Abstract
In most placental mammals, SRY is a single-copy gene located on the Y chromosome and is the trigger for male sex determination during embryonic development. Here, we present comparative genomic analyses of SRY (705 bp) along with the adjacent noncoding 5′ flank (997 bp) and 3′ flank (948 bp) in 36 species of the cat family Felidae. Phylogenetic analyses indicate that the noncoding genomic flanks and SRY closely track species divergence. However, several inconsistencies are observed in SRY. Overall, the gene exhibits purifying selection to maintain function (ω = 0.815) yet SRY is under positive selection in two of the eight felid lineages. SRY has low numbers of nucleotide substitutions, yet most encode amino acid changes between species, and four different species have significantly altered SRY due to insertion/deletions. Moreover, fixation of nonsynonymous substitutions between sister taxa is not consistent and may occur rapidly, as in the case of domestic cat, or not at all over long periods of time, as observed within the Panthera lineage. The former resembles positive selection during speciation, and the latter purifying selection to maintain function. Thus, SRY evolution in cats likely reflects the different phylogeographic histories, selection pressures, and patterns of speciation in modern felids.
IN placental mammals, SRY (sex-determining region on the Y chromosome), termed Sry in the mouse, is the male-determining gene (Gubbay et al. 1990; Sinclair et al. 1990) and functions in early development to trigger a cascade of gene expression that result in testis formation (Capel 1998; Swain and Lovell-Badge 1999). Comparative genomic mapping indicates that SRY/Sry became Y specific ∼170 MYA in a common ancestor to marsupial and placental mammals (Foster et al. 1992) and is mapped consistently to the nonrecombining region of the Y chromosome (NRY) in multiple placental mammal taxa (Yang et al. 1993; Lahn and Page 1997; Glaser et al. 1999; Murphy et al. 1999; Tilford et al. 2001; Liu et al. 2002; Skaletsky et al. 2003). The NRY was originally presumed to be a “functional wasteland,” but subsequent gene mapping studies in human and mouse and the full-length sequence of human Y chromosome suggest otherwise (Bachtrog and Charlesworth 2001; Skaletsky et al. 2003). In the absence of recombination, male-specific genes in this region, such as SRY, are predicted to be under strong purifying selection to maintain function (Graves 1995, 1998a).
Purifying selection in SRY is indicated by its conserved structure and function across placental mammalian orders. In primates (Whitfield et al. 1993), rodents (Tucker and Lundrigan 1993), artiodactyls (Cheng et al. 2001; Quilter et al. 2002), and carnivores (Murphy et al. 1999; Olivier et al. 1999) SRY is a single-copy, intronless gene that encodes a protein with a conserved high mobility group (HMG) box. The HMG box is a transcription factor shared by autosomal paralogues in the SOX gene family. In humans, most mutations within this critical HMG region of SRY alter the DNA-binding ability of the protein, resulting in gonadal dysgenesis of a female phenotype with a male genotype (Gubbay et al. 1992; Hawkins et al. 1992; Haqq and Donahoe 1998; Murphy et al. 2001). The importance of SRY in male sex determination is further underscored by studies of sex chromosomal abnormalities that involve either translocation of SRY to the X chromosome, resulting in 46, XX true hermaphrodites, or the loss of SRY in 46, XY phenotypic females (Gubbay et al. 1990; Koopman et al. 1991; Haqq and Donahoe 1998).
Although these and other studies affirm SRY as the vital, male-determining gene, there are a few, but notable exceptions. For example, Sry is completely missing in two species of vole (Just et al. 1995). Likewise, studies with marsupials indicate that Sry is not critical to male determination (Graves 2001) and that Atry may fulfill that function (Pask et al. 2000). Additional studies in rodents describe gene amplification resulting in multiple copies of Sry in Old World murine species (Nagamine 1994) and New World Akodon (Cricetidae) species (Bianchi et al. 1993; Bianchi 2002), as well as in six African murine species (Lundrigan and Tucker 1997).
SRY exhibits significant sequence differences across mammalian orders, suggesting that there is an unusual level of divergence despite its conserved gene structure (Delbridge and Graves 1999; O'Neill and O'Neill 1999; Graves 2001, 2002; Marshall Graves 2002a,b). An excess of nonsynonymous substitutions observed in the non-HMG box regions of the gene in rodents (Tucker and Lundrigan 1993) and primates (Whitfield et al. 1993) was viewed as evidence for positive selection during speciation. Subsequently, this result has been tempered by likelihood-based analyses that describe minimal selection in SRY in New World primates (Moreira 2002) and four closely related Mus species (Jansa et al. 2003).
Here we investigate the evolutionary patterns of SRY diversification within 36 species of the cat family Felidae and compare and contrast rates of change between coding and noncoding adjacent flanking regions of the Y chromosome. Our results indicate that SRY is under purifying selection to maintain function, yet several sites are under positive selection in a pattern that is correlated with speciation and divergence events in Felidae.
MATERIALS AND METHODS
DNA specimens:
DNA was purified from blood or skin cell fibroblasts from at least one male of 36 species of Felidae representing eight defined evolutionary lineages (supplemental Table 1 at http://www.genetics.org/supplemental/). With few exceptions, multiple male individuals were sampled for each species within each lineage. No males were available for the bay cat (Pardofelis badia) of the bay cat lineage and Andean mountain cat (Leopardus jacobitus) of the ocelot lineage.
PCR amplification:
Primers were designed to span the entire coding region (hereafter termed SRY) and the adjacent 5′ and 3′ genomic flanking regions (supplemental Table 2 at http://www.genetics.org/supplemental/). Primers were designed from a SRY clone (3.5 kb) isolated from a λ-phage library constructed from testis tissue of domestic cat (King 1996). This single clone contained the contiguous sequence of the 5′ genomic flank, SRY, and the 3′ genomic flank from domestic cat. The Y chromosome specificity of all PCR reactions was confirmed by the presence of the expected product in males and not in females. Sequences are deposited in GenBank with accession nos. DQ095160–DQ095195.
PCR reactions contained 125 ng total genomic DNA, 50 mm KCl, 10 mm Tris–HCl (pH 8.3), 1.5–2 mm MgCl2, 0.2 mm dNTPs, 0.2 mm of each primer, and 2.5 units Taq polymerase in a total volume of 25 μl. Specific conditions for PCR amplification of SRY started with a 3-min hot start at 95°, followed by 40 cycles of a 15-sec 95° denaturing step, a 30-sec 50° annealing step, and a 1-min 72° extension step. A final extension at 72° for 5 min completed the program.
PCR amplification conditions for the 3′ and 5′ flanking regions used a touchdown format. Each reaction ran for 40 cycles and was composed of an initial hot start of 3 min at 95°, followed by 15 sec at 95°, 30 sec at 58°–48° with a 2° drop every fourth cycle, and 1 min at 72°, ending with a final extension of 72° for 5 min. Reactions were run in either an ABI (Columbia, MD) Perkin-Elmer (Norwalk, CT) 9700 or in a Biometra T1 thermocycler. Products were visualized on 1% agarose gels run in 1× TAE buffer at 120 V for 60 min using 5 μl of 5 mg/ml ethidium bromide in the gel and in the buffer. Products were cleaned using Microcon Centricon filters or Microcon PCR filters and sequenced using ABI BigDye sequencing reagents and ABI automated sequence machine models 377, 3700, and 3730.
Phylogenetic analyses of nucleotide sequences from SRY and genomic flanks in 36 species of Felidae:
Sequences were compiled using Sequencher (version 4.1; Gene Codes, Ann Arbor, MI) and aligned using Clustal X (Thompson et al. 1997). Ambiguous sites and all SINE retroelements were removed from the alignment prior to phylogenetic analysis. The program ModelTest (Posada and Crandall 1998) was used to determine the optimal model of substitution for phylogenetic analyses.
Each of the three genomic regions (SRY, 5′ flank, and 3′ flank) was analyzed separately as well as in a combined data set for phylogenetic reconstruction. Three different optimality criteria of minimum-evolution (ME), maximum-likelihood (ML), and maximum-parsimony (MP) methods were used to analyze the data as implemented by PAUP* (Swofford 1998). Specific models for the ML and ME analyses for SRY, 5′ flank, and 3′ flank were based on the results of ModelTest. For SRY, the HKY + γ substitution model was used with estimated nucleotide frequencies of A = 0.27214, C = 0.27828, G = 0.26847, and T = 0.18111; a transition:transversion ratio = 1.2107; and γ = 2.435. For the 5′ flanking region the substitution model of GTR + γ was selected and used estimated nucleotide base frequencies of A = 0.2690, C = 0.1640, G = 0.2488, and T = 0.3182; a rate matrix of AC = 1.1579, AG = 2.0069, AT = 0.1925, CG = 2.1002, CT = 3.2506, and GT = 1.0; and γ = 0.5483. Likewise, a GTR + γ model was selected for the 3′ flanking region and used estimated nucleotide base frequencies of A = 0.2753, C = 0.2376, G = 0.1920, and T = 0.2950; a rate matrix of AC = 0.7582, AG = 2.5104, AT = 0.4245, CG = 1.5787, CT = 1.8718, and GT = 1.0; and γ = 0.1901. The combined data set (SRY, 3′ flank, and 5′ flank) was analyzed using likelihood settings from the best-fit model TVM + γ with base frequencies of A = 0.2679, C = 0.2240, G = 0.2361, and T = 0.2720. The rate matrix was AC = 0.9736, AG = 2.4559, AT = 0.3694, CG = 1.7034, CT = 2.4559, and GT = 1.0 with γ = 0.7039.
Optimal trees based on nucleotide data were obtained using heuristic searches implemented in PAUP*. Conditions for the ML analysis included starting trees obtained by stepwise addition and branch swapping using the tree-bisection-reconnection (TBR) algorithm. Specific conditions for the ME heuristic search included starting trees obtained by neighbor joining, the TBR branch-swapping algorithm, and no collapsing of zero-length branches. The heuristic search with MP coded gaps as “missing,” with stepwise addition of taxa and TBR branch swapping. Support for specific clades was assessed by bootstrap analysis using identical settings established for each method of phylogenetic reconstruction and retention of node bootstrap values >50%. For the ME and MP analyses, 1000 iterations of bootstrap were performed with heuristic tree searches employing the TBR branch-swapping algorithm. With the ML bootstrap, 100 iterations were implemented using nearest-neighbor interchange for branch swapping. A fourth method utilizing a Bayesian approach for computing clade credibility values for nodes within the tree was performed by the program MrBayes (version 3.1.2) (Huelsenbeck and Ronquist 2001). Specific parameters included: (1) four Markov chains were run for 1,000,000 generations, (2) empirical estimates of stability-likelihood values were used to set the burn in at 30,000 generations, and (3) trees were sampled every 20 generations. Two runs were performed to confirm the stability of posterior probabilities and parameters.
Additional analyses were conducted on SRY sequences after translation into amino acids. Phylogenetic trees were derived from the amino acid residue data using two algorithms of distance-based and maximum-likelihood analyses. The Pam–Dayhoff model of amino acid substitution was used to construct a neighbor-joining tree using the program PHYLIP version 3.5 (Felsenstein 1993). Bootstrap analyses consisting of 1000 repetitions were conducted for the distance-based analysis using PHYLIP. The maximum-likelihood analyses of amino acids used the Pam–Dayhoff model of substitution as implemented by ProtML in the program MOLPHY version 2.3 (Adachi and Hasegawa 1996).
Testing for selection within the SRY gene in Felidae:
Selection can be measured in coding nucleotide sequences by ω, which is equal to dN/dS, where dN is the proportion of nonsynonymous substitutions present from all possible nonsynonymous sites and dS is the number of synonymous substitutions for all possible synonymous sites. Neutral evolution is represented by ω = 1, compared with ω >1, which signifies Darwinian positive selection leading to fixation of new, more favorable mutations, and ω < 1, which indicates purifying selection to remove deleterious mutations.
Selection was estimated using the maximum-likelihood approach implemented by the CODEML subroutine of PAML version 3.15 (Yang 1997). The input tree for PAML was based on the resolved cat family phylogeny of Johnson et al. (2006). Variable ω among sites within SRY was tested using M0 (one rate), M1a (nearly neutral), M2a (selection), M7 (β), and M8 (β and ω) models. Tests of significance used the log-likelihood ratio test (LRT) of 2Δλ = 2 (l1 − l0), with likelihood scores of models M2a vs. M1a and M8 vs. M7, with 2 d.f. Model M8a was used as an alternative null distribution to correct for false positives (Swanson et al. 2003) and was compared against M8 with 1 d.f. and the χ2 P-value adjusted by dividing by 2. Sites under positive selection were then identified by Bayes' empirical Bayes' (BEB) posterior probability for models M2a and M8 with significant LRT values.
SRY was tested for differences in selection among the recognized lineages within the cat family using the revised branch-site model A (Zhang et al. 2005) as implemented in PAML. In this model specific lineages in the cat phylogeny are designated as either foreground or background. Sites within the foreground lineage are tested for positive selection. The model has four site classes: class 0 are sites that are under purifying selection (0 < ω < 1) throughout the tree; class 1 sites are neutral throughout the tree (ω = 1); class 2a sites are undergoing purifying selection in the background and positive selection in the foreground lineage; and class 2b sites are neutral in the background, but positively selected in the foreground lineage. The LRT used test 2, which compared model A against model A with ω2 = 1 fixed as the null model and critical χ2-values of 5 and 1% of 3.84 and 5.99, respectively. The BEB procedure was used for identifying sites under positive selection in the foreground lineage with significant LRTs.
RESULTS
Three contiguous regions spanning the 5′ genomic flank (1038 bp), the SRY coding region (706 bp), and the 3′ genomic flank (1213 bp) of the Y chromosome were sequenced in 36 species of cat (supplemental Figure 1 at http://www.genetics.org/supplemental/). For subsequent analyses, 304 bp were identified as phylogenetically ambiguous (uncertainty within the alignment) and removed in the final alignment (supplemental Figure 2). None of the ambiguous sites occur within SRY.
SRY had fewer nucleotide substitutions relative to its adjacent noncoding flanks (Table 1). Across SRY, there were 85 (12%) variable sites, of which 34 were parsimony informative. If partitioned into functional domains, the HMG box of SRY was slightly more conserved with 8.5% variable sites compared with the amino (15.9%) and carboxyl (12.7%) region of the gene. The adjacent noncoding genomic regions differed from SRY with 17.6% of the sites within the 5′ flank and 19.8% of the 3′ flank exhibiting variation in Felidae.
TABLE 1.
Phylogenetic parameters of the SRY gene and adjacent noncoding genomic flanks in Felidae
| Genomic region | No. BPa | No. variable sites | % variable sites | No. PI sitesb | % PI sites | MP tree length | CIc | No. homoplasies | No. species defining auto apomorphiesd | Lineage support: ME/MP/ML | No. lineages recovered | Trees |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ genomic flank | 997 | 176 | 17.6 | 81 | 46.0 | 216 | 0.88 | 19 | 129 | ME < 50–100% | 6 of 8 | Supplemental Figure 3e |
| MP < 50–100% | ||||||||||||
| ML < 50–100% | ||||||||||||
| SRY | 706 | 85 | 12 | 34 | 40.0 | 91 | 0.9560 | 4 | 54 | ME 57–100% | 6 of 8 | Figure 2A |
| MP 52–100% | ||||||||||||
| ML 59–100% | ||||||||||||
| NH3 terminus | 157 | 25 | 15.9 | 10 | 40.0 | NA | NA | 3 | 17 | NA | NA | |
| HMG box | 234 | 20 | 8.5 | 8 | 40.0 | NA | NA | 0 | 13 | NA | NA | |
| COOH terminus | 315 | 40 | 12.7 | 16 | 40.0 | NA | NA | 1 | 24 | NA | NA | |
| 3′ genomic flank | 948 | 188 | 19.8 | 81 | 46.0 | 221 | 0.9050 | 18 | 144 | ME < 68–100% | 8 of 8 | Supplemental Figure 4e |
| MP < 64–100% | ||||||||||||
| ML < 73–100% | ||||||||||||
| Combined | 2653 | 449 | 16.9 | 230 | 46.0 | 534 | 0.8914 | 47 | 338 | ME 79–100% | 8 of 8 | Figure 3 |
| MP 88–100% | ||||||||||||
| ML 94–100% |
Ambiguous sites excluded, see supplemental Figure 2 at http://www.genetics.org/supplemental/.
Parsimonious informative sites.
Consistency index.
Excluding indels.
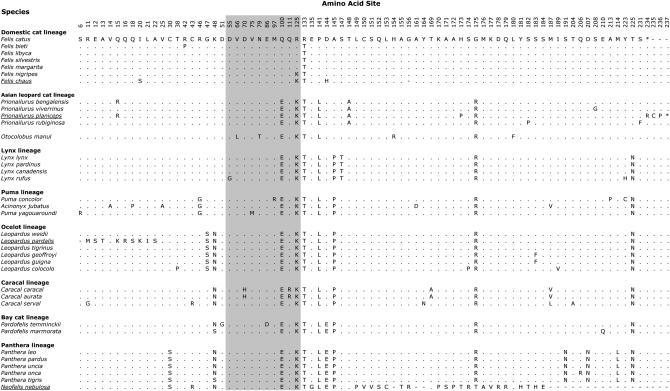
Within Felidae, SRY exhibited four insertion/deletion events (indels) unique to different species that changed the length of the expected protein product of 234 amino acids. First, a single-base-pair insertion at nucleotide position 64 in ocelot (L. pardalis) (supplemental Figure 2 at http://www.genetics.org/supplemental/, site 1062) resulted in a frameshift mutation creating multiple stop codons within the protein. However, if translation initiation began at an alternate ATG at positions 22–24 (supplemental Figure 2, sites 1021–1023; Figure 1, sites 6–22), a complete SRY protein was generated, although shortened by seven residues in the amino region. This frameshift insertion in ocelot SRY was confirmed in multiple unrelated males (N = 24). Second, a single-base-pair deletion at position 437 in the carboxyl region (supplemental Figure 2, site 1436) of clouded leopard (Neofelis nebulosa) resulted in a truncated SRY protein of 172 residues (Figure 1) and was observed in all males examined (N = 4). Third, SRY of flat-headed cat (Prionailurus planiceps) encoded a protein extended by three amino acids (N = 2). The substitution of TAG to CAG of the terminal codon extended the reading frame into the first nine nucleotides of 3′ genomic flank (supplemental Figure 2, sites 1706–1714; Figure 1, site 234). Fourth, an in-frame triplet deletion at sites 458–460 that encodes a glycine residue (supplemental Figure 2, sites 1456–1458) in jungle cat (Felis chaus) resulted in a SRY protein shortened by one amino acid (N = 2) (Figure 1, site 156).
Figure 1.—
Alignment of the 80 variable amino acid residues within SRY in 36 species of Felidae. Taxa are organized by recognized felid lineage (O'Brien and Johnson 2005; Johnson et al. 2006). The shaded area defines variable sites within the conserved HMG box. If these alignments are corrected for ocelot and clouded leopard (see text), the total number of variable sites is reduced from 80 to 61. Underlined species have indels within SRY. Asterisk denotes stop codon. Dash refers to insertion/deletion event.
Variable amino acid sites within SRY were not distributed equally among the functional domains of the protein (Figure 1). The HMG box was most conserved with 12.8% (10/78) variable amino acid sites. Correcting for contiguous changes brought about by the indels in ocelot and clouded leopard SRY to a single event in each species, the estimates for variable sites were 28.8% (15/52) and 29.8% (31/104) for the amino and carboxyl regions, respectively.
Phylogenetic analyses of SRY in Felidae:
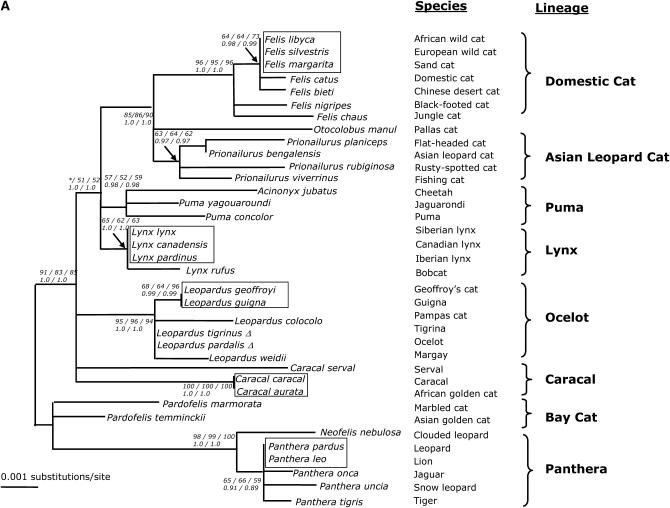
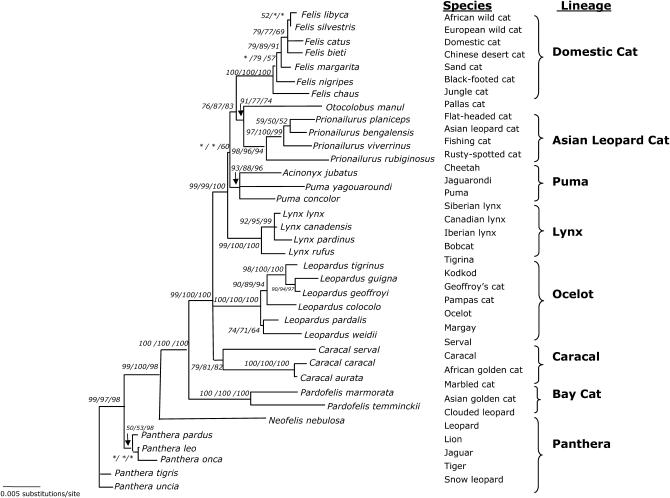
SRY sequences were informative in recovering the expected lineages within felid evolution (Figure 2A). Three lineages (domestic cat, ocelot, and panthera) were strongly supported with bootstrap values >94% (MP/ME/ML) and Bayesian posterior probabilities of 1.0. Moderate support for the higher-order clade uniting Pallas cat with the sister lineages of domestic cat and Asian leopard cat was obtained with bootstrap values >85% and Bayesian posterior probability of 1.0 (Figure 2A). The node for the inclusion of Caracal serval within the caracal lineage was not recovered and the bay cat lineage was collapsed into a polytomy.
Figure 2.—
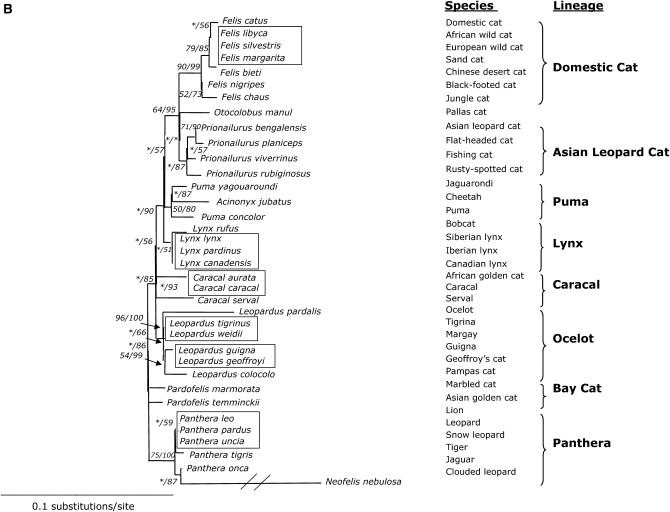
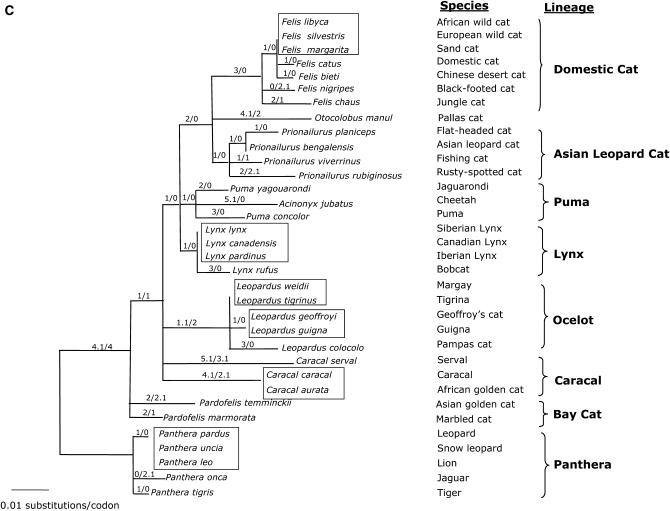
(A) Phylogenetic tree of the SRY coding region (706 bp) evolution in 36 species of Felidae. Shown is the maximum-likelihood tree derived by a heuristic search using the tree-bisection-reconnection branch-swapping method (−Ln likelihood score = 1636.78568, 14,430 rearrangements tried). Nearly identical topologies to this tree were recovered using minimum-evolution (ME) (HKY85 model of substitution) and maximum-parsimony (MP) algorithms with identical conditions for conducting the heuristic searches. Phylogenetic analyses using ME resulted in 1080 equivalent trees (tree score = 0.12879, 15,566,734 rearrangements tried). Phylogenetic analysis using MP recovered 8 equivalent trees (length = 91 steps, 112,348 rearrangements tried). Bayesian posterior probabilities were estimated from 1,000,000 generations with trees sampled every 20 generations and burn in was empirically determined at 50,000 generations. Numbers in italics reflect bootstrap proportion support for adjacent nodes as ME (1000 iterations)/MP (1000 iterations)/ML (100 iterations) with Bayesian posterior probability values below (two iterations). Boxes indicate identical SRY sequences between related species within the designated lineage. Δ indicate species that are identical except for insertion/deletions. Asterisk indicates bootstrap proportions <50%. (B) Phylogenetic tree of SRY based on amino acid sequences (234 aa). Shown is the maximum-likelihood tree derived by MOLPHY (Adachi and Hasegawa 1996). Numbers in italics on each branch are bootstrap proportions in support of the adjacent node ME (1000 iterations)/ML (500 iterations). Asterisk denotes nodes with <50% bootstrap support. Boxes cluster species with identical SRY sequences within each lineage. (C) Phylogenetic tree of SRY codons in 34 species of Felidae (234 aa). Shown is the maximum-likelihood tree derived using CODEML of PAML (Yang 1997). Identical species were removed to meet requirements of PAML, but added later (boxes). Numbers on each branch are no. nonsynonymous substitutions/no. synonymous substitutions estimated using the free-ratio model. Branch length is no. substitutions/codon. Ocelot (Leopardus pardalis) and clouded leopard (Neofelis nebulosa) were omitted from analysis due to frameshift mutations incompatible with PAML program.
Overall, the majority of substitutions within SRY were diagnostic for species-level identification. Under the MP optimality criteria (whereby the most parsimonious tree has the shortest number of steps), 59% of the character changes (54 of 91) used to build the tree were species specific (Table 1). The frequency of convergent, reversal, or parallel substitutions (homoplasies) was extremely low within the cat SRY phylogeny. The MP tree had a markedly high consistency index CI = 0.9560 (excluding uninformative sites CI = 0.8974). However, there were instances where SRY was identical between multiple species within a lineage on the nucleotide level (boxes in Figure 2A). Indels were ignored in this analysis, and thus the SRY of tigrina (L. tigrina) appeared identical to that of ocelot (L. pardalis) but actually differed by the single-base-pair insertion in L. pardalis described previously.
The phylogenetic tree based on amino acid residues from translated SRY sequences was highly similar to the nucleotide phylogeny (Figure 2B). Both the distance-based and ML analyses supported the same higher-order node observed in the nucleotide phylogeny uniting domestic cat lineage, Asian leopard cat lineage, and Pallas cat. In the ocelot lineage, L. tigrina had the same SRY protein as L. weidii rather than the shortened version of L. pardalis. In the panthera lineage, Panthera uncia had the identical amino acid sequence as P. leo and P. pardus. The unusually long branch length of N. nebulosa reflected an altered amino acid sequence due to the single-nucleotide base pair frameshift in the carboxyl region, as described above. The bay cat lineage and caracal lineage were not fully resolved.
Testing for selection in SRY in Felidae:
Each codon within SRY was tested for a higher than expected ratio of dN/dS (ω) across felid species. Due to indel substitutions incompatible with PAML requirements, L. pardalis and N. nebulosa were omitted from this analysis. The one-ratio model M0, which assumes all sites have an equivalent ω (ωo), indicated SRY was under purifying selection (ωo = 0.815). However, tests using models M2a and M8 estimated that 4.1% of the sites were under positive selection with ω1 = 5.98942 (M2a) and ωs= 5.99456 (M8) (Table 2). The BEB procedure identified six candidate codons for positive selection, although none were significant at P < 0.05 (Table 2). Codon 47, which substitutes the amino acid glycine with serine in all species within the ocelot lineage, had the highest posterior P-value (P = 0.91, M2a; P = 0.94, M8).
TABLE 2.
Tests for selection among codons of SRY in Felidae using models implemented in PAML 315 using the likelihood method (see materialsandmethods)
| Criteria | Model | Parameter estimates | Ln likelihood | LRT | Selected codon, BEB posterior probabilitya |
|---|---|---|---|---|---|
| Among | M0 | κ = 2.11751 | −1624.383 | NA | NA |
| sites | (one ratio) | ω0 = 0.81556 | |||
| M1a | κ = 2.04609 | −1623.179 | NA | NA | |
| (nearly neutral) | ω0 = 0.21097 (p0 = 0.38216) | ||||
| ω1 = 1.00000 (p1 = 0.61784) | |||||
| M2a | κ = 2.11857 | −1619.815 | M2a vs. M1a | R46G (0.829) | |
| (selection) | ω0 = 0.63684 (p0 = 0.95861) | 6.729, P = 0.034, 2 d.f. | G47S (0.910) | ||
| ω1 = 1 (p1 = 0) | K125R (0.736) | ||||
| ω2 = 5.98942 (p2 = 0.04139) | D144E (0.569) | ||||
| M187V, L (0.656) | |||||
| M7 | κ = 2.06047 | −1623.196 | NA | NA | |
| (β-distribution, neutral) | β-distribution | ||||
| p = 0.13573, q = 0.03540 | |||||
| M8 | κ = 2.11873 | −1619.819 | M8 vs. M7 | R46G (0.870) | |
| (β-distribution, selection) | ωs = 5.99456 (p1 = 0.04130) | 6.754, P = 0.034, 2 d.f. | G47S (0.940) | ||
| β-distribution | K125R (0.815) | ||||
| p0 = 0.95870, p = 99.0, q = 56.358 | D144E (0.694) | ||||
| M187V, L (0.759) | |||||
| Y223H, C (0.588) | |||||
| M8a | κ = 2.04630 | −1623.181 | M8 vs. M8a | NA | |
| (β-distribution, ω = 1, fixed) | ω1 = 1.00000 (p1 = 0.61804) | 6.724, P = 0.0047, 1 d.f. | |||
| β-distribution | |||||
| p0 = 0.38196, p = 26.636, q = 99.0 |
P, proportion of sites; κ, transition:transversion ratio; ω = dN/dS for each proportion of sites; NA, not applicable;
Corresponding codon (see Figure 1) of 36-species alignment.
Branch-site model A indicated that positively selected codons occur within species from two of the eight cat lineages. In this model, each lineage was tested for positive selection (foreground ω) against the remaining phylogeny (background ω). The domestic cat lineage and the puma lineage were the only two with a significant LRT (Table 3). BEB identified 1 codon in the domestic cat lineage and 11 codons in the puma lineage to be under positive selection (Table 3, Figure 1). Of these, codons 75, 97, and 125 were located in the HMG box (Figure 1). Of the 6 codons identified (albeit with no statistical support) by among-sites models M2a and M8 (Table 2), codons 187 and 223 were also identified by the branch-site model A to be under positive selection in the puma lineage and codon 125 was identified in the domestic cat lineage. Thus, it is likely that the among-sites models were significantly different from the one-ratio M0 due to positive selection present in SRY from the domestic cat and puma lineages and not across all cat species.
TABLE 3.
Test of selection among lineages using revised branch-site model A and test 2 as implemented in PAML 315 (see materialsandmethods)
| Parameter estimates
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Model A | Site class | p | ω0 background | ω1 foreground | Ln likelihood, LRT test 2 | Selected codon, posterior P | ||
| Domestic cat lineage | κ = 2.0844 | 0 | 0.42368 | 0.27811 | 0.27811 | −1618.646 | K125R (0.943) | |
| 1 | 0.57024 | 1 | 1 | −1623.128 (Null) | ||||
| 2a | 0.00259 | 0.27811 | 74.02392 | |||||
| 2b | 0.00349 | 1 | 74.02392 | LRT = 8.965, P < 0.1 | ||||
| Asian leopard cat lineage | κ = 2.04134 | 0 | 0.11785 | 0.12684 | 0.12684 | −1622.952 | ||
| 1 | 0.18680 | 1 | 1 | −1622.952 (Null) | ||||
| 2a | 0.26899 | 0.12684 | 1 | |||||
| 2b | 0.42637 | 1 | 1 | LRT = 0, NS | ||||
| Ocelot lineagea | κ = 2.04608 | 0 | 0.38217 | 0.21098 | 0.21098 | −1623.179 | ||
| 1 | 0.61783 | 1 | 1 | −1623.179 (Null) | ||||
| 2a | 0.00000 | 0.21098 | 1 | |||||
| 2b | 0.00000 | 1 | 1 | LRT = 0, NS | ||||
| Puma lineage | κ = 1.99378 | 0 | 0 | 0.16775 | 0.16775 | −1618.396 | S6R** | M97R** |
| 1 | 0 | 1 | 1 | −1621.544 (Null) | V14A** | A161D** | ||
| 2a | 0.49690 | 0.16775 | 999.000 | Q18P** | M187V* | |||
| 2b | 0.50310 | 1 | 999.000 | LRT = 6.295, P > 0.01 | V25A** | A213P** | ||
| R46G* | Y223C* | |||||||
| V75M** | ||||||||
| Lynx lineage | κ = 2.0609 | 0 | 0.39158 | 0.22067 | 0.22067 | −1621.591 | ||
| 1 | 0.57691 | 1 | 1 | −1623.132 (Null) | ||||
| 2a | 0.01235 | 0.22067 | 84.78123 | |||||
| 2b | 0.01816 | 1 | 84.78123 | LRT = 3.082, NS | ||||
| Caracal lineage | κ = 2.04609 | 0 | 0.38216 | 0.21097 | 0.21097 | −1623.179 | ||
| 1 | 0.31784 | 1 | 1 | −1623.179 (Null) | ||||
| 2a | 0 | 0.21097 | 1 | |||||
| 2b | 0 | 1 | 1 | LRT = 0, NS | ||||
| Bay cat lineage | κ = 2.04401 | 0 | 0 | 0.22458 | 0.22458 | −1622.007 | ||
| 1 | 0 | 1 | 1 | −1623.139 (Null) | ||||
| 2a | 0.40068 | 0.22458 | 277.069 | |||||
| 2b | 0.59932 | 1 | 277.069 | LRT = −2.263, NS | ||||
| Panthera lineagea | κ = 2.04609 | 0 | 0.38216 | 0.21097 | 0.21097 | −1623.179 | ||
| 1 | 0.61784 | 1 | 1 | −1623.179 (Null) | ||||
| 2a | 0 | 0.21097 | 1 | |||||
| 2b | 0 | 1 | 1 | LRT = 0, NS | ||||
κ, transition/transversion ratio; NS, not significant results of Ln likelihood-ratio test; ω0, background ratio of dN/dS; ω1, lineage-specific ratio of dN/dS. *Posterior P > 0.95%, **Posterior P > 0.99%.
Neofelis nebulosa and Leopardus pardalis were omitted due to frameshift mutations incompatible with PAML.
Felid phylogeny defined by the adjacent 5′ genomic flank of SRY:
Phylogenetic inferences based on sequence from the 5′ flanking region upstream exhibited several differences from the SRY tree. Except for F. catus and F. silvestris within the domestic cat lineage, all other species were identified by unique substitutions (supplemental Figure 3 at http://www.genetics.org/supplemental/). Also, the caracal lineage that was collapsed in the SRY tree was recovered in the 5′ flank. A greater number of homoplasies were present in the 5′ flank tree (CI = 0.8796, or, excluding uninformative sites, CI = 0.7778) compared with the SRY tree (Table 1). Four homoplasies were on the branch forming the bay cat [branch length (BrL) = 15] lineage and one on the ocelot lineage (BrL = 5) branch. Even with an increase in variable sites (17.4%) and the improved resolution of individual species, the phylogeny was less resolved than the SRY tree in the following ways: (1) the puma lineage was collapsed into a polytomy, (2) the higher-order node that united the Pallas cat with the two sister lineages of domestic cat and Asian leopard cat was collapsed, and (3) P. rubiginosa, a member of the Asian leopard cat lineage, was placed erroneously in the lynx lineage.
Phylogenetic analyses of the 3′ flanking region adjacent to SRY in Felidae:
Sequences from the adjacent 3′ flanking region to SRY were highly informative in recovering the expected phylogeny (supplemental Figure 4 at http://www.genetics.org/supplemental/). With the greatest proportion of variable sites (19.8%) of the three genomic regions examined, the 3′ flank phylogeny recovered all eight lineages with moderate to high measures of support. In the MP analysis (CI = 0.9050; corrected for uninformative characters, CI = 0.8019), species were defined by multiple autoapomorphic substitutions with very few homoplasies (Table 1). In the case of F. libyca and F. silvestris, these two species appear identical except for three separate indels at positions 1756, 2423, and 2459 of the combined alignment (supplemental Figure 2 at http://www.genetics.org/supplemental/). Diagnostic substitutions defining each of the monophyletic lineages had very few homoplasies. No homoplasies were located on the branches leading to domestic cat lineage (BrL = 4), puma lineage (BrL = 2), and bay cat lineage (BrL = 7), and one homoplasy occurred in panthera (BrL = 12), ocelot (BrL = 8), and Asian leopard cat lineages (BrL = 5) (data not shown).
Combined data analysis:
The combined sequences spanning the 5′ flank, SRY, and the 3′ flank (2651 bp) provided the most accurate reconstruction of the cat family phylogeny. Moderate to high support occurred for each of the eight monophyletic lineages (Figure 3). Within each lineage, the relative branching order among species did not vary among the MP, ME, and ML analyses (Figure 3, Table 1). Two ambiguous intralineage nodes were not resolved in the phylogeny. The first was the relative branching order between puma–cheetah–jaguarondi in the puma lineage. The second was the relative associations between lion, leopard, and jaguar in the panthera lineage. Consistent with the trees from each separate analysis, the combined tree was marked by few homoplasies (CI = 0.8912; excluding noninformative characters, CI = 0.7836). Very few homoplasies occurred in the monophyletic branches defining each lineage. Three homoplasies occur on the branches leading to lynx (BrL = 11) and bay cat (BrL = 21) lineages, one for domestic cat (BrL = 12) and ocelot (BrL = 16) lineages, and zero homoplasies for panthera (BrL = 28), Asian leopard cat (BrL = 7), puma (BrL = 3) and caracal (BrL = 4) lineages. Of the total tree length (534 steps), 338 substitutions (63%) were diagnostic for species identification (Table 1).
Figure 3.—
Phylogenetic tree based on total combined data in 36 species of Felidae (2651 bp). Shown is the maximum-likelihood (ML) tree derived by a heuristic search using the tree-bisection-reconnection branch-swapping method (−Ln likelihood score = 7497.62678, 13,442 rearrangements tried). Nearly identical topologies to this tree were recovered using minimum-evolution (ME: GTR model of substitution) and maximum-parsimony (MP) algorithms with identical conditions for conducting the heuristic searches. Phylogenetic analyses using ME resulted in two equivalent trees (tree score = 0.20710, 13,201 rearrangements tried). Phylogenetic analysis using MP recovered 1869 equivalent trees (length = 529 steps, 25,092,482 rearrangements tried). Bayesian posterior probabilities were estimated from 1,000,000 generations with trees sampled every 20 generations and burn in was empirically determined at 50,000 generations. All nodes within the phylogeny had Bayesian probability values of 1.0 (two iterations). Numbers in italics reflect bootstrap proportion support for adjacent nodes as ME (1000 iterations)/MP (1000 iterations)/ML (100 iterations). Asterisk denotes bootstrap support <50.
DISCUSSION
In the cat family, SRY encodes a protein of 234 amino acid residues and is partitioned into an amino region of 52 amino acids, followed by the HMG box of 78 amino acids and the terminal carboxyl region of 104 amino acids. The length of the HMG box is identical among most mammalian species with the exception of rodent taxa (data not shown), indicating functional constraints for this critical structure. In Felidae, the percentage of variable nucleotide sites within the HMG box (8.5%) is slightly less than those within the amino (15.9%) and carboxyl (12.7%) regions. However, as observed with primates (Whitfield et al. 1993; Moreira 2002), rodents (Tucker and Lundrigan 1993), and marsupials (O'Neill et al. 1997), translation to amino acids reveals the felid HMG box to be most conserved, with 12.8% variable sites vs. 28.8 and 29.8% for the amino and carboxyl regions, respectively.
Several cat species exhibit indels within the amino and carboxyl domains. In ocelot (L. pardalis) the SRY transcript is predicted to begin at an alternate downstream ATG to encode a protein shortened by seven amino acids. In the flat-headed cat (P. planiceps) and jungle cat (F. chaus) the carboxyl domain of SRY is altered by indels. A single-base-pair insertion truncates the protein to 172 amino acids in the clouded leopard (N. nebulosa). Sequences from multiple individuals confirm these changes as fixed within each of the four species. A recent study has shown that the clouded leopard samples herein represent two deeply divergent, geographically disparate groups that separated 1.4 MYA (Buckley-Beason et al. 2007), yet still retain this altered SRY protein. Similar incidences of indels in primates (Moreira 2002) and the genus Mus (Tucker and Lundrigan 1993, 1995; Jansa et al. 2003) suggest a general relaxation of functional constraints within the carboxyl domain of SRY in mammals.
Comparing the SRY coding region with adjacent flanks offers additional insights into the mode and tempo of change within this important sex-determining gene. Both the 5′ and the 3′ noncoding genomic regions had greater numbers of variable sites. Yet, the 5′ flank phylogeny is the least resolved compared against SRY and 3′ flank phylogenies. Scanning the domestic cat sequence for transcription factors using TFSEARCH (Akiyama 1998; Heinemeyer et al. 1998) indicated that the 5′ flank had nearly four times (30 sites within 926 bp) the number of putative transcription sites than the 3′ flank (5 of 963 bp). In addition, comparison with primates, cow, and pig (Veitia et al. 1997) indicates that the felid SRY shares the same transcription start site, located ∼100 bp upstream of the starting ATG codon, as well as the two Sp1 motifs in a similar location (supplemental Figure 2 at http://www.genetics.org/supplemental/).
Reconstruction of felid evolution based on the entire genetic sequence spanning the 5′ flank, SRY, and the 3′ genomic flank is remarkably informative. With two exceptions reflecting minor rearrangements between closely related species, this region of the NRY (2651 bp) recovers the same species associations as those based on 23 kb of data (O'Brien and Johnson 2005; Johnson et al. 2006). Overall, this high level of phylogenetic fidelity is consistent with previous research of Y-linked genes in Felidae (Pecon Slattery and O'Brien 1998; Pecon-Slattery et al. 2004a).
Selection in SRY during Felidae evolution:
Using Felidae as a reference species tree, and being aided by robust estimates of species divergence times (O'Brien and Johnson 2005; Johnson et al. 2006), provides a comprehensive depiction of SRY evolution. Estimating an average substitution rate of 0.07% per site per MY (supplemental Table 3 at http://www.genetics.org/supplemental/), SRY accumulates mutations slowly and with little phylogenetic “noise.” An average ratio of dN:dS substitutions ω = 0.815 across Felidae is consistent with expectations for male-specific genes within the NRY (Graves 1995, 1998b) and indicates that SRY is under purifying selection to maintain function in the absence of recombination.
Yet, there are indications that SRY evolution is not consistently under purifying selection in Felidae. For example, SRY exhibits a fivefold difference in average substitution rates among the eight lineages (supplemental Table 3 at http://www.genetics.org/supplemental/). Further, seven pairs of closely related species have the same SRY sequence. In five cases, the species share identical nucleotide sequence (Figure 2A) with the remainder sharing identical amino acid sequence (Figure 2B). There is a large discrepancy between divergence times for these species, ranging from 0.74 MY (Geoffroy's cat–kodkod) to 2.8 MY (lion–leopard) (O'Brien and Johnson 2005; Johnson et al. 2006). Therefore, if SRY is evolving in the same way between species, then identical sequences would have been expected to be shared between all species with divergence times <2.8 MY, a result not observed.
Differences in SRY evolution may be attributed to relaxation of functional constraints leading to either neutral or positive selection within species. Among sites models M2a and M8 were both significant and estimated that 4.1% of the SRY codons were positively selected. Six codons are identified as possibly being under positive selection, but were not supported by significant posterior probability; therefore, among sites models were inconclusive in defining patterns of selection in SRY.
In contrast, models that allowed specific sites within the eight lineages to be under differential selection indicated that positive selection occurred in the puma and domestic cat lineages. The puma lineage arose ∼4.9 MYA (O'Brien and Johnson 2005; Johnson et al. 2006) and is composed of puma, cheetah, and jaguarondi. Marked by long branch lengths in multiple studies using different genetic markers (Johnson and O'Brien 1997; Pecon Slattery and O'Brien 1998; Pecon-Slattery et al. 2004b; O'Brien and Johnson 2005; Johnson et al. 2006) these three species are among the oldest in the cat family, having divergence times estimated between 4.92 and 4.17 MYA (Johnson et al. 2006). Of the 11 codons under positive selection (Table 3), 10 were unique to one of the three species within the puma lineage (Figure 1). Positively selected codon 75 (jaguarondi) and codon 97 (puma) are located in the conserved HMG box. Notably, codon 75 (Figure 1) corresponds to codon 78 in humans that causes 46, XY pure gonadal dysgenesis if altered by point mutations (Haqq and Donahoe 1998; Assumpcao et al. 2002). In the domestic cat lineage, codon 125, also within the HMG box, is under positive selection in five species that last shared a common ancestor ∼2.49 MYA (Johnson et al. 2006). Thus, adaptive evolution in SRY is evident within portions of the felid phylogeny with positive selection observed in some species in the highly conserved HMG region. Moreover, SRY from ocelot and clouded leopard, the two species not included in the PAML analysis due to frameshift mutations incompatible with the program, may have acquired these profound changes due to relaxed functional constraints.
Conclusions:
Consistent with purifying selection, SRY evolves slowly and with extremely low levels of noise in the cat family. However, the majority of substitutions are amino acid altering, some result in truncation of the SRY protein, and others are positively selected in specific cat lineages.
The fixation of nonsynonymous substitutions between sister taxa is not consistent and may occur rapidly, as in the case of domestic cat, or not at all over long periods of time, as observed within the panthera lineage. The former resembles positive selection during speciation and the latter purifying selection to maintain function. In addition, identical SRY sequences occurring between species indicates that the gene is not directly involved with speciation. These inconsistencies in SRY evolution are intriguing and may be clarified by defining these changes in SRY against the unique evolutionary and phylogeographic history of different cat species. In addition, further investigation in NRY genes will determine if this pattern is unique to SRY or is indicative of the evolutionary history of the Y chromosome in general.
Acknowledgments
All tissue samples were collected in full compliance with specific Federal Fish and Wildlife permits from the Convention of International Trade in Endangered Species of Wild Flora and Fauna: Endangered and Threatened species, Captive bred issued to the National Cancer Institute (NCI)–National Institutes of Health (NIH) (S.J.O., principal officer) by the U.S. Fish and Wildlife Services of the Department of the Interior. This publication has been funded in whole or in part with Federal funds from the NCI–NIH, under contract no. N01-CO-12400. This research was supported (in part) by the Intramural Research Program of the NIH, NCI, Center for Cancer Research. Additional support for V.K. was from the Marshall Aid Commemoration Commission, the Medical Research Council, and the Wellcome Trust.
References
- Adachi, J., and M. Hasegawa, 1996. MOLPHY: programs for molecular phylogenetics based on maximum likelihood. Comput. Sci. Monogr. 28: 1–150. [Google Scholar]
- Akiyama, Y., 1998. TFSEARCH: Searching Transcription Factor Binding Sites (http://www.cbrc.jp/research/db/tfsearch.html).
- Assumpcao, J. G., C. E. Benedetti, A. T. Maciel-Guerra, G. Guerra Jr., M. T. Baptista et al., 2002. Novel mutations affecting SRY DNA-binding activity: the HMG box N65H associated with 46,XY pure gonadal dysgenesis and the familial non-HMG box R30I associated with variable phenotypes. J. Mol. Med. 80: 782–790. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., and B. Charlesworth, 2001. Towards a complete sequence of the human Y chromosome. Genome Biol. 2: REVIEWS1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, N. O., 2002. Akodon sex reversed females: the never ending story. Cytogenet. Genome Res. 96: 60–65. [DOI] [PubMed] [Google Scholar]
- Bianchi, N. O., M. S. Bianchi, G. Bailliet and A. de la Chapelle, 1993. Characterization and sequencing of the sex determining region Y gene (Sry) in Akodon (Cricetidae) species with sex reversed females. Chromosoma 102: 389–395. [DOI] [PubMed] [Google Scholar]
- Buckley-Beason, V., W. Johnson, W. G. Nash, R. Stanyon, J. Menninger et al., 2007. Moelcular evidence for species-level distinctions in clouded leopards (Neofelis nebulosa). Curr. Biol. 16: 2371–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel, B., 1998. Sex in the 90s: SRY and the switch to the male pathway. Annu. Rev. Physiol. 60: 497–523. [DOI] [PubMed] [Google Scholar]
- Cheng, H., H. Shi, R. Zhou, Y. Guo, L. Liu et al., 2001. Characterization of Bovidae sex-determining gene SRY. Genet. Sel. Evol. 33: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbridge, M. L., and J. A. Graves, 1999. Mammalian Y chromosome evolution and the male-specific functions of Y chromosome-borne genes. Rev. Reprod. 4: 101–109. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1993. PHYLIP (Phylogenetic Inference Package). University of Washington, Seattle.
- Foster, J. W., F. E. Brennan, G. K. Hampikian, P. N. Goodfellow, A. H. Sinclair et al., 1992. Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials. Nature 359: 531–533. [DOI] [PubMed] [Google Scholar]
- Glaser, B., D. Myrtek, Y. Rumpler, K. Schiebel, M. Hauwy et al., 1999. Transposition of SRY into the ancestral pseudoautosomal region creates a new pseudoautosomal boundary in a progenitor of simian primates. Hum. Mol. Genet. 8: 2071–2078. [DOI] [PubMed] [Google Scholar]
- Graves, J. A., 1995. The evolution of mammalian sex chromosomes and the origin of sex determining genes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 350: 305–312. [DOI] [PubMed] [Google Scholar]
- Graves, J. A., 1998. a Evolution of the mammalian Y chromosome and sex-determining genes. J. Exp. Zool. 281: 472–481. [PubMed] [Google Scholar]
- Graves, J. A., 1998. b Interactions between SRY and SOX genes in mammalian sex determination. BioEssays 20: 264–269. [DOI] [PubMed] [Google Scholar]
- Graves, J. A., 2001. From brain determination to testis determination: evolution of the mammalian sex-determining gene. Reprod. Fertil. Dev. 13: 665–672. [DOI] [PubMed] [Google Scholar]
- Graves, J. A., 2002. Evolution of the testis-determining gene–the rise and fall of SRY. Novartis Found. Symp. 244: 86–101, 203–206, 253–257. [PubMed] [Google Scholar]
- Gubbay, J., J. Collignon, P. Koopman, B. Capel, A. Economou et al., 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346: 245–250. [DOI] [PubMed] [Google Scholar]
- Gubbay, J., N. Vivian, A. Economou, D. Jackson, P. Goodfellow et al., 1992. Inverted repeat structure of the Sry locus in mice. Proc. Natl. Acad. Sci. USA 89: 7953–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haqq, C. M., and P. K. Donahoe, 1998. Regulation of sexual dimorphism in mammals. Physiol. Rev. 78: 1–33. [DOI] [PubMed] [Google Scholar]
- Hawkins, J. R., A. Taylor, P. N. Goodfellow, C. J. Migeon, K. D. Smith et al., 1992. Evidence for increased prevalence of SRY mutations in XY females with complete rather than partial gonadal dysgenesis. Am. J. Hum. Genet. 51: 979–984. [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel et al., 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26: 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Jansa, S. A., B. L. Lundrigan and P. K. Tucker, 2003. Tests for positive selection on immune and reproductive genes in closely related species of the murine genus mus. J. Mol. Evol. 56: 294–307. [DOI] [PubMed] [Google Scholar]
- Johnson, W. E., E. Eizirik, J. Pecon-Slattery, W. J. Murphy, A. Antunes et al., 2006. The late Miocene radiation of modern Felidae: a genetic assessment. Science 311: 73–77. [DOI] [PubMed] [Google Scholar]
- Johnson, W. E., and S. J. O'Brien, 1997. Phylogenetic reconstruction of the Felidae using 16S rRNA and NADH-5 mitochondrial genes. J. Mol. Evol. 44(Suppl. 1): S98–S116. [DOI] [PubMed] [Google Scholar]
- Just, W., W. Rau, W. Vogel, M. Akhverdian, K. Fredga et al., 1995. Absence of Sry in species of the vole Ellobius. Nat. Genet. 11: 117–118. [DOI] [PubMed] [Google Scholar]
- King, V. E., 1996. Evolution of SRY and Y Chromosome in the Family Felidae. Ph.D. Thesis, Trinity College, University of Cambridge, Cambridge, UK.
- Koopman, P., J. Gubbay, N. Vivian, P. Goodfellow and R. Lovell-Badge, 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351: 117–121. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1997. Functional coherence of the human Y chromosome. Science 278: 675–680. [DOI] [PubMed] [Google Scholar]
- Liu, W. S., P. Mariani, C. W. Beattie, L. J. Alexander and F. A. Ponce De Leon, 2002. A radiation hybrid map for the bovine Y chromosome. Mamm. Genome 13: 320–326. [DOI] [PubMed] [Google Scholar]
- Lundrigan, B. L., and P. K. Tucker, 1997. Evidence for multiple functional copies of the male sex-determining locus, Sry, in African murine rodents. J. Mol. Evol. 45: 60–65. [DOI] [PubMed] [Google Scholar]
- Marshall Graves, J. A., 2002. a Sex chromosomes and sex determination in weird mammals. Cytogenet. Genome Res. 96: 161–168. [DOI] [PubMed] [Google Scholar]
- Marshall Graves, J. A., 2002. b The rise and fall of SRY. Trends Genet. 18: 259–264. [DOI] [PubMed] [Google Scholar]
- Moreira, M. A., 2002. SRY evolution in Cebidae (Platyrrhini: Primates). J. Mol. Evol. 55: 92–103. [DOI] [PubMed] [Google Scholar]
- Murphy, E. C., V. B. Zhurkin, J. M. Louis, G. Cornilescu and G. M. Clore, 2001. Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol. 312: 481–499. [DOI] [PubMed] [Google Scholar]
- Murphy, W. J., S. Sun, Z. Q. Chen, J. Pecon-Slattery and S. J. O'Brien, 1999. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 9: 1223–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine, C. M., 1994. The testis-determining gene, SRY, exists in multiple copies in Old World rodents. Genet. Res. 64: 151–159. [DOI] [PubMed] [Google Scholar]
- O'Brien, S. J., and W. E. Johnson, 2005. Big cat genomics. Annu. Rev. Genomics Hum. Genet. 6: 407–429. [DOI] [PubMed] [Google Scholar]
- O'Neill, M. J., and R. J. O'Neill, 1999. Whatever happened to SRY? Cell. Mol. Life Sci. 56: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, R. J., M. D. Eldridge, R. H. Crozier and J. A. Graves, 1997. Low levels of sequence divergence in rock wallabies (Petrogale) suggest a lack of positive directional selection in SRY. Mol. Biol. Evol. 14: 350–353. [DOI] [PubMed] [Google Scholar]
- Olivier, M., M. Breen, M. M. Binns and G. Lust, 1999. Localization and characterization of nucleotide sequences from the canine Y chromosome. Chromosome Res. 7: 223–233. [DOI] [PubMed] [Google Scholar]
- Pask, A., M. B. Renfree and J. A. Marshall Graves, 2000. The human sex-reversing ATRX gene has a homologue on the marsupial Y chromosome, ATRY: implications for the evolution of mammalian sex determination. Proc. Natl. Acad. Sci. USA 97: 13198–13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon Slattery, J., and S. J. O'Brien, 1998. Patterns of Y and X chromosome DNA sequence divergence during the Felidae radiation. Genetics 148: 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon-Slattery, J., A. J. Pearks Wilkerson, W. J. Murphy and S. J. O'Brien, 2004. a Phylogenetic assessment of introns and SINEs within the Y chromosome using the cat family Felidae as a species tree. Mol. Biol. Evol. 21: 2299–2309. [DOI] [PubMed] [Google Scholar]
- Pecon-Slattery, J., A. J. Pearks Wilkerson, W. J. Murphy and S. J. O'Brien, 2004. b Phylogenetic assessment of introns and SINEs within the Y chromosome using the cat family felidae as a species tree. Mol. Biol. Evol. 21: 2299–2309. [DOI] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Quilter, C. R., S. C. Blott, A. J. Mileham, N. A. Affara, C. A. Sargent et al., 2002. A mapping and evolutionary study of porcine sex chromosome genes. Mamm. Genome 13: 588–594. [DOI] [PubMed] [Google Scholar]
- Sinclair, A. H., P. Berta, M. S. Palmer, J. R. Hawkins, B. L. Griffiths et al., 1990. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346: 240–244. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., T. Kuroda-Kawaguchi, P. J. Minx, H. S. Cordum, L. Hillier et al., 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837. [DOI] [PubMed] [Google Scholar]
- Swain, A., and R. Lovell-Badge, 1999. Mammalian sex determination: a molecular drama. Genes Dev. 13: 755–767. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., R. Nielsen and Q. Yang, 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 20: 18–20. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 1998. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer Associates, Sunderland, MA.
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilford, C. A., T. Kuroda-Kawaguchi, H. Skaletsky, S. Rozen, L. G. Brown et al., 2001. A physical map of the human Y chromosome. Nature 409: 943–945. [DOI] [PubMed] [Google Scholar]
- Tucker, P. K., and B. L. Lundrigan, 1993. Rapid evolution of the sex determining locus in Old World mice and rats. Nature 364: 715–717. [DOI] [PubMed] [Google Scholar]
- Tucker, P. K., and B. L. Lundrigan, 1995. The nature of gene evolution on the mammalian Y chromosome: lessons from Sry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 350: 221–227. [DOI] [PubMed] [Google Scholar]
- Veitia, R. A., M. Fellous and K. McElreavey, 1997. Conservation of Y chromosome-specific sequences immediately 5′ to the testis determining gene in primates. Gene 199: 63–70. [DOI] [PubMed] [Google Scholar]
- Whitfield, L. S., R. Lovell-Badge and P. N. Goodfellow, 1993. Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature 364: 713–715. [DOI] [PubMed] [Google Scholar]
- Yang, H., R. Fries and G. Stranzinger, 1993. The sex-determining region Y (SRY) gene is mapped to p12-p13 of the Y chromosome in pig (Sus scrofa domestica) by in situ hybridization. Anim. Genet. 24: 297–300. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Zhang, J., R. Nielsen and Z. Yang, 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22: 2472–2479. [DOI] [PubMed] [Google Scholar]