000). These results suggest the entire homeodomain is masked by the N terminus, preventing both DNA binding and nuclear localization in the absence of Hth.
We have shown that deletion of the polyalanine residues that separate PBC-A and PBC-B also deregulates NLS activity, suggesting that the NLS mask has been compromised by this deletion. We favor the idea that the polyalanine stretch is not, itself, the mask but instead functions to correctly position the mask. Unlike the nonpolar polyalanine sequence in Exd, NLS masks are usually composed of acidic residues or are created by phosphorylation (Kaffman and O'Shea 1999). However, apparently unlike Pbx1 (Kilstrup-Nielsen et al. 2003), we have found no evidence that Exd is phosphorylated when cytoplasmic (our unpublished observations), suggesting that Exd's subcellular localization may not be regulated by this post-translational modification. When we disrupt both cytoplasmic localization systems (the NES and the NLS mask), myc–GFP–ExdNES,ΔAla is mostly nuclear, but residual cytoplasmic staining suggests that the polyalanine deletion does not fully disable the NLS mask. Alternatively, there may be additional mechanisms built into Exd that promote its cytoplasmic localization. Experiments by Huang et al. (2003) demonstrate that nonmuscle myosin binds the N terminus of Exd and promotes cytoplasmic localization. Indeed, Exd subcellular localization is deregulated in embryos mutant for the Drosophila homolog of the nonmuscle myosin, zipper. This phenotype, however, is more subtle than the engineered mutations that affect the nuclear transport of Exd described here. Therefore, while nonmuscle myosin may refine the localization equilibrium by retaining Exd in the cytoplasm, we suggest that the regulated activities of the NES and NLSs are the principal determinants of the subcellular localization of Exd.
A model of the subcellular localization of Exd:
On the basis of these observations, we propose the following model of Exd subcellular localization. By immunostain, Exd is detected in the cytoplasm of any cell that does not express hth. To accomplish this, we suggest that the NLSs are inhibited by an NLS mask, while the NES promotes the cytoplasmic localization of the protein. Disruption of the NES allows Exd to accumulate in the nucleus, suggesting that Exd+ enters the nucleus in the absence of Hth, but normally is rapidly exported. We suggest that, in distal cells, the affinity of CRM1 for Exd is greater than the affinity of importin-α for Exd due to the NLS mask, thus biasing the protein toward cytoplasmic localization.
In proximal cells of the leg disc, the transcription factor Hth induces the nuclear localization of Exd. Although Hth also has a NLS in its homeodomain, expression of the HM domain of Hth, which does not have a NLS, can also induce the complete nuclear localization of Exd (Ryoo et al. 1999; Jaw et al. 2000; Noro et al. 2006). This demonstrates that, upon binding the HM domain, both the NLS mask and the NES in Exd are disabled. The equally matched strengths of Exd's NES and NLSs, as shown by the mixed localization of Exd178–300, may facilitate additional regulation by the N-terminal mask. If, instead, one of the nuclear transport signals dominated in Exd, phosphorylation or other covalent modifications might be required to regulate these signals. Instead, the regulated subcellular localization of Exd appears to be realized through an Hth-dependent conformational change that shifts the localization equilibrium from the cytoplasm to the nucleus.
Nuclear localization of Exd is not sufficient for transcriptional regulation:
Although exd is transcribed throughout the imaginal discs, mutant analyses revealed that exd is required only in proximal portions of the appendages (González-Crespo and Morata 1995; Rauskolb et al. 1995). Consistent with these genetic analyses, Exd was found to be nuclear in proximal cells and cytoplasmic in distal cells, suggesting that Exd activity is regulated by subcellular localization (Mann and Abu-Shaar 1996; Aspland and White 1997). On the basis of these observations, it was proposed that the cytoplasmic localization of Exd prevents inappropriate gene regulation by this transcription factor.
By manipulating the transport signals in Exd, we have been able to artificially promote Exd's nuclear localization in the absence of Hth. Surprisingly, predominantly nuclear-localized forms of Exd (myc–GFP–ExdNES or myc–GFP–ExdNES,ΔAla) do not produce any detectable gain-of-function phenotypes when expressed in distal, non-Hth-expressing cells. However, although myc–GFP–ExdNES appears to be inactive in the absence of Hth, we find that it can rescue exd− phenotypes in the proximal leg, strongly suggesting that the protein retains biological activity. Together, our results suggest that Hth not only is required to promote Exd's nuclear localization but also is required for Exd to function as a transcription factor. One possible explanation for these observations is that Hth provides an essential domain that is required for the recruitment of transcriptional coactivators or corepressors. Alternatively, we cannot rule out the possibility that these artificially nuclear-localized forms of Exd are in a conformation that inhibits efficient DNA binding. This inhibition, however, would have to be strong because these forms are inactive in distal appendage cells even when expressed at high concentrations with a variety of strong Gal4 drivers (data not shown).
Given Exd's lack of activity in the absence of Hth, why has this system evolved to maintain Exd's default localization in the cytoplasm? One possibility is that our functional assays in the adult appendages are too crude to detect a subtle—yet important—effect of having Exd in the nucleus in the absence of Hth. Perhaps in other cell types not examined here, nuclear Exd in the absence of Hth would be deleterious. We also note that the system is engineered to create a constant nuclear ratio of Hth:Exd. In wild-type animals, Exd's nuclear localization is fully dependent on Hth. Reciprocally, Hth is unstable in the absence of Exd. As a result, the amount of Hth in the cell governs how much Exd is in the nucleus. Further, cytoplasmic Exd may be necessary to maintain Hth levels and to build the required number of Hth–Exd dimers in the cell. The alternative situation, in which Exd was constitutively nuclear, would not allow for such a tight control over the stoichiometry of nuclear Hth and Exd. Although we have not observed phenotypes resulting from too much nuclear Exd in proximal cells, we again cannot rule out that there are defects that are not measured by our experiments. Finally, as mentioned above, it is also possible that the forms of nuclear-localized Exd that we created are in a conformation that cannot bind DNA and/or recruit required transcriptional cofactors. According to this scenario, binding of Hth to Exd may perform at least three functions: (1) Hth changes Exd's conformation to promote nuclear localization (by relieving the NLS mask and inhibiting NES activity); (2) Hth changes Exd's conformation to allow it to bind to DNA and/or recruit transcriptional cofactors; and (3) Hth directly recruits transcriptional cofactors that are required for Exd to function as a transcription factor.
Acknowledgments
We thank Claude Desplan, Cynthia Kenyon, and Gary Struhl for reagents and members of the Mann lab and Laura Johnston for advice and encouragement throughout this project. We thank Barbara Noro for comments on the manuscript. This work was supported by a grant from the National Institutes of Health to R.S.M.
References
- Abu-Shaar, M., and R. S. Mann, 1998. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development 125: 3821–3830. [DOI] [PubMed] [Google Scholar]
- Abu-Shaar, M., H. D. Ryoo and R. S. Mann, 1999. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspland, S. E., and R. A. H. White, 1997. Nucleocytoplasmic localization of extradenticle protein is spatially regulated throughout development of Drosophila. Development 124: 741–747. [DOI] [PubMed] [Google Scholar]
- Azpiazu, N., and G. Morata, 2002. Distinct functions of homothorax in leg development in Drosophila. Mech. Dev. 119: 55–67. [DOI] [PubMed] [Google Scholar]
- Berthelsen, J., C. Kilstrup-Nielsen, F. Blasi, F. Mavilio and V. Zappavigna, 1999. The subcellular localization of PBX1 and Exd proteins depend on nuclear import and export signals and is modulated by association with PREP1 and Hth. Genes Dev. 13: 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Bürglin, T. R., and G. Ruvkun, 1992. New motif in PBX genes. Nat. Genet. 1: 319–320. [DOI] [PubMed] [Google Scholar]
- Calvo, K. R., P. Knoepfler, S. McGrath and M. P. Kamps, 1999. An inhibitory switch derepressed by Pbx, Hox and Meis/Prep1 partners regulates DNA-binding by Pbx1 and E2a-Pbx1 and is dispensable for myeloid immortalization by E2a-Pbx1. Oncogene 18: 8033–8043. [DOI] [PubMed] [Google Scholar]
- Fontes, M. R. M., T. Teh and B. Kobe, 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-a. J. Mol. Biol. 297: 1183–1194. [DOI] [PubMed] [Google Scholar]
- Fontes, M. R. M., T. Teh, D. Jans, R. I. Brinkworth and B. Kobe, 2003. Structural basis for the specificity of bipartite nuclear localization sequence binding to importin-α. J. Biol. Chem. 278: 27981–27987. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., M. Ohno, M. Yoshida and I. W. Mattaj, 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 9: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., I. Gotoh, Y. Gotoh and E. Nishida, 1996. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH3-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem. 271: 20024–20028. [DOI] [PubMed] [Google Scholar]
- Gill, G., 2003. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13: 108–113. [DOI] [PubMed] [Google Scholar]
- Glazov, E. A., M. Phesant, E. A. McGraw, G. Bejerano and J. S. Mattick, 2005. Ultraconserved elements in insect genome: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 15: 800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Crespo, S., and G. Morata, 1995. Control of Drosophila adult pattern by extradenticle. Development 121: 2117–2125. [DOI] [PubMed] [Google Scholar]
- Green, N. C., I. Rambaldi, J. Teakles and M. S. Featherstone, 1998. A conserved C-terminal domain in Pbx increases DNA binding by the Pbx homeodomain and is not a primer site of contact for the YPWM motif of HoxA1. J. Biol. Chem. 273: 13273–13279. [DOI] [PubMed] [Google Scholar]
- Huang, H., M. Paliouras, I. Rambaldi, P. Lasko and M. Featherstone, 2003. Nonmuscle myosin promotes cytoplasmic localization of PBX. Mol. Cell. Biol. 23: 3636–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaw, T. J., L.-R. You, P. S. Knoepfler, L.-C. Yao, C.-Y. Pai et al., 2000. Direct interaction of two homeoproteins, Homothorax and Extradenticle, is essential for EXD nuclear localization and function. Mech. Dev. 91: 279–291. [DOI] [PubMed] [Google Scholar]
- Kaffman, A., and E. K. O'Shea, 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15: 291–339. [DOI] [PubMed] [Google Scholar]
- Kilstrup-Nielsen, C., M. Alessio and V. Zappavigna, 2003. PBX1 nuclear export is regulated independently of PBX1-MEINOX interaction by PKA phosophorylation of the PBC-B domain. EMBO J. 22: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant, E., C.-Y. Pai, R. Sharf, N. Halachmi, Y. H. Sun et al., 1998. dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning the embryonic PNS. Development 125: 1037–1048. [DOI] [PubMed] [Google Scholar]
- Kutay, U., and S. Güttinger, 2005. Leucine-rich nuclear export signals: born to be weak. Trends Cell Biol. 15: 2005. [DOI] [PubMed] [Google Scholar]
- la Cour, T., L. Kiemer, A. Mølgaard, R. Gupta, K. Skriver et al., 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17: 527–536. [DOI] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 1999. Mosaic analysis with repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Lu, Q., and M. P. Kamps, 1996. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: proposal for a model of a Pbx-Hox-DNA complex. Mol. Cell. Biol. 16: 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, R. S., and M. Abu-Shaar, 1996. Nuclear import of the homeodomain protein Extradenticle in response to Wg and Dpp signalling. Nature 383: 630–633. [DOI] [PubMed] [Google Scholar]
- Mann, R. S., and G. Morata, 2000. The developmental and molecular biology of genes that subdivide the body of Drosophila. Annu. Rev. Cell Dev. Biol. 16: 243–271. [DOI] [PubMed] [Google Scholar]
- Mosammaparast, N., and L. F. Pemberton, 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14: 547–556. [DOI] [PubMed] [Google Scholar]
- Neuteboom, S. T. C., and C. Murre, 1997. Pbx raises the DNA binding specificity but not the selectivity of antennapedia Hox proteins. Mol. Cell. Biol. 19: 4696–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noro, B., J. Culi, D. J. McKay, W. Zhang and R. S. Mann, 2006. Distinct funtions of homeodomain-containing and homeodomain-less isoforms encoded by homothorax. Genes Dev. 20: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltenburg, L. T. C., and C. Murre, 1997. Specific residues in the Pbx homeodomain differentially modulate the DNA-binding activity of Hox and Engrailed proteins. Development 124: 1089–1098. [DOI] [PubMed] [Google Scholar]
- Rauskolb, C., K. M. Smith, M. Peifer and E. Wieschaus, 1995. extradenticle determines segmental identities throughout Drosophila development. Development 121: 3663–3673. [DOI] [PubMed] [Google Scholar]
- Rieckhof, G. E., F. Casares, H. D. Ryoo, M. Abu-Shaar and R. S. Mann, 1997. Nuclear translocation of Extradenticle requires homothorax, which encodes an Extradenticle-related homeodomain protein. Cell 91: 171–183. [DOI] [PubMed] [Google Scholar]
- Ryoo, H. D., T. Marty, F. Casares, M. Affolter and R. S. Mann, 1999. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle target. Development 126: 5137–5148. [DOI] [PubMed] [Google Scholar]
- Saleh, M., H. Huang, N. C. Green and M. S. Featherstone, 2000. A conformational change in Pbx1A is necessary for its nuclear localization. Exp. Cell Res. 260: 105–115. [DOI] [PubMed] [Google Scholar]
- Spek, E. J., A. Olson, Z. Shi and N. R. Kallenbach, 1999. Alanine is an intrinsic a-helix stabilizing amino acid. J. Am. Chem. Soc. 121: 5571–5572. [Google Scholar]
- Whitmarsh, A. J., and R. J. Davis, 2000. Regulation of transcription factor function by phosphorylation. Cell. Mol. Life Sci. 57: 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., and J. Massagué, 2004. Nucleocytoplasmic shuttling of signal transducers. Nat. Rev. Mol. Cell Biol. 5: 1–11. [DOI] [PubMed] [Google Scholar]