Abstract
Apparent full-length cDNA sequences coding for manganese superoxide dismutase (HvMnSOD) and extracellular superoxide dismutase (HvEC-SOD) were isolated from Hydra vulgaris in order to understand their expression and 3D structures; and explore their possibility of being used as for biomarkers for environmental stress and toxicity. The deduced HvMnSOD protein consists of 219 amino acids of which first 21 amino acids constitute a presumed mitochondria-targeting signal peptide whereas HvEC-SOD protein consists of 189 amino acids of which first 19 amino acids constitute a presumed signal peptide. Molecular model generated for HvMnSOD displayed the N-terminal long alpha antiparallel hairpin and the C-terminal mixed alpha/beta fold characteristic of MnSODs and that for HvEC-SOD displayed the characteristic CuZnSOD beta-barrel fold. Hydrae subjected to thermal, starvation, metal and oxidative stress responded by regulating MnSOD and EC-SOD mRNA transcription. These results indicated that these genes are involved in the cellular stress response and (anti)oxidative processes triggered by stressor and contaminant exposure. Hence the expression of these SODs in hydra may have potential as molecular biomarkers for assessing stress, toxicity and pro-oxidant quality of chemicals and aquatic environmental quality.
Keywords: MnSOD, EC-SOD, Homology modeling, Gene expression, Molecular biomarker
1. Introduction
Reactive oxygen species (ROS) are produced as by-products of aerobic metabolism or exposure to pro-oxidants. They become toxic or lethal when they damage nucleic acids, proteins and membrane lipids (Halliwell and Gutteridge, 1999). In order to resist these potentially damaging oxygen species, aerobic organisms have developed an enzymatic protection system consisting of antioxidant enzymes: superoxide dismutases (SODs, EC 1.15.1.1), glutathione peroxidases and catalases. SOD activity reduces the superoxide radical O2·— into hydrogen peroxide and molecular oxygen (Fridovich, 1995); and peroxidases and catalases reduce hydrogen peroxide into water and oxygen (Halliwell and Gutteridge, 1999).
There are four types of SODs identified in eukaryotic cells and they differ by the metallic ion present at the their active site: (1) manganese-containing SOD (MnSOD) synthesized in the cytosol and imported post-translationally into the mitochondrial matrix, (2) copper/zinc-containing intracellular SOD (CuZnSOD) found in the cytosol and also in chloroplasts of plants, algae, etc., (3) copper/zinc-containing extracellular SOD (EC-SOD) secreted by many types of cells and anchored to the plasma membrane or circulate in the extracellular fluids (Stallings et al., 1984; Halliwell and Gutteridge, 1999), and (4) iron containing SOD (FeSOD). MnSODs found in eukaryotes are homologous to iron-dependent bacterial SODs (FeSOD) (Stallings et al., 1984; Fridovich 1995). FeSODs are also present within the chloroplasts of some plants (Bannister et al., 1987). Though these enzymes catalyze the same reaction, CuZnSODs are structurally distinct from MnSODs and FeSODs. MnSODs and FeSODs share a high degree of amino acid sequence and structural homologies (Stallings et al., 1984; Halliwell and Gutteridge, 1999). Also the two forms of CuZnSOD share high sequence and structural homology.
Genes encoding MnSOD (Ditlow et al., 1982; Henkle-Duhrsen et al., 1995; Jones et al., 1995) and EC-SOD (Hjalmarsson et al., 1987; Rodney et al., 1997; Liddell and Knox, 1998) enzymes have been identified in many organisms. Also a large number of bacterial FeSOD genes have been identified (Campbell and Laudenbach, 1995; Tsolis et al., 1995). SODs have been used as biomarkers of cellular stress and toxicity in different aquatic organisms such as mollusk, cnidarians, fish; etc. (Niyogi et al., 2001; Li et al., 2003; Richier et al., 2003; Zhang et al., 2004).
Among cnidarians CuZnSODs are reported in antarctic soft coral Alcyonium paessleri (an RT-PCR fragment, accession number: AAG10392) and sea anemone Anemonia viridis (accession number: AY532063, Plantivaux et al., 2004). Also an extracellular CuZnSOD has been reported in cnidarian Anemonia viridis (accession number: AY532063, Plantivaux et al., 2004). An expressed sequence tag (EST) is reported from the hydra, Hydra magnipapillata, (accession number gi|57833081) coding for a CuZnSOD that is similar to the N-terminal end of Caenorhabditis elegans Cu-Zn superoxide dismutase (P34697). Also an EST is reported from H. magnipapillata (accession number gi|57833081) coding for a MnSOD that is similar to the N-terminal end of Equus caballus manganese superoxide dismutase (Q9XS41). Many studies have indicated the presence and importance of MnSOD along with the other forms of SODs in several cnidarians (Dykens and Shick, 1982; Dykens and Shick, 1984; Brown et al., 2002; Downs et al., 2002; Richier et al., 2003;Plantivaux et al., 2004). For example, the very efficient defense system against reactive oxygen species developed by the symbiotic corals and sea anemones is attributed to the presence and function of SODs in them (Dykens and Shick, 1982; Dykens and Shick, 1984).
Hydra, a fresh water cnidarian, is used as an environmental toxicological model to study the acute and chronic toxicity effects of environmental toxicants (Johnson et al., 1982; Lum et al., 2003). Unable to move (= sessile), all of the cells of hydrae are in close proximity to the aqueous medium and the immediate environment. This characteristic enables hydra to be very sensitive and susceptible to minute amounts of environmental toxicants (Lum et al., 2003). The changes in external gross morphology, anatomy, and physiology are useful as markers of toxicity or toxicity end points in the hydra bioassays (Beach et al., 1998; Pollino and Holdway, 1999; Holdway et al., 2001; Karntanut and Pascoe, 2002). It may be postulated that the detection of SOD messages in hydra can constitute an early-warning marker for the presence of potentially deleterious agents in water, and thus may in general act as a potential sentinel animal species. Because H. vulgaris are sensitive to a variety of compounds, the detection of SOD messages could be applied as a prescreening tool in determining the relative toxicity of many toxicants, and new compounds that are to be screened for toxicity. The overwhelming majority of existing substances and most of the new ones appearing in commerce annually can be evaluated for their potential health hazards.
In this research, the molecular analysis of a manganese superoxide dismutase (HvMnSOD) and an extracellular CuZnSOD (HvEC-SOD) in cnidarian, H. vulgaris, is described. The expression of hydra MnSOD and EC-SOD mRNA is assayed with respect to different environmental contaminant challenge (i.e., arsenic, cadmium, selenium, zinc and copper) and stressors (both oxidative and non-oxidative). Furthermore, suitable homology based structural model of HvMnSOD and HvEC-SOD is also presented to assist in understanding the structural and functional evolutionary significance of the protein. The molecular analysis of hydra MnSOD and EC-SOD is the first reported for a diploblastic hydrozoan organism and represents an important step in the evolutionary study of these enzymes.
2. Materials and methods
2.1. Hydra culture
Hydra vulgaris (formerly known as Hydra attenuata) were originally obtained from E. Marshall Johnson, Jefferson Medical College (Philadelphia, PA, USA). H. vulgaris were maintained in shallow dishes at 18 °C in a medium containing 1 mM CaCl2.2H2O, 0.012 mM EDTA, and 0.458 mM TES (N-tris(hydroxymethyl)-methyl-2-amino-ethanesulfonic acid, sodium salt) buffer (pH 7.0). Daily, hydrae were fed with brine shrimp (Artemia nauplii) hatched in a solution of 1 % sodium chloride and treated with iodine (40 μg ml-1). Hydrae were maintained free from bacterial and fungal contamination and were not fed for 24 h before initiating the experiments. Deionized water was used throughout this portion of the study (Mayura et al., 1991).
2.2. RNA isolation and clean up
Total RNA was extracted from hydra by application of 2 ml of TRIzoL® reagent (Invitrogen, USA) to approximately 20 mg of fresh tissue, according to manufacturer’s instructions. The RNA was quantified by ultraviolet absorbance at 260 nm. Integrity of the total RNA was confirmed by 1 % formaldehyde agarose gel electrophoresis. The RNA isolated was cleaned up from contaminating DNA using RNeasy Mini Kit (Invitrogen, USA) following the manufacturer’s instruction.
2.3. Oligonucleotides
All oligonucleotides were procured from Integrated DNA Technology Inc. (IA, USA). AP2 primer (5'-GATCAGGACGTTCGTTTGAGd(T)17-3') was used for 5'-RACE (Rapid Amplification of cDNA Ends) experiments. The oligo(dT) bifunctional primer N was supplied in a cDNA preparation kit (Amersham Biosciences, USA) and was used for 3'-RACE experiments.
MnSOD protein sequences from diverse organisms retrieved from the NCBI protein database were aligned using program ClustalW (Fig. 1) resulted in identifying the conserved regions in MnSODs. Oligonucleotides DF and DR were designed corresponding to the amino acids FNGGGHIN and VWEHAYY (Table 1).
Fig. 1.

Multiple sequence alignment of MnSOD proteins performed using the program ClustalW. Accession numbers of the proteins as extracted using the Entrez web service are: AAF57955 (Drosophila melanogaster), AAP35613 (Homo sapiens), CAA39937 (Rattus norvegi cus), AAQ63483 (Xenopus laevis), Hydra vulgaris (DQ286038), CAB02913 (Caenorhabditis elegans), NP989542 (Gallus gallus), CAI11702 (Danio rerio), and X03951 (Escherichia coli). The properties of amino acids are identified by different shadings as given in the legend. The stretches of amino acids chosen for designing primers are underlined.
Table 1.
Oligonucleotides Used in the Study
| Oligonucleotides | Sequence |
|---|---|
| AP2 | 5’-GATCAGGACGTTCGTTTGAGd(T)17-3’ |
| AF | 5’-AAGCTCTTCCCTCGAAGAATC-3’ |
| AR | 5’-CCAAAATAGATCCTCCGATCC-3’ |
| Oligonucleotides exclusively used in the cloning of HvMnSOD cDNA | |
| DF | 5’-TTCAATGGWGGWGGRCAYATYA-3’ |
| DR | 5’-TAGTAMGCRTGCTCCCAMAC-3’ |
| F | 5’-TTCAATGGAGGTGGGCACATCA-3’ |
| R | 5’-TAGTACGCATGCTCCCAAAC-3’ |
| R2 | 5’-TTCATTGCCTCAAAAGACCC-3’ |
| R3 | 5’-ATGGAATGATTGATGTGCCC-3’ |
| Oligonucleotides exclusively used in the cloning of HvEC-SOD cDNA | |
| F1 | 5’-CATGGTTTTCATATCCA-3’ |
| R1 | 5’-TGCTTTCCCCAAATCATC-3’ |
| R4 | 5’-TTGCACCACTCCATCTTTACC-3’ |
| R5 | 5’-CGTTATCTGCTTTCCCCAAA-3’ |
*Mix base definitions: W=A, T; R=A, G; Y=C, T; M=A, C
The GenBank was searched for the cnidaria/hydra CuZnSOD. The CuZnSOD coded by the H. magnipapillata EST (gi|57833081) had similar conserved amino acid residues as identified by multiple sequence alignment of several CuZnSODs (Fig. 4). Hence, a pair of primes F1 and R1 (Table 1) was designed from the EST sequence that corresponded to the conserved amino acid residues (HGFHIH and DDLGKA) to clone a fragment of CuZnSOD from H. vulgaris. Other H. vulgaris gene specific primers used in the experiments are also given in Table 1.
Fig. 4.
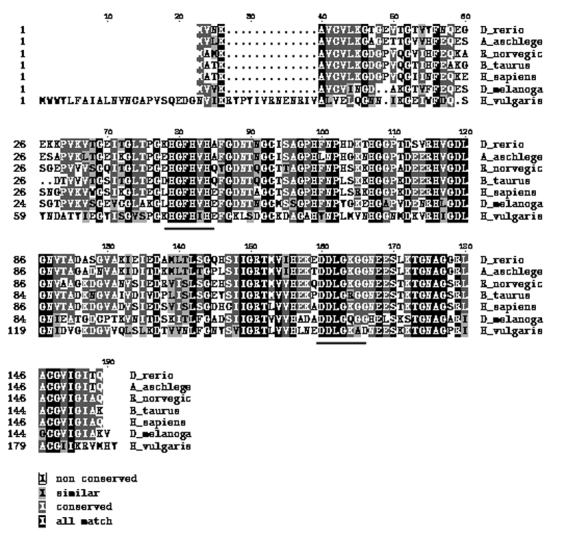
Multiple sequence alignment of CuZnSOD proteins performed using the program ClustalW. Accession numbers from the proteins as extracted using the Entrez web service are: Hydra vulgaris (DQ286039), Danio rerio (Y12236), Acanthopagrus schlegelii (AJ000249), Rattus norvegicus (Y00404), Bos taurus (M81129), Homo sapiens (X02317) and Drosophila melanogaster (Y00367). The properties of different amino acids are identified by shading as given in the legend. The stretches of amino acids chosen for designing primers are underlined.
2.4. Cloning and identification of partial fragments of H. vulgaris MnSOD and CuZnSOD cDNA
All polymerase chain reaction (RT-PCR and RACE-PCR) experiments were performed using Taq DNA polymerase (Invitrogen, USA) and a thermal cycler (MJ Research, USA).
RNA (5 μg) was reverse-transcribed to cDNA at 37 °C for 60 min using the oligo(dT) bifunctional primer N (Table 1) and the AMV RT supplied in the cDNA synthesis kit (Amersham Biosciences, USA). The first-strand cDNA was amplified using the primer pairs: (i) DF and DR for cloning and identifying partial fragments of H. vulgaris MnSOD cDNA (Table 1); and (ii) F1 and R1 for cloning and identifying partial fragments of H. vulgaris CuZnSOD cDNA. The PCR was performed for 30 cycles, consisting of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 1 min and a final extension at 72 °C for 10 min. The resultant PCR products was subcloned into the pCR®II-TOPO® vector using a TA cloning kit (Invitrogen, USA) according to the manufacturer’s instructions. Multiple independent clones were sequenced using automated methods (DNA Technologies Lab, Department of Veterinary Pathobiology, Texas A&M University) on an ABI PRISM™ 310 Genetic Analyzer (PE Biosystems, USA) using a Big-Dye sequencing kit (PE Biosystems) and M13 primers. The identity of the clones was evaluated by matching the sequences to the nucleotide/protein sequences available at the GenBank. The cloned sequences constituted residues 322 to 623 in the HvMnSOD nucleotide sequence shown in Fig. 2; and residues 368 to 628 in the HvEC-SOD nucleotide sequence shown in Fig. 5.
Fig. 2.
Nucleotide and deduced amino acid sequences of H. vulgaris MnSOD. The deduced amino acid sequence is shown in single-letter code above the nucleotide sequence. The nucleotide and amino acid sequences are numbered from the 5'-end of the 853 bp cDNA sequence, and from the N-terminal start codon methionine, respectively. The first residue of the presumed matured protein aspargine (N) is shown bold face. Amino acid residues corresponding to the metal-binding ligands are bold face and underlined. The asterisk denotes the translation stop signal.
Fig. 5.
Nucleotide and deduced amino acid sequences of H. vulgaris EC-SOD. The deduced amino acid sequence is shown in single-letter code above the nucleotide sequence. The nucleotide and amino acid sequences are numbered from the 5'-end of the 927 bp cDNA sequence, and from the N-terminal start codon methionine, respectively. The first residue of the presumed matured protein glutamate (E) is shown bold face. Amino acid residues corresponding to the metal-binding ligands are underlined. The asterisk denotes the translation stop signal.
2.5 3'-RACE of the HvMnSOD and HvEC-SOD cDNA
First-strand cDNA prepared above was amplified using the oligo(dT) bifunctional primer and a gene-specific primer F (Table 1) (complementary to nucleotide (nt) 322 to 343, Fig. 2) for 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 3 min in order to clone the 3'-end of the HvMnSOD cDNA. In order to clone the 3'-end of the HvEC-SOD cDNA, the first-strand cDNA prepared above was amplified using the oligo(dT) bifunctional primer N and a gene-specific primer F1 (Table 1) (complementary to nucleotide (nt) 368 to 384, Fig. 5) for 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 3 min. The products were subcloned and sequenced as described above. The identity of the clones was evaluated by matching the sequences to the nucleotide/protein sequences available at the GenBank.
2.6. 5'-RACE of the HvMnSOD and HvEC-SOD cDNA
In order to clone the 5'-end of the HvMnSOD cDNA, the template cDNA was synthesized using a primer R2 (Table 1) (complementary to nt 430 to 449) and Superscript II (Invitrogen, USA), followed by dA tailing of the cDNA using dATP and terminal transferase (Invitrogen, USA) using standard procedures (Sambrook et al., 1989). The first strand cDNA was amplified using an oligo(dT) anchor primer AP2 and the gene-specific primer R2, and the first-round PCR products were reamplified using the PCR anchor primer AP2 and another gene-specific primer R3 (Table 1) (complementary to nt 334 to 353). Each PCR was performed with an initial amplification of 95 for °C for 5 min, 48 °C for 5 min and 72 °C for 5 min followed by 20 cycles of 95 °C for 40 s, 48 °C for 1 min and 72 °C for 3 min with a final extension of 10 min at 72 °C.
For cloning the 5'-end of the HvEC-SOD cDNA, the template cDNA was synthesized using a primer R4 (complementary to nucleotide (nt) 509-529). The first strand cDNA was amplified using oligo(dT) anchor primer AP2 and the gene-specific primer R4, and the first-round PCR products were amplified using the PCR anchor primer AP2 and another gene-specific primer R5 (Table 1) (complementary to nt 616 to 635).
The PCR products were subcloned and sequenced as described above. The identity of the clones was confirmed by matching the sequences to the nucleotide/protein sequences available at the GenBank.
2.7. GenBank accession numbers
The full-length sequences of the HvMnSOD and HvEC-SOD mRNA are available in the GenBank databases under the accession numbers DQ286038 and DQ286039 respectively.
2.8. General bioinformatic analyses
Conceptual translation of the full-length cDNA was performed using the program SIXFRAME (http://biologyworkbench.ucsd.edu). Homology to other MnSOD and CuZnSOD/EC-SOD genes and proteins were identified by using the BLAST program with default settings (http://www.ncbi.nlm.nih.gov/BLAST). The identification of HvMnSOD and HvEC-SOD domains were performed by use of the SMART program (http://smart.embl-heidelberg.de/) using default pattern definitions. Prosite (http://us.expasy.org/prosite/) identification of glycosylation, phosphorylation, myristilation and amidation sites was performed by use of the ExPASy program using default settings. The isoelectric point (pI) and the molecular weight (Mw) of the HvMnSOD and HvEC-SOD were calculated using the ExPasy (http://us.expasy.org/tools/pi_tool.html) program. The prediction of the presence and location of signal peptide cleavage sites in amino acid sequence of HvMnSOD and HvEC-SOD was done using the program SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/). The secondary structure analysis of the HvMnSOD transit peptide was done using the program GOR4 (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html). An axial projection of the transit peptide residues on a helical wheel was done using the web server (http://bioinf.man.ac.uk/~gibson/HelixDraw/helixdraw.html). Several MnSOD and CuZnSOD domain-containing sequences retrieved from the National Center for Biotechnology Information (NCBI) Entrez Web service were aligned to each other using the web program T-coffe http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi or ClustalW (http://www.ch.embnet.org/software/ClustalW.html).
2.9. Threading
Five web-available threading methods used to find the structure templates for the HvMnSOD and HvEC-SOD homology modeling were as follows: TOPITS (http://dodo.cpmc.columbia.edu/predictprotein/); HMM (http://www.cse.ucsc.edu/research/compbio/HMM-apps/HMM-applications.html); 3D-JIGSAW (http://www.bmm.icnet.uk/~3djigsaw/); 3D-PSSM (http://www.sbg.bio.ic.ac.uk/~3dpssm/); and HFR (http://www.cs.bgu.ac.il/~bioinbgu/). Through the threading experiments on the Web sites, several templates for the query sequence were found. These templates were further screened using the SWISS-MODEL Protein Modeling Server (Guex and Peitsch, 1997) on the Web. Protein 1LUV (Hearn et al., 2003) was selected as the structure template for the query sequence HvMnSOD and protein 1N18 (Cardoso et al., 2002) was selected as the structure template for the query sequence HvEC-SOD. Hence 3D models of the HvMnSOD and HvEC-SOD protein were built based on the coordinates of 1LUV: A (1LUV.pdb) and 1N18 (1N18.pdb) respectively. Respective models retrieved are shown in Fig. 3 and Fig. 6.
Fig. 3.
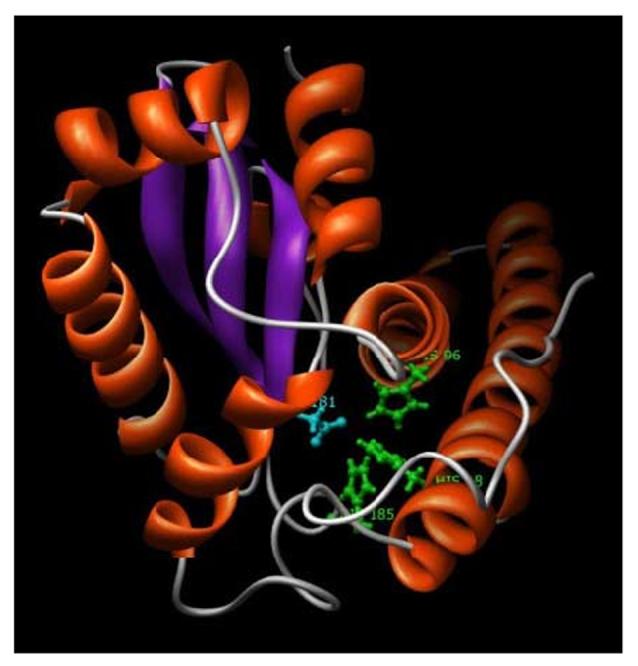
A 3D model of the H. vulgaris MnSOD. HvMnSOD model was generated based on the crystal structure of human manganese superoxide dismutase (PDB code: 1LUV:A). The ligands of the metal manganese are His-48, His-96, His-185 (green colored residues) and Asp-181 (cyan colored residue). N-terminal long alpha antiparallel hairpin and the C-terminal mixed alpha/beta fold characteristic of MnSODs are shown in the figure. The figure displayed was drawn using the program Chimera.
Fig. 6.
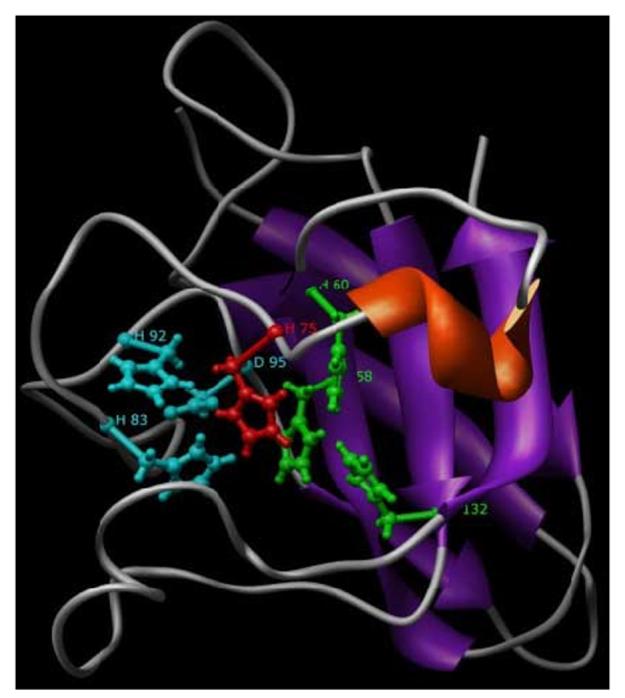
A 3D model of the H. vulgaris EC-SOD. HvEC-SOD model was generated based on the crystal structure of human CuZnSOD (PDB code: 1N18). The ligands of the metal copper are His-58, His-60, His-132 (green colored residues) and His-75 (red colored residue); that of zinc are His-83, His-92, Asp-95 (cyan colored residue) and His-75 (red colored residue). Conserved beta-barrel fold typical of CuZnSODs is shown in the figure. The figure displayed was drawn using the program Chimera.
2.10. Analysis of the models
The overall stereo-chemical quality of the final models was assessed by the program PROCHECK (http://biotech.ebi.ac.uk:8400/cgi-bin/sendquery) (Laskowski et al., 1994). A secondary structure and Ramachandran plot of the model were also drawn. The structural quality of the model was also verified using the program Verify-3D (http://nihserver.mbi.ucla.edu/Verify_3D/) that measures the compatibility of a protein model with its sequence where the values are calculated using 3D profile (Bowie et al., 1991).
2.11. Stress treatment
2.11.1. Heat treatment
Hydrae were incubated at 18 (control temperature), 30 and 37 (maximum induction temperature) °C for 1 h and 6 h in 5 ml hydra media.
2.11.2. Metal treatment
Hydrae were incubated with 100 ppm of CuSO4.5H2O, ZnCl2, CdCl2.2.5H2O, K2Cr2O7, As2O3, Na2HAsO4 or Na2SeO3 for 1 h and 6 h in 5 ml hydra media. Hence the concentration of Cu (II), Zn (II), Cd (II), Cr (VI), As (III), As (V), and Se (IV) (active ingredients) were 0.4 mM (= 25.4 ppm), 0.73 mM (= 47.9 ppm), 0.43 mM (= 49.2 ppm), 0.33 mM (= 17.6 ppm), 0.506 mM (= 37.8 ppm), 0.302 mM (= 24.0), and 0.57 mM (= 45.6 ppm) respectively.
2.11.3. Oxidative stress treatment
Hydrae were incubated in 300 ppm (= 9.79 mM) H2O2, 100 ppm (= 0.38mM) of paraquat and 100 ppm (= 1.538 mM) sodium azide in 5 ml hydra media for 1 or 6 h.
2.11.4. Starvation treatment
For starvation experiment hydrae were unfed for 5 days
In all cases, treatments were carried out in triplicate in 14 ml polypropylene round-bottom tubes (Becton-Dickinson, NJ, USA) containing approximately a pool of 500 hydrae in 5 ml hydra media. At the end of the treatment, hydrae were collected by centrifugation at 7,500 Xg for 5 min. The supernatants were discarded. Then hydrae were washed once in 5 ml of 0.05 M PBS for 5 min at 7,500 Xg. The supernatants were discarded as before and the animals were homogenized immediately in 3 ml of TRIzoL®.
2.12. Expression analysis of HvMnSOD and HvEC-SOD mRNA in hydrae
Total RNA was extracted from TRIzoL® (Invitrogen, USA) treated whole hydrae and cleaned as before using RNeasy Mini Kit (Invitrogen, CA, USA) following procedures previously described. For RT-PCR, 5 μg of total cleaned RNA was reverse transcribed using 500 ng of oligo(dT)12-18 primer (Invitrogen, USA) and 200 units of the Superscript II enzyme (Invitrogen, USA) for 50 min at 42 °C. The reaction was inactivated by heating the mixture at 70 °C for 15 min. PCR assays were designed to normalize HvMnSOD and HvEC-SOD mRNA expression levels to actin transcription rate. Two μl of first strand cDNA (from 25 μl of reverse transcription mix) were diluted 100 times prior to PCR amplification. PCR was carried out in 50 μl total volume of 1 × PCR buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCl), 2 mM MgCl2, 0.1 mM each of dNTPs, 10 pmol each of primers, and 0.5 U Taq DNA polymerase (Invitrogen, USA) using 2 μl of (1:100) diluted RT-product. Actin mRNA was amplified using AF and AR primers; HvMnSOD mRNA was amplified using F and R primers; and HvEC-SOD mRNA was amplified using F1 and R1 primers. Cycling profile after initial denaturation at 94 °C for 4 min was 30 cycles of amplification as follows: denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 45 s. Equal amounts (9.5 μl) of RT-PCR reactions were loaded on standardized 2 % agarose gels containing 0.1 μg/ml ethidium bromide. The gel images were digitalized by a gel documentation system (Kodak Laboratories, USA) (Fig. 7A, 8A, 9A and 10A) and HvMnSOD and HvMnSOD mRNA bands were quantified by NIH ImageJ software (Fig. 7B, 8B, 9B, 10B). Respective mRNA levels were normalized to the control (18 °C) after the normalization to actin mRNA levels.
Fig. 7.
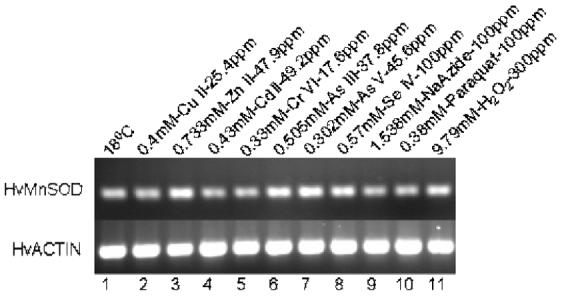
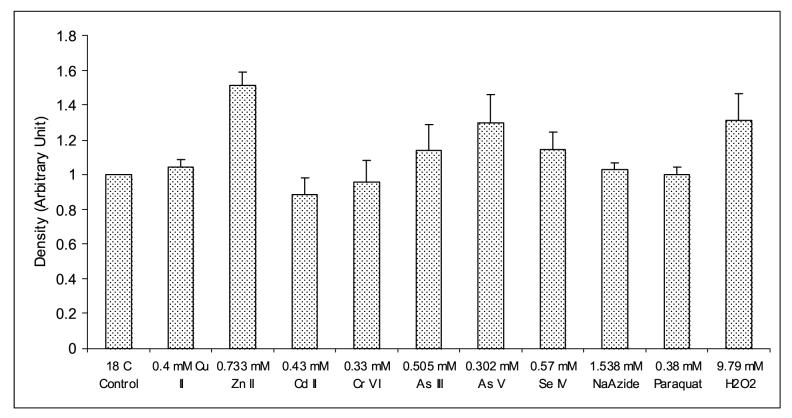
Expression analysis of MnSOD mRNA from H. vulgaris exposed to metal and oxidative stress for 1 h. (A) A representative agarose gel image of the expression of HvMnSOD mRNA due to different stress conditions: Metal stress for 1 h (lanes 2-8) and oxidative stress for 1 h (lanes 9-11). The expression HvMnSOD mRNA is compared to that of actin. Detected HvMnSOD mRNA bands were quantified by NIH ImageJ software. HvMnSOD mRNA levels were normalized to the control (18 °C) after the normalization to actin. Data represent mean ± S.D. (n=3). Error bars in the graphs indicate standard deviation (S.D.) of the mean.
Fig. 8.
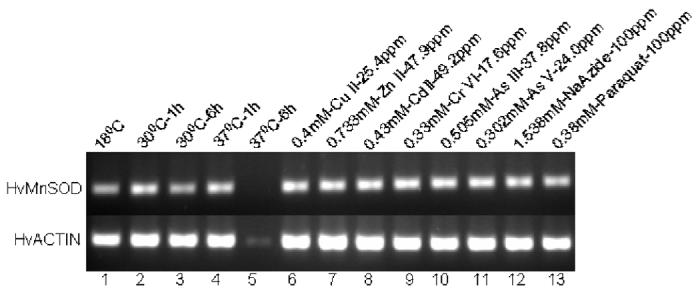
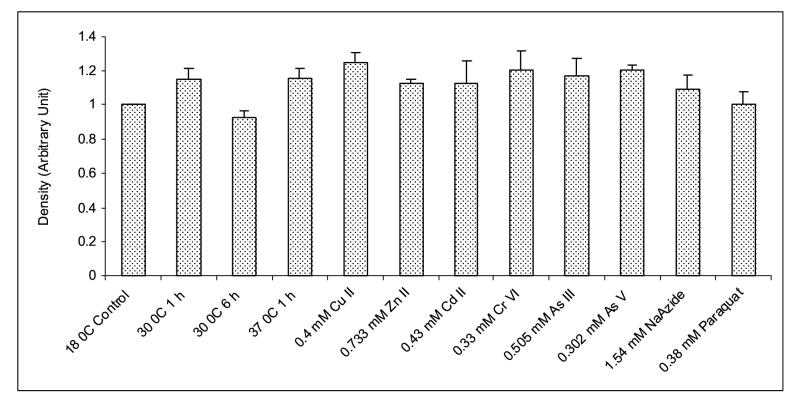
Expression analysis of MnSOD mRNA from H. vulgaris exposed to thermal, metal and oxidative stress. (A) A representative agarose gel image of the expression of HvMnSOD mRNA due to different stress conditions: Thermal stress (lanes 2-5), metal stress for 6 h (lanes 6-11), and oxidative stress for 6 h (lanes 12-13). The expression HvMnSOD mRNA is compared to that of actin. (B) Detected HvMnSOD mRNA bands were quantified by NIH ImageJ software. HvMnSOD mRNA levels were normalized to the control (18 °C) after the normalization to actin. HvMnSOD mRNA expression in hydra was drastically reduced when exposed to 37 °C for 6 h and hence its expression couldn’t be quantified. Data represent mean ± S.D. (n=3). Error bars in the graphs indicate standard deviation (S.D.) of the mean.
Fig. 9.
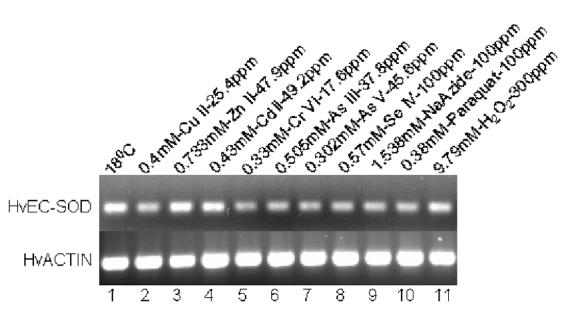
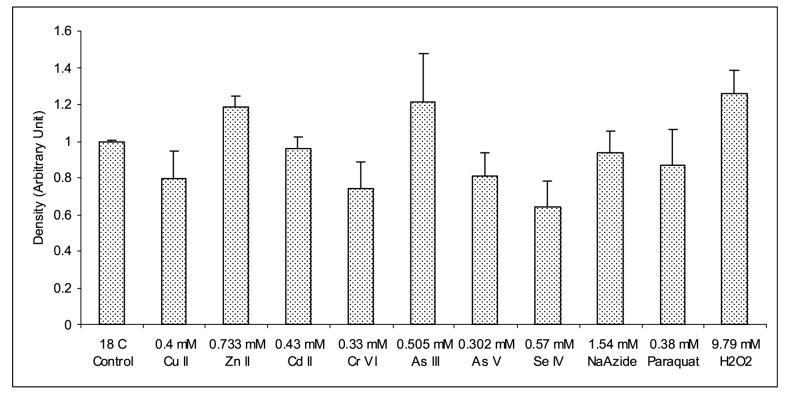
Expression analysis of EC-SOD mRNA from H. vulgaris exposed to metal and oxidative stress for 1 h. (A) A representative agarose gel image of the expression of HvEC-SOD mRNA due to different stress conditions: Metal stress for 1 h (lanes 2-8) and oxidative stress for 1 h (lanes 9-11). The expression HvEC-SOD mRNA is compared to that of actin. (B) Detected HvEC-SOD mRNA bands were quantified by NIH ImageJ software. HvEC-SOD mRNA levels were normalized to the control (18 °C) after the normalization to actin. Data represent mean ± S.D. (n=3). Error bars in the graphs indicate standard deviation (S.D.) of the mean.
Fig. 10.
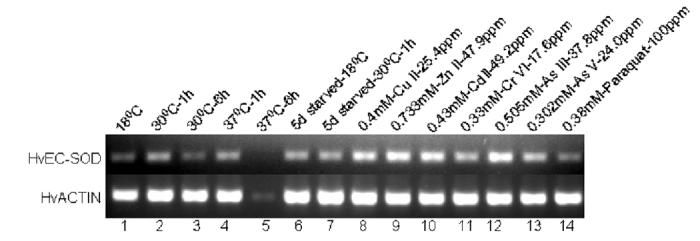
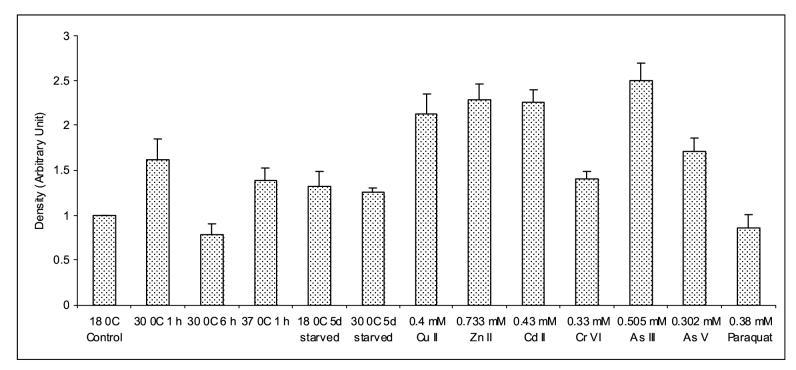
Expression analysis of EC-SOD mRNA from H. vulgaris exposed to thermal, starvation, metal and oxidative stress. (A) A representative agarose gel image of the expression of HvEC-SOD mRNA under different stress conditions: Thermal stress (lanes 2-5), starvation stress (lanes 6-7), metal stress for 6 h (lanes 8-13) and oxidative stress for 6 h (lane 14). The expression HvEC-SOD mRNA is compared to that of actin. (B) Detected HvEC-SOD mRNA bands were quantified by NIH ImageJ software. HvEC-SOD mRNA levels were normalized to the control (18 °C) after the normalization to actin. HvECSOD mRNA expression in hydra was drastically reduced when exposed to 37 °C for 6 h and hence its expression couldn’t be quantified. Data represent mean ± S.D. (n=3). Error bars in the graphs indicate standard deviation (S.D.) of the mean.
2.13. Data analysis
Data were presented (Fig. 7B, 8B, 9B, 10B) as mean ± S.D. (n=3). Error bars in the graphs (Fig. 7B, 8B, 9B, 10B) indicate standard deviation (S.D.) of the mean. For statistical analysis of the data, Student’s t test was used. All statistical comparisons made are with respect to the controls. Statistically significant results were defined as those with a p-value of less than 0.05. The statistical package SAS (version 9.0) (SAS Institute, Inc., Cary, NC) was used to analyze the data.
3. Results
3.1. Cloning and analysis of HvMnSOD cDNA
Comparative analysis of amino acid sequences of MnSODs from several species revealed that stretches of amino acids are highly conserved among them and multiple sequence alignment (Fig. 1) led to the identification two stretches of conserved amino acids: FNGGGHIN and VWEHAYY. A pair of degenerate primers (DF and DR) was designed corresponding to these two conserved stretches of amino acids (Table 1) and was used in RT-PCR experiments to amplify a segment of HvMnSOD. The amplified cDNA product was subcloned and sequenced. Initial sequence analysis of the cloned fragment indicated the presence of an open reading frame (ORF) encoding a polypeptide with a high degree of similarity to MnSOD of many organisms.
The full-length cDNA sequence encoding the putative HvMnSOD was cloned employing the 5'-/3'-RACE methods using gene specific primers designed from the above cloned segment (Table 1). To confirm that the cDNA and PCR cloning products are indeed from the same gene, the full-length ORFs are cloned by PCR and sequenced (data not shown). The nucleotide and the predicted amino acid sequences of the HvMnSOD cDNA are shown in Fig. 2. The putative transcription initiation site found by 5'-RACE is located at nucleotide (nt) position 1 (Fig. 2). The initiation site of translation was placed at nt 61, inferred by conceptual translation of the cDNA sequence in all three reading frames and alignment with the known sequences of MnSOD proteins available in the GenBank database. The putative HvMnSOD cDNA (Fig. 2) is found to contain a 657 bp ORF and an in-frame TGA stop codon at the 3'-end of the coding region. The ORF is flanked a 117 bp 3'-untranslated region followed by the putative poly(A) tail. Three polyadenylation consensus sequences (5'-AATAA-3') are located between the stop codon and the poly(A) tail. The cDNA sequence also contained a splice leader (nt 7 to 52) in the 5'-untranslated region and belonged to the splice leader B (SL-B) category. The H. vulgaris SL sequence was 100 % identical to SL sequences of H. vulgaris mRNA for cAMP response element binding protein (X83872) (E < 10-16) or H. littoralis Pax-A mRNA (U96193) (E < 10-16) or H. vulgaris 5S ribosomal RNA gene (AF217320) (E < 10-16).
As shown in Fig. 2, the deduced protein is composed of 219 amino acid residues. The theoretical isoelectric point (pI) and molecular weight (Mw) of the protein sequence are calculated to be 9.02 and 24348.75 Da, respectively. The predicted amino acid sequence of the HvMnSOD gene exhibited the characteristic motifs of the MnSOD family. The consensus sequence DXWEH observed in MnSODs is located between the above mentioned D-181 and H-185 and is identified as DVWEH in HvMnSOD. Also the sequence LPEL, resembling the peptide LPDL conserved at the N-terminal region in most of eukaryotic MnSODs, is present in HvMnSOD. Four residues known to be involved in metal binding in Mn or Fe SODs are found in the HvMnSOD amino acid sequence (H-48, H-96, D-181 and H-185) (Fig. 2, Fig. 3). A single potential N-glycosylation site (NXS/T) (p=0.5342) is also found at N-95 (NHS) in HvMnSOD (Fig. 2). A putative mitochondrial transit peptide consisting of residues 1-21 (MFSFGIHRLSVFRKISRIAFA) is also identified in HvMnSOD by program Signal 3.
Analysis of residues 1-21 of the HvMnSOD mitochondrial transit peptide showed that residues 9-15 (LSVFRKI) likely to form an α-helix. An axial projection of amino acid residues 9-15 on a helical wheel showed that the helix forms an amphiphilic structure. Hydrophobic residues (L-9, V-11, F-12, I-15) are clustered on one side of the helix, and a polar residue (ser-10) along with two positively charged (R-13, K-14) residues are found on the opposite side of the helix.
The HvMnSOD deduced amino acid sequence was aligned by the ClustalW method with several Mn- and Fe-SODs (data not shown) and an alignment of HvMnSOD with representative MnSODs is shown in Fig. 1. The absolutely conserved residues in all these sequences are (numbering is based on HvMnSOD amino acid sequence) H-48, Y-56, H-96, D-181, W-183, E-184, H-185 and Y-188. HvMnSOD protein sequence when compared to the proteins of GenBank database using the program BLAST gave significant similarity scores with MnSODs of Callinectes sapidus (82 %), Biomphalaria glabrata (82 %), Xenopus laevis (82 %), Homo sapiens (81 %), Rattus norvegicus (80 %), Mus musculus (78 %), C. elegans (78 %), D. melanogaster (76 %); etc.), and FeSODs of Propionibacterium shermanii (70 %), Cinnamomum camphora (68 %), Tetrahymena pyriformis (67 %), Mycobacterium tuberculosis (67 %); etc. Also HvMnSOD protein sequence when compared (using BLAST) to the deduced protein sequence of an EST reported from H. magnipapillata (accession number gi|57833081) coding for a MnSOD gave a significant similarity score (98 %).
So, identification of DVWEH and LPEL motif, mitochondria transit peptide, conserved metal binding residues and homology to other MnSOD proteins strongly suggested that this protein is a MnSOD. Overall, these results suggested that the putative HvMnSOD of the H. vulgaris possessed the essential properties of MnSOD family.
3.2. Cloning and analysis of HvEC-SOD cDNA
Prior to cloning of the HvEC-SOD, a number of CuZnSODs from diverse organisms were aligned. Comparative analysis of amino acid sequences of CuZnSODs from several species showed that stretches of amino acids are highly conserved among CuZnSODs (Fig. 4). Also, prior observations suggest that proteins between H. magnipapillata and H. vulgaris share high sequence similarity. Therefore, a pair of primers (F1 and R1) was designed that corresponded to the stretches of conserved amino acids (HGFHIH and DDLGKA) among several CuZnSODs (also the same conserved residues are identified in an EST encoding CuZnSOD from H. magnipapillata). Primers F1 and R1 were used in RT-PCR experiments to amplify a segment of HvEC-SOD and the amplified cDNA product was subcloned and sequenced. Initial sequence analysis indicated that the presence of an ORF encoding a polypeptide with a high degree of similarity to CuZnSOD of many organisms.
The full-length cDNA sequence encoding the putative HvEC-SOD was cloned using the 5'-/3'-RACE method using gene specific primers designed from the above cloned segment (Table 1). To confirm that the cDNA and PCR cloning products are indeed from the same gene, the full-length ORFs are cloned by PCR and sequenced (data not shown). The putative transcription initiation site found by 5'-RACE is located at nucleotide (nt) position 1 (Fig. 5). The initiation site of translation was placed at nt 141, inferred by conceptual translation of the sequence in all three reading frames and alignment with the known sequences of CuZnSOD and EC-SOD proteins available in the GenBank database. The putative HvEC-SOD cDNA (Fig. 5) is shown to contain a 567 bp ORF and an in-frame TGA stop codon at the 3'-end of the coding region. The ORF is flanked by a 218 bp 3'-untranslated region followed by the putative poly(A) tail.
The predicted amino acid sequence of the HvEC-SOD cDNA is shown in Fig. 5. The deduced protein is composed of 189 amino acid residues. The theoretical isoelectric point (pI) and molecular weight (Mw) of the protein are calculated to be 6.22 and 20959.73 Da, respectively. An N-glycosylation site is predicted on residue 143 (NYSV) (p= 0.7296). The predicted HvEC-SOD amino acid sequence exhibited the characteristic copper zinc superoxide dismutase signature GNAGpRiACgiI (residues 172-183) of the CuZnSOD family. Seven residues known to be involved in metal binding in CuZnSODs are also found in the HvEC-SOD amino acid sequence. The residues required for coordinating copper are identified as: H-80, -82, -97, and -154 and for zinc are H-97, -114, -105 and D-117 with H-97 being the common residue for coordination. The same residues are identified as H-58, -60, -75, and 132 for catalytic copper and H-75, -83, -92 and D-95 for ligand zinc in Fig. 6. Also the residues responsible for oligomerization of CuZnSODs are identified by multiple sequence alignment as G-71, V-72, H-77, C-91, R-113, G-119, I-178 and C-180. The arginine residue likely involved in the positioning of the substrate (superoxide) occurs at position 177. The two cysteines believed to be form a disulfide bond are: C-91 and C-180. Analysis of protein sequence (by Signal 3 and SMART) showed that residues 1-19 of the HvEC-SOD might form a signal peptide (p=0.933).
The HvEC-SOD deduced amino acid sequence was aligned by the ClustalW method with several CuZn- and EC-SODs (data not shown) and an alignment of HvEC-SOD with representative CuZnSODs is shown in Fig. 4. The absolutely conserved residues in these sequences are (numbering based on H. vulgaris amino acid sequence): G-71, H-80, H-82, C-91, H-97, N-99, P-100, H-105, H-114, G-116, D-117, N-120, L-140, G-148, V-152, H-154, D-158, D-159, L-160, G-161, S-168, G-172, A-174, G-175, R-177 and C-190. The HvEC-SOD protein when compared to the proteins of GenBank database using the BLAST program gave significant similarity scores with intracellular CuZnSODs of Biomphalaria glabrata (71 %), Lymnaea stagnalis (72 %), Caretta caretta (74 %), Cavia porcellus (71 %), Macaca mulatta (73 %), Canis familiaris (71 %), Mus musculus (71 %), Equus caballus (69 %), A viridis (70%), etc.; and extracellular CuZnSODs of Acanthocheilonema viteae (61 %), Brugia pahangi (64 %), Lasius niger (64 %), Onchocerca volvulus (65 %), A. viridis (67%); etc. Also HvEC-SOD protein sequence when compared (using BLAST) to the deduced protein sequences of ESTs reported from H. magnipapillata (gi|57833081) and A. paessleri (AAG10392) coding for CuZnSODs gave a significant similarity score of 97 and 72 % respectively.
Hence identification of copper zinc superoxide dismutase signature GNAGpRiACgiI (residues 172-183), presence of signal peptide and conserved metal binding, and homology to other CuZn- and EC-SOD proteins strongly suggested that this protein is an extracellular SOD. Overall, these results suggested that the putative HvEC-SOD of the H. vulgaris identified possessed the essential properties of an EC-SOD and can be classified accordingly.
3.3. The structural models of HvMnSOD and HvEC-SOD
By threading analysis it was found that the human manganese superoxide dismutase (PDB code: 1LUV) and human copper zinc superoxide dismutase (PDB code: 1N18.pdb) were the best templates for homologous modeling of the HvMnSOD and HvEC-SOD target proteins respectively. The respective target and template proteins shared a high degree of homology across their entire length (Fig. 1, Fig. 4)
The comparative stereo-chemical analysis of the φ-ψ plots (Ramachandran diagram) of the model and template (HvMnSOD vs 1LUV: A; HvEC-SOD vs 1N18: A) is shown in Table 2. The analysis for the HvMnSOD model (Table 2) presents 90.6 % of residues in the most favorable, 8.8 % of residues in additional allowed regions, 0.6 % of the residues in generously allowed regions, and 0.0 % of the residues in disallowed regions. Similarly the analysis for the HvEC-SOD model (Table 2) presents 86.1 % of residues in the most favorable, 12.9 % of residues in additional allowed regions, 1.0 % of the residues in generously allowed regions, and 0.0 % of the residues in disallowed regions. These results indicated that the molecular models presented here have good overall stereo-chemical qualities. A Verify-3D run on each model also showed a good stereo-chemical quality of the models (data not shown).
Table 2.
Comparison of the Ramachandran Plot Statistics between the Model and the Template
| Model/Template | Region of the Ramachandran plot (%) | |||
|---|---|---|---|---|
| Most favorable | Additional allowed | Generously allowed | Disallowed | |
| Model HvMnSOD vs Template | 1LUV:A | |||
| HvMnSOD | 90.6 | 8.8 | 0.6 | 0.0 |
| 1LUV:A | 91.0 | 8.1 | 0.9 | 0.0 |
| Model HvEC-SOD vs Template | 1N18 | |||
| HvEC-SOD | 86.1 | 12.9 | 1.0 | 0.0 |
| 1N18 | 88.8 | 11.2 | 0.0 | 0.0 |
The HvMnSOD model (Fig. 3) displayed active sites having conserved metal-binding residues (H-48, H-96, D-181, and H-185) including shells of aromatic residues (Y-31, Y-33, Y-56, F-88, F-99, W-100, W-145, W-147, W-183, Y-187, Y-188, Y-191, Y-198, F-203) that surrounded the active-site metal and its ligands. N-terminal domain is a long alpha antiparallel hairpin and the C-terminal domain is a mixed alpha/beta fold (Fig. 3). Characteristically the model generated for HvEC-SOD (Fig. 6) displayed a six-stranded beta sandwich (conserved β-barrel fold). The HvEC-SOD model displayed active sites having conserved metal-binding residues: H-58, H-60, H-132 (green colored residues) and H-75 for copper; and H-83, H-92, D-95 (cyan colored residue) and H-75 (red colored residue) zinc. The HvEC-SOD model also revealed clear evolutionary conservation of the active-site and β-barrel fold of the hydra enzyme with the much-studied eukaryotic (E-class) intracellular CuZnSODs.
3.4. HvMnSOD and HvEC-SOD mRNA expression analysis
The expression patterns of the HvMnSOD and HvEC-SOD mRNA before and after stressor exposure were investigated in whole organisms by RT-PCR experiments (Fig. 7-10). For RT-PCR gene-specific primers were used. The results obtained demonstrate the variable effects of different treatments on HvMnSOD and HvEC-SOD mRNA expression, though transcription of the actin DNA was almost constant before and after exposure to stressors.
The expression HvMnSOD mRNA was induced when hydrae were exposed to Zn (II) (p<0.001) and As (V) (p<0.039) at the tested concentrations for 1 h (Fig. 7A and 7B) whereas exposure to Cu (II) (p<0.001), Zn (II) (p<0.001), Cr (VI) (p<0.001), As (III) (p<0.043) and As (V) (p<0.001) at the tested concentrations for 6 h increased the HvMnSOD mRNA expression (Fig. 8A and 8B). Six-hour exposure to Cu (II), Zn (II), Cd (II), Cr (IV), As (III) and As (V) (Fig. 8A and 8B) enhanced the expression of HvMnSOD mRNA.
One-hour exposure of hydrae to Zn (II) (p<0.007) (Fig. 9A and 9B) induced the expression of HvEC-SOD mRNA where as Cr (VI) (p<0.038) and Se (IV) (p<0.011) treatments suppressed the expression of HvEC-SOD mRNA. When hydrae were exposed to the metal toxicants for 6 h (Fig. 10A and 10B) Cu (II) (p<0.001), Zn (II) (p<0.001), Cd (II) (p<0.001), Cr (IV) (p<0.001), As (III) (p<0.001) and As (V) (p<0.001) induced the expression of HvEC-SOD mRNA at the treated concentrations.
Exposure to sodium azide (1.538 mM) and paraquat (0.38mM) for both 1- and 6-h (Fig. 7A and 7B; Fig. 8A and 8B) didn’t affect the expression of HvMnSOD mRNA. One-hour exposure of hydrae to H2O2 (9.79 mM) (p<0.027) (Fig. 7A and 7B) induced HvMnSOD mRNA expression.
Exposure of hydrae to 0.38 mM paraquat for 1 or 6 h treatment didn’t affect the expression of EC-SOD mRNA (Fig. 9A and 9B; Fig 10A and 10B). One-hour exposure of hydrae to 9.79 mM H2O2 (p<0.024) (Fig. 9A and 9B) induced HvEC-SOD mRNA expression.
Exposure of hydrae to temperatures of both 30 (p<0.005) and 37 (p<0.003) °C for 1 h (Fig. 8A and 8B) induced the MnSOD mRNA expression although 6 h exposure to 30 (p<0.011) °C reduced MnSOD mRNA expression (Fig. 8A and 8B) as compared to 18 °C (control). However, HvMnSOD mRNA expression was drastically reduced when animals were exposed to 37 °C for 6 h (Fig. 8A and 8B); and hence its expression couldn’t be quantified.
Similarly, thermal stress for 1 h at 30 (p<0.001) and 37 (p<0.001) °C (Fig. 10A and 10B) enhanced the expression of HvEC-SOD mRNA as compared to 18 °C although thermal stress for 6 h at 30 (p<0.027) °C reduced the expression of HvEC-SOD mRNA as compared to control at 18 °C. However, HvEC-SOD mRNA expression was drastically reduced when animals were exposed to 37 °C for 6 h (Fig. 10A and 10B). Enhanced expression of HvEC-SOD mRNA was observed both at 18 (p<0.028) and 30 (p<0.001) °C for hydra starved for 5 days as compared to the control (Fig. 10A and 10B).
4. DISCUSSION
Two superoxide dismutase (SOD) genes are cloned and characterized from H. vulgaris in this study. Heterologously designed degenerate primers combined with 5'-/3'-RACE experiments are used to clone full-length cDNAs of HvMnSOD and HvEC-SOD. To confirm that the cDNA and PCR cloning products are indeed from the same gene, the full-length ORFs are cloned by PCR and sequenced (data not shown).
Superoxide dismuatses (SODs) are the first line of defense against toxic intracellular radicals produced during normal cellular metabolism or under oxidative stress (Fridovich, 1995). MnSODs located in the mitochondrial matrix scavenge superoxide anions and, typically, active as homotetramers (Hearn et al., 2003). The finding of a putative mitochondrial transit peptide in HvMnSOD like most eukaryotic MnSODs entails its localization to mitochondria and similar role.
Although mitochondrial transit peptides from MnSODs are not very similar, targeting sequences are generally rich in positively charged and hydroxylated amino acids and lack acidic amino acids (Roise et al., 1988; Hartl and Neupert, 1990). The hydra MnSOD transit peptide contained hydrophobic residues (2 ala, 4 phe, 3 ile, 1 leu, 1 met, 1 val), basic residues (1 his, 1 lys,, 3 arg); and polar residues (1 gly, 3 ser). It lacked acidic residues. It also possessed the amphiphilic structural characteristics typical of mitochondrial-targeting sequences where residues 9-15 (LSVFRKI) are likely to form an α helix; hydrophobic residues (L-9, V-11, F-12, I-15) one face of the helix and a polar residue (ser-10) along with two positively charged (arg-13, lys-14) residues on the other face of the helix.
Messenger RNAs have been identified that can receive either splice leader A or B (SL-A or -B), although the impact of the two different SLs on the function of the mRNA is not known (Stover and Steele, 2001). To date, SL addition has been identified in four metazoan phyla (Nematoda, Platyhelminthes, Chordata and Cnidaria) and in one unicellular eukaryotic phylum (Sarcomastigophora). No evidence of SL addition has been detected in any intensively studied plants, fungi, insects, echinoderms, or vertebrates (Stover and Steele, 2001). So the presence of SL-B addition in the HvMnSOD is another addition to the repertoire of mRNAs that receive splice leaders in cnidaria.
Until now, EC-SODs have been identified only in platyhelminths (Hong et al., 1992), nematodes (Fujii et al., 1998), arthropods (Johansson et al., 1999; Brouwer at al., 2003), mammals (Petersen et al., 2004) and cnidarians (Plantivaux et al., 2004). Therefore HvEC-SOD would be the first EC-SOD described in hydra (hydrozoans). The presence of a putative glycosylation site is another characteristic of an EC-SOD (Halliwell and Gutteridge, 1999), has also been found in HvEC-SOD sequence. Consequently, this extracellular enzyme could be released into the coelenteric cavity and/or be trapped within the mucus produced by hydra. A similar role has been suggested for EC-SOD in A. viridis (Plantivaux et al., 2004). It could also be bound to the cell surface as found in the crustacean Pacifastacus leniusculus (Johansson et al., 1999) or associated with the collagen present in the mesoglea, reducing its oxidative fragmentation, as demonstrated for human EC-SODs (Petersen et al., 2004).
Crystallographic studies with human MnSOD and E. coli FeSOD have shown that metal-binding sites of these two enzymes are located at three histidine residues and one aspartate residue (Carlioz et al., 1988; Borgstahl et al., 1992). The corresponding residues in H. vulgaris MnSOD are identified as H-48, H-96, D-181 and H-185 (Fig. 3). These residues, as well as the sequence DXWEH including D-181 and H-185, are conserved in all MnSODs so far described (Fig. 1). Also potential N-glycosylation site, NXS/T, located at H-96 in HvMnSOD is also conserved in all other eukaryotic species except S. cerevisiae (Ditlow et al., 1982). The structural model of H. vulgaris EC-SOD revealed structural near-identity and clear evolutionary conservation of the active-site and β-barrel fold of the hydra enzyme with the eukaryotic (E-class) intracellular CuZnSODs, exemplified in bovine, yeast, human and frog (Tainer et al., 1982; Ogihara et al., 1996). HvEC-SOD has metal binding residues in the same sequence and structural order. The catalytic mechanism of CuZnSOD, established by numerous structural and spectroscopic studies (Tainer et al., 1982; Hart et al., 1999) requires the successive reduction of tetrahedral Cu (II) to trigonal planar Cu (I) by superoxide, forming molecular oxygen, followed by re-oxidation of Cu (I) to Cu (II) by a second superoxide and 2H+, forming hydrogen peroxide. This transition involves the breaking and reforming of a bond from the solvent-accessible copper to the bridging histidine residue (H-75), which is also a ligand for the buried active-site zinc. The bridging histidine residue (H-75) and five additional histidine residues and one aspartate residue complete the structurally conserved active-site of hydra enzyme HvEC-SOD (Fig. 6).
Superoxide dismutases have been proposed as a possible tool for biomonitoring environmental toxicants and stress (Niyogi et al., 2001, Li et al., 2003; Richier et al., 2003; Zhang et al., 2004). Their differential expression as compared to normal expression pattern indicates exposure to cellular stress and adverse cellular effects, thus serving as biomarkers of these effects. Many studies have identified the enzymatic activities and expression of MnSOD protein upon exposure to stressors and metals. MnSODs are inducible enzymes and its expression is regulated by many agents, including tumor necrosis factor, in terleukin-1, interleukin-6, lipopolysaccharide, phorbol esters and inhibitors of protein synthesis (Visner et al., 1990; Fujii and Taniguchi, 1991; Fujii et al., 1994).
Few studies conducted on other organisms other than mammals are available; and results observed in the current study are comparable to them if a direct relation between transcriptional activity and enzymatic activity is assumed which is often the case unless other regulatory mechanisms such as reduced mRNA stability, translational feed back inhibition; etc. exist and are important.
To test whether the two superoxide dismutases identified in the current study behaved in a similar way, hydrae were exposed to a variety of toxicants and treatments and their mRNA expression was studied by RT-PCR using actin gene as a control. Although induction of both superoxide dismutases mRNA was detectable over a temperature range of 30-37 °C, the level of synthesis depended on the stress temperature and exposure duration.
Lee and Gu (2003) used a recombinant bioluminescent Escherichia coli strain, (sodA::luxCDABE), containing the promoter for the manganese superoxide dismutase (sodA) gene fused to the Vibrio fischeri luxCDABE operon, and characterized with regard to redox-cycling agents, such as paraquat and chromium. Both strongly induced a sodA-regulated response in dose-dependent manners, resulting in an increase of the bioluminescence. Similarly, Rohrdanz and Kahl (1998) studied the effect of hydrogen peroxide (H2O2) on the expression of different antioxidant enzymes in primary rat hepatocytes and showed that MnSOD mRNA levels were induced after exposure to H2O2. Similar observations are made in the current study that H2O2 (exposure to 9.79 mM for 1 h) and chromium (exposure to 0.33 mM for 6 h) induced the expression of HvMnSOD mRNA. As expected, hydra responded to oxidative stress by inducing MnSOD mRNA. Similar mechanisms and response that protects organisms from deleterious effects of reactive oxygen species (ROS) formed due to exposure to oxidants or as a natural consequence of metabolism are well conserved in prokaryotes and eukaryotes (Angelova et al., 2005). Metals are also known to influence the oxidative status of organisms, and antioxidant enzymes have been often proposed as biomarkers of effect (Geret et al., 2002).
Morales et al. (2004) observed in fish (Dentex dentex) liver that prolonged starvation (for 5 weeks) increased superoxide dismutase (SOD) activity and concluded that prolonged starvation leads to a pro-oxidant situation and oxidative stress. Similarly in this study it is observed that HvECSOD mRNA levels are increased due to starvation.
Downs et al. (2001) demonstrated that CuZnSOD activity was 2.3 times greater when the tropical coral Montastraea faveolata was heat-stressed than in controls. Similarly, Brown et al. (2002) showed a higher increase in CuZnSOD activity after a thermal stress on another tropical coral, Goniastrea aspera. Induction of CuZnSOD has also been observed in other invertebrates. For example, an inducible isoform of CuZnSOD from the blue mussel, Mytilus edulis, has been shown to be overexpressed in mussels collected in polluted sites or exposed to copper (Manduzio et al., 2003). The total lack of expression of EC-SOD when exposed to 37 °C for 6 h may be due to that the biochemical machinery at the base of the oxidative stress response is compromised.
Atsushi et al. (2002) analyzed the expression of CuZnSOD and ECSOD genes in 0.5 mM H2O2-treated D. discoideum cells by RT-PCR. The expression levels of CuZnSOD and EC-SOD increased (three-fold) following the treatment. Results showed that the gene for CuZnSOD was induced by H2O2 exposure but not by UV irradiation, whereas gene for EC-SOD was induced both by H2O2 exposure and UV irradiation. Similarly, Stralin and Marklund (1994) exposed two fibroblast lines to a wide concentration range of oxidizing agents (xanthine oxidase plus hypoxanthine, paraquat, pyrogallol, alpha-naphthoflavone, 2+ ions, Cu2+ ions, buthionine sulphoximine, hydroquinone, catechol, Fe diethylmaleate, t-butyl hydroperoxide, cumene hydroperoxide, selenite, citiolone and high oxygen partial pressure) for periods of up to 4 days in order to determine the effect of oxidative stress on expression of extracellular superoxide dismutase and other SODs (EC-SOD). Under no condition was there any evidence of EC-SOD induction. Instead, the agents uniformly, dose-dependently and continuously reduced EC-SOD expression. Stralin and Marklund (1994) indicated that the effect may have been due to toxicity. Increased expression of EC-SOD in Hydra vulgaris under certain treatments like Zn, Cd, Cr, etc. suggests that one function of the HvEC-SOD is to shield the hydra against exogenous superoxides.
The concentration of some of the toxicants chosen was based on previous scientific reports on whole organism based bioassay reports in hydra (Beach et al., 1998; Pollino and Holdway, 1999; Holdway et al., 2001; Karntanut and Pascoe, 2002). In the context of known inducers (metals and oxidative stressors) not provoking a stress response especially in the case of HvEC-SOD at the tested concentration offers few possibilities. It may be due to that the doses tested couldn’t provoke a response or the doses tested cause toxicity impairing the transcriptional ability of H. vulgaris. In order to draw firm conclusions regarding the expression of HvMnSOD and HvEC-SOD under metal and oxidative stress quantitative dose-response studies are being planned. Also the preliminary gene expression data presented here gives further basis to conduct dose-response and dose-equivalent studies to elucidate contaminant specific gene expression and mechanistic basis of HvMnSOD and HvEC-SOD expression in hydra. The expression pattern of HvMnSOD when compared to that of HvEC-SOD show that HvEC-SOD may not be very well associated with environmental contaminant exposure or the doses tested and duration of exposure wasn’t sufficient enough to provoke a response or there exist entirely different mechanism of HvEC-SOD gene expression.
Thus HvMnSOD mRNA is highly inducible under stress and exposure to toxicants; and the deduced protein sequence has similar topological features and domains of higher model organisms. These features of HvMnSOD combined with the ease of conducting hydra bioassay offer an opportunity to use H. vulgaris as a model system to study the pro-oxidant properties of chemicals and environmental toxicants. Based on the evidence provided here, further study is warranted to determine how hydra regulates its EC-SOD expression with respect to tissue specificity, regulatory elements, and both intrinsic and extrinsic stimuli. Further research should also focus on whether a CuZnSOD exist in hydra and how it would compare with its EC-form.
Acknowledgements
This work is funded by USAID TAM50 and NIEHS P42-ES04917.
Abbreviations:
- H.
Hydra
- Hv
Hydra vulgaris
- cDNA
DNA complementary to RNA
- PCR
polymerase chain reaction
- RT-PCR
reverse transcription-polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- ORF
open reading frame
- UTR
untranslated region
- nt
nucleotide
- aa
amino acid
- bp
base pairs
- kDa
kilodalton(s)
- dNTP
deoxyribonucleoside triphosphate
- EDTA
ethylenediaminetetraacetic acid
- ppm
parts per million
- Cu
copper
- Zn
zinc
- Cd
cadmium
- Cr
chromium
- As
arsenic
- Se
selenium
- Na
sodium
- H2O2
hydrogen peroxide
- EST
expressed sequence tag
- SL1
trans spliced leader
- SOD
superoxide dismutase(s)
- SOD
gene(s), cDNA or mRNA encoding SOD
- MnSOD
manganese SOD
- FeSOD
iron SOD
- Cu/ZnSOD
copper/zinc SOD
- EC-SOD
extracellular SOD
- MnSOD
gene, cDNA or mRNA encoding manganese SOD
- FeSOD
gene, cDNA or mRNA encoding iron SOD
- Cu/ZnSOD
gene cDNA or mRNA encoding copper/zinc SOD
- EC-SOD
gene, cDNA or mRNA encoding extracellular SOD
- ROS
reactive oxygen species
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angelova MB, Pashova SB, Spasova BK, Vassilev SV, Slokoska LS. Oxidative stress response of filamentous fungi induced by hydrogen peroxide and paraquat. Mycol. Res. 2005;109:150–158. doi: 10.1017/s0953756204001352. [DOI] [PubMed] [Google Scholar]
- Atsushi T, Yuya A, Ken-Ichi K, Hiroo Y. Copper/zinc superoxide dismutases in Dictyostelium discoideum: Amino acid sequences and expression kinetics. J. Biochem. Mol. Biol. Biophys. 2002;6:215–220. doi: 10.1080/10258140290018711. [DOI] [PubMed] [Google Scholar]
- Bannister JV, Bannister WH, Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Crit. Rev. Biochem. 1987;22:111–180. doi: 10.3109/10409238709083738. [DOI] [PubMed] [Google Scholar]
- Beach MJ, Pascoe D. The role of Hydra vulgaris (Pallas) in assessing the toxicity of fresh water pollutants. Water Res. 1998;32:101–106. [Google Scholar]
- Borgstahl GEO, Parge HE, Hickey MJ, Beyer WF, Hallewell RA, Tainer TA. The structure of human mitochondrial manganese superoxide dismutase reveals a novel tetrameric interface of two 4-helix bundles. Cell. 1992;71:107–118. doi: 10.1016/0092-8674(92)90270-m. [DOI] [PubMed] [Google Scholar]
- Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Brouwer M, Hoexum BT, Grater W, Brown-Peterson N. Replacement of a cytosolic copper/zinc superoxide dismutase by a novel cytosolic manganese superoxide dismutase in crustaceans that use copper haemocyanin for oxygen transport. Biochem. J. 2003;374:219–228. doi: 10.1042/BJ20030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BE, Downs CA, Dunne RP, Gibb SW. Exploring the basis of thermotolerance in the reef coral Goniastrea aspera. Mar. Ecol. Prog. Ser. 2002;242:119–129. [Google Scholar]
- Campbell WS, Laudenbach DE. Characterization of four superoxide dismutase genes from a filamentous cyanobacterium. J. Bacteriol. 1995;177:964–972. doi: 10.1128/jb.177.4.964-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso RMF, Thayer MM, Didonato M, Lo TP, Bruns CK, Getzoff ED, Tainer JA. Insights into Lou Gehrig’s disease from the structure and instability of the A4V mutant of Cu, Zn superoxide dismutase. J. Mol. Biol. 2002;324:247–256. doi: 10.1016/s0022-2836(02)01090-2. [DOI] [PubMed] [Google Scholar]
- Carlioz A, Ludwig ML, Stallings WC, Fee JA, Steinman HM, Touati D. Iron superoxide dismutase: Nucleotide sequence of the gene from Escherichia coli K12 and correlations with crystal structures. J. Biol. Chem. 1998;263:1555–1562. [PubMed] [Google Scholar]
- Ditlow C, Johansen JT, Martin BM, Svendsen I. The complete amino acid sequence of manganese-superoxide dismutase from Saccharomyces cerevisiae. Carlsberg Res. Commun. 1982;47:81–91. [Google Scholar]
- Downs CA, Fauth JE, Halas JC, Dustan P, Bemiss J, Woodley CM. Oxidative stress and seasonal coral bleaching. Free Radic. Biol. Med. 2002;33:533–543. doi: 10.1016/s0891-5849(02)00907-3. [DOI] [PubMed] [Google Scholar]
- Downs CA, Mueller E, Phillips S, Fauth JE, Woodley CM. A molecular biomarker system for assessing the health of coral Montastraea faveolata during heat stress. Mar. Biotechnol. 2001;2:533–544. doi: 10.1007/s101260000038. [DOI] [PubMed] [Google Scholar]
- Dykens JA, Shick JM. Oxygen production by endosymbiotic algae controls superoxide dismutase activity in their animal host. Nature. 1982;297:579–585. [Google Scholar]
- Dykens JA, Shick JM. Photobiology of the symbiotic sea anemone, Anthopleura elegantissima defenses against photodynamic effects, and seasonal photoacclimatization. Biol. Bull. 1984;167:683–697. doi: 10.2307/1541419. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- Fujii J, Nakata T, Miyoshi E, Ikeda Y, Taniguchi N. Induction of manganese superoxide dismutase mRNA by okadaic acid and protein synthesis inhibitors. Biochem. J. 1994;301:31–34. doi: 10.1042/bj3010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii J, Taniguchi N. Phorbol ester induces manganese-superoxide dismutase in tumor necrosis factor-resistant cells. J. Biol. Chem. 1991;266:23142–23146. [PubMed] [Google Scholar]
- Fujii M, Ishii N, Joguchi A, Yasuda K, Ayusawa D. A novel superoxide dismutase gene encoding membrane-bound and extracellular isoforms by alternative splicing in Caenorhabditis elegans. DNA Res. 1998;5:25–30. doi: 10.1093/dnares/5.1.25. [DOI] [PubMed] [Google Scholar]
- Geret F, Serafim A, Barreira L, Bebianno MJ. Effect of cadmium on antioxidant enzyme activities and lipid peroxidation in the gills of the clam Ruditapes decussatus. Biomarkers. 2002;7:242–256. doi: 10.1080/13547500210125040. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford University Press; Oxford, England: 1999. [Google Scholar]
- Hart PJ, Balbirnie MM, Ogihara NL, Nersissian AM, Weiss MS, Valentine JS, Eisenberg D. A structure-based mechanism for copper-zinc superoxide dismutase. Biochemistry. 1999;38:2167–2178. doi: 10.1021/bi982284u. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Neupert W. Protein sorting to mitochondria: Evolutionary conservations of folding and assembly. Science. 1990;247:930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- Hearn AS, Stroupe ME, Cabelli DE, Ramilo CA, Luba JP, Tainer JA, Nick HS, Silverman DN. Catalytic and structural effects of amino acid substitution at histidine 30 in human manganese superoxide dismutase: Insertion of valine C gamma into the substrate access channel. Biochemistry. 2003;42:2781–2789. doi: 10.1021/bi0266481. [DOI] [PubMed] [Google Scholar]
- Henkle-Duhrsen K, Tawe W, Warnecke C, Walter RD. Characterization of the manganese superoxide dismutase cDNA and gene from the human parasite Onchocerca volvulvus. Biochem. J. 1995;308:441–446. doi: 10.1042/bj3080441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjalmarsson K, Marklund SL, Engström A, Edlund T. Isolation and sequence of complementary DNA encoding human extracellular superoxide dismutase. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6340–6344. doi: 10.1073/pnas.84.18.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdway DA, Lok K, Semaan M. The acute and chronic toxicity of cadmium and zinc to two hydra species. Environ. Toxicol. 2001;16:557–565. 2001. [PubMed] [Google Scholar]
- Hong Z, Kosman DJ, Thakur A, Rekosh D, LoVerde PT. Identification and purification of a second form of Cu/Zn superoxide dismutase from Schistosoma mansoni. Infect. Immun. 1992;60:3641–3651. doi: 10.1128/iai.60.9.3641-3651.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MW, Holmblad T, Thornqvist PO, Cammarata M, Parrinello N, Soderhall K. A cell-surface superoxide dismutase is a binding protein for peroxinectin, a cell-adhesive peroxidase in crayfish. J. Cell Sci. 1999;112:917–925. doi: 10.1242/jcs.112.6.917. [DOI] [PubMed] [Google Scholar]
- Johnson EM, Gorman RM, Gabel BE, George ME. The Hydra attenuata system for detection of teratogenic hazards. Teratog. Carcinog. Mutagen. 1982;2:263–276. doi: 10.1002/1520-6866(1990)2:3/4<263::aid-tcm1770020308>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Jones PL, Kucera G, Gordon H, Boss JM. Cloning and characterization of the murine manganous superoxide dismutase-encoding gene. Gene. 1995;153:155–161. doi: 10.1016/0378-1119(94)00666-g. [DOI] [PubMed] [Google Scholar]
- Karntanut W, Pascoe D. The toxicity of copper, cadmium and zinc to four different Hydra (Cnidaria. Hydrozoa) Chemosphere. 2002;47:1059–1064. doi: 10.1016/s0045-6535(02)00050-4. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthurm MW, Smith DK, Jones DT, Hutchinson EG, Morris AL, Naylor D, Moss DS, Thornton JM. PROCHECK v.3.0-Program to check the stereochemistry quality of protein structures-Operating instructions. 1994. [Google Scholar]
- Lee HJ, Gu MB. Construction of a soda:: luxCDABE fusion Escherichia coli: Comparison with a katG fusion strain through their responses to oxidative stresses. Appl. Microbiol. Biotechnol. 2003;60:577–580. doi: 10.1007/s00253-002-1168-4. [DOI] [PubMed] [Google Scholar]
- Li WM, Yin DQ, Zhou Y, Hu SQ, Wang LS. 3,4-dichloroaniline-induced oxidative stress in liver of crucian carp (Carassius auratus) Ecotoxicol. Environ. Saf. 2003;56:251–255. doi: 10.1016/s0147-6513(02)00117-3. [DOI] [PubMed] [Google Scholar]
- Liddell S, Knox DP. Extracellular and cytoplasmic Cu/Zn superoxide dismutases from Haemonchus contortus. Parasitology. 1998;116:383–394. doi: 10.1017/s0031182098002418. [DOI] [PubMed] [Google Scholar]
- Lum KT, Huebner HJ, Li Y, Phillips TD, Raushel FM. Organophosphate nerve agent toxicity in Hydra attenuata. Chem. Res. Toxicol. 2003;16:953–957. doi: 10.1021/tx034047k. [DOI] [PubMed] [Google Scholar]
- Manduzio H, Monsinjon T, Rocher B, Leboulenger F, Galap C. Characterization of an inducible isoform of the Cu/Zn superoxide dismutase in the blue mussel Mytilus edulis. Aquat. Toxicol. 2003;64:73–83. doi: 10.1016/s0166-445x(03)00026-2. [DOI] [PubMed] [Google Scholar]
- Mayura K, Smith EE, Clement BA, Phillips TD. Evaluation of the developmental toxicity of chlorinated phenols utilizing Hydra attenuata and postimplantation rat embryos in culture. Toxicol. Appl. Pharmacol. 1991;108:253–266. doi: 10.1016/0041-008x(91)90116-v. [DOI] [PubMed] [Google Scholar]
- Morales AE, Perez-Jimenez A, Hidalgo MC, Abellan E, Cardenete G. Oxidative stress and antioxidant defenses after prolonged starvation in Dentex dentex liver. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2004;139:153–161. doi: 10.1016/j.cca.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Niyogi S, Biswas S, Sarker S, Datta AG. Seasonal variation of antioxidant and biotransformation enzymes in barnacle, Balanus balanoides, and their relation with polyaromatic hydrocarbons. Mar. Environ. Res. 2001;52:13–26. doi: 10.1016/s0141-1136(00)00257-9. [DOI] [PubMed] [Google Scholar]
- Ogihara NL, Parge HE, Hart PJ, Weiss MS, Goto JJ, Crane BR, Tsang J, Slater K, Roe JA, Valentine JS, Eisenberg D, Tainer JA. Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase. Biochemistry. 1996;35:2316–2321. doi: 10.1021/bi951930b. [DOI] [PubMed] [Google Scholar]
- Petersen SV, Oury TD, Ostergaard L, Valnickova Z, Wegrzyn J, Thogersen IB, Jacobsen C, Bowler RP, Fattman CL, Crapo JD, Enghild JJ. Extracellular superoxide dismutase EC-SOD binds to type I collagen and protects against oxidative fragmentation. J. Biol. Chem. 2004;279:13705–13710. doi: 10.1074/jbc.M310217200. [DOI] [PubMed] [Google Scholar]
- Plantivaux A, Furla P, Zoccola D, Garello G, Forcioli D, Richier S, Merle PL, Tambutte E, Tambutte S, Allemand D. Molecular characterization of two CuZn-superoxide dismutases in a sea anemone. Free Radic. Biol. Med. 2004;37:1170–1181. doi: 10.1016/j.freeradbiomed.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Pollino CA, Holdway D. A. Potential of two hydra species as standard toxicity test animals. Ecotoxicol. Environ. Saf. 1999;43:309–316. doi: 10.1006/eesa.1999.1796. [DOI] [PubMed] [Google Scholar]
- Richier S, Merle PL, Furla P, Pigozzi D, Sola F, Allemand D. Characterization of superoxide dismutases in anoxia- and hyperoxia-tolerant symbiotic cnidarians. Biochim. Biophys. Acta. 2003;1621:84–91. doi: 10.1016/s0304-4165(03)00049-7. [DOI] [PubMed] [Google Scholar]
- Rodney JF, Guan J, Seldin MF, Oury TD, Enghild JJ, Crapo JD. Mouse extracellular superoxide dismutase: Primary structure, tissue-specific gene expression, chromosomal localization, and lung in situ hybridization. Am. J. Respir. Cell. Mol. Biol. 1997;17:393–403. doi: 10.1165/ajrcmb.17.4.2826. [DOI] [PubMed] [Google Scholar]
- Rohrdanz E, Kahl R. Alterations of antioxidant enzyme expression in response to hydrogen peroxide. Free Radic. Biol. Med. 1998;24:27–38. doi: 10.1016/s0891-5849(97)00159-7. [DOI] [PubMed] [Google Scholar]
- Roise D, Theiler F, Horvath SJ, Tomich JM, Richards JH, Allison DS, Schatz G. Amphiphilicity is essential for mitochondrial presequence function. EMBO J. 1988;7:649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor; New York: 1989. [Google Scholar]
- Stallings WC, Pattridge KA, Strong RK, Ludwig ML. Manganese and iron superoxide dismutases are structural homologs. J. Biol. Chem. 1984;259:10695–10699. [PubMed] [Google Scholar]
- Stover NA, Steele RE. Trans-spliced leader addition to mRNAs in a cnidarian. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5693–5698. doi: 10.1073/pnas.101049998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stralin P, Marklund SL. Effects of oxidative stress on expression of extracellular superoxide dismutase, CuZn-superoxide dismutase and Mn-superoxide dismutase in human dermal fibroblasts. Biochem. J. 1994;298:347–352. doi: 10.1042/bj2980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainer JA, Getzoff ED, Beem KM, Richardson JS, Richardson DC. Determination and analysis of the 2 Å structure of copper, zinc superoxide dismutase. J. Mol. Biol. 1982;160:181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- Tsolis RM, Baumler AJ, Heffron F. Role of Salmonella typhimurium Mn-superoxide dismutase SodA in protection against early killing by J774 macrophages. Infect. Immun. 1995;63:1739–1744. doi: 10.1128/iai.63.5.1739-1744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visner GA, Dougall WC, Wilson JM, Burr IA, Nick HS. Regulation of manganese superoxide dismutase by lipopolysaccharide, interleukin-1, and tumor necrosis factor: Role in the acute inflammatory response. J. Biol. Chem. 1990;265:2856–2864. [PubMed] [Google Scholar]
- Zhang J, Shen H, Wang X, Wu J, Xue Y. Effects of chronic exposure of 2,4-dichlorophenol on the antioxidant system in liver of freshwater fish Carassius auratus. Chemosphere. 2004;55:167–74. doi: 10.1016/j.chemosphere.2003.10.048. [DOI] [PubMed] [Google Scholar]