Abstract
DNA is methylated at the dinucleotide CpG in genomes of a wide range of plants and animals. Among animals, variable patterns of genomic CpG methylation have been described, ranging from undetectable levels (e.g., in Caenorhabditis elegans) to high levels of global methylation in the vertebrates. The most frequent pattern in invertebrate animals, however, is mosaic methylation, comprising domains of methylated DNA interspersed with unmethylated domains. To understand the origin of mosaic DNA methylation patterns, we examined the distribution of DNA methylation in the Ciona intestinalis genome. Bisulfite sequencing and computational analysis revealed methylated domains with sharp boundaries that strongly colocalize with ∼60% of transcription units. By contrast, promoters, intergenic DNA, and transposons are not preferentially targeted by DNA methylation. Methylated transcription units include evolutionarily conserved genes, whereas the most highly expressed genes preferentially belong to the unmethylated fraction. The results lend support to the hypothesis that CpG methylation functions to suppress spurious transcriptional initiation within infrequently transcribed genes.
The overall pattern of DNA methylation varies in different organisms. In mammalian cells the pattern consists of long tracts of DNA in which CpGs are methylated to a high average level (∼80%) punctuated by short unmethylated regions called “CpG islands” (Bird 1986, 2002). CpG islands are GC-rich in base composition and have a CpG approximately every 10 base pairs. By contrast, the methylated majority of the genome is relatively AT-rich and has methyl-CpGs approximately every 100 base pairs. The rarity of CpGs in bulk genomic DNA is due to the mutability of 5-methylcytosine (m5C), which tends to mutate to T (Bird 1980; Duncan and Miller 1980). CpG islands avoid CpG loss by remaining free of methylation in the germ line at least.
Unlike vertebrates, whose genomes are predominantly heavily methylated, many invertebrate genomes contain roughly comparable amounts of methylated and unmethylated DNA (Tweedie et al. 1997). Early studies of the sea urchin genome detected relatively long tracts of stably methylated DNA interspersed with equally long tracts of unmethylated DNA (Bird et al. 1979). More recently, the first mosaic methylation pattern was mapped in sea squirt Ciona intestinalis, an invertebrate member of the chordate phylum, using methylation-sensitive restriction enzymes (Simmen et al. 1999). As in the sea urchin, about half of the C. intestinalis genome consists of heavily methylated domains, whereas the other half is DNA methylation-free. Analysis of three ∼30-kb domains at low resolution suggested that these domains are interspersed, switching between methylation states many times on each chromosome. Interestingly, the locations of methylated and unmethylated patches correlated with CpG-deficient and CpG-normal domains in the genomic sequence respectively. Methylated patches were often associated with genes (Tweedie et al. 1997; Simmen et al. 1999) as in vertebrate genomes, but long unmethylated domains in the sea squirt genome had no obvious counterpart in mammals and their relationship to genes was unclear.
Comparative studies have indicated that most invertebrates, selected from diverse phyla, showed similar mosaic DNA methylation patterns (Tweedie et al. 1997). Invertebrates with very low or undetectable levels of CpG methylation (e.g., Drosophila melanogaster, Caenorhabditis elegans) were in the minority according to this survey. As invertebrates account for >95% of animal species, it follows that the mosaic DNA methylation pattern exemplified by C. intestinalis is the most abundant configuration in the animal kingdom. Given limited understanding of the origin of DNA methylation patterns in mammals, we decided to further study mosaic methylation in C. intestinalis. Our results demonstrate a close correlation between patches of DNA methylation and gene transcription units.
Results and Discussion
Methylated domains with sharp intragenic boundaries
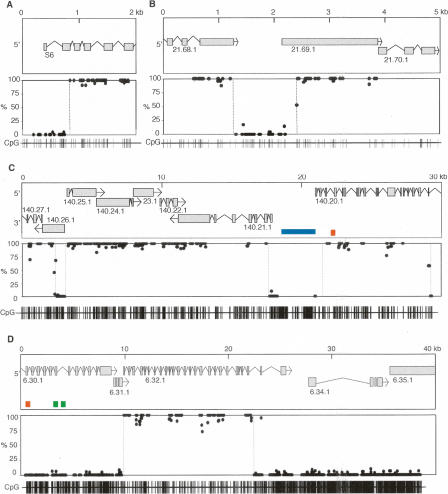
To obtain more detailed information about the C. intestinalis DNA methylation, we profiled all CpG sites in defined genomic segments by bisulfite sequencing analysis (Feil et al. 1994; Rakyan et al. 2004). Our analysis took advantage of the genome sequence of C. intestinalis and recent accurately annotated gene models that are supported by a comprehensive EST analysis to eliminate most pseudogenes (Satou et al. 2005). We predominantly analyzed sperm DNA, as previous studies have not uncovered changes in DNA methylation patterns during development of mosaically methylated genomes (Bird et al. 1979; Simmen et al. 1999). In addition, bisulfite analysis of six randomly selected C. intestinalis genes showed indistinguishable methylation patterns in genomic DNA of sperm, gastrula embryos, and adult muscle (data not shown). Bisulfite sequencing of the 40S ribosomal protein S6 gene (Fig. 1A), which has been shown by methylation-sensitive restriction enzymes to be methylated in several invertebrate species (Tweedie et al. 1997), and a random 5-kb domain from chromosome 9q (Fig. 1B) showed sharp transitions between essentially 100% methylated domains (MDs) and unmethylated domains (UMDs). Both UMDs in Figures 1A and B resemble promoter-associated CpG islands of vertebrate genomes (Grunau et al. 2000). Two additional unmethylated CpG island-like domains located at sites of predicted divergent promoters are present in a longer 30-kb profile of chromosome 7q (Fig. 1C). The 4.3-kb UMD between genes 140.20.1 and 140.21.1 contains an inserted 2.4-kb repetitive foldback element (Simmen and Bird 2000) (see below). In contrast to these unmethylated CpG islands, transcription units in all three regions are heavily methylated. These overall patterns closely resemble typical regions of vertebrate DNA, although levels of vertebrate CpG methylation are usually <100%. In contrast, a 40-kb domain of chromosome 4q gave a different CpG methylation pattern (Fig. 1D). Most of the region is unmethylated, including four genes, but a discrete MD covers the majority of the transcription unit of gene 6.32.1.
Figure 1.
A mosaic pattern of methlyated domains (MDs) and unmethylated domains (UMDs) in the C. intestinalis genome. Bisulfite sequencing showed the DNA methylation status at every CpG site in ribosomal protein S6 (scaffold 13: 227234–231211) (A), a randomly selected region of chromosome 9q (2421536–2429959) (B), a region of chromosome 7q corresponding to 29,300 bp of cosmid insert cos41 (C), and a region of chromosome 4q corresponding to 38,500 bp of cosmid insert cos2 (D). The top panels show gene annotation and plots of CpG methylation are shown below. Each dot represents a specific CpG corresponding to the CpG map at the bottom of each figure. MD/UMD boundaries are shown by broken lines. Repetitive sequences Cifo-1 (blue), Cimi-1 (green), and Cics-1 (orange) are shown.
All ten DNA methylation transitions seen in these analyses occurred within transcription units and in each case the change from UMD to MD was abrupt, occurring within a few base pairs. In two cases, a single CpG site showed intermediate levels of DNA methylation (Fig. 1B, gene 21.69.1, and Fig. 1C, gene 140.26.1), suggesting that these sites are within the transition region. Similar results were observed in Scottish C. intestinalis (Fig. 1) and Pacific C. intestinalis, which represents a subspecies (Suzuki et al. 2005). Boundaries were less precise in Pacific specimens, but the transition from UMD to MD was completed within 500 bp and occurred within predicted transcription units (Supplemental Fig. 2B,C). We were unable to detect common boundary DNA sequence motifs by computational analysis of the primary DNA sequence.
MDs colocalize with a subset of genes
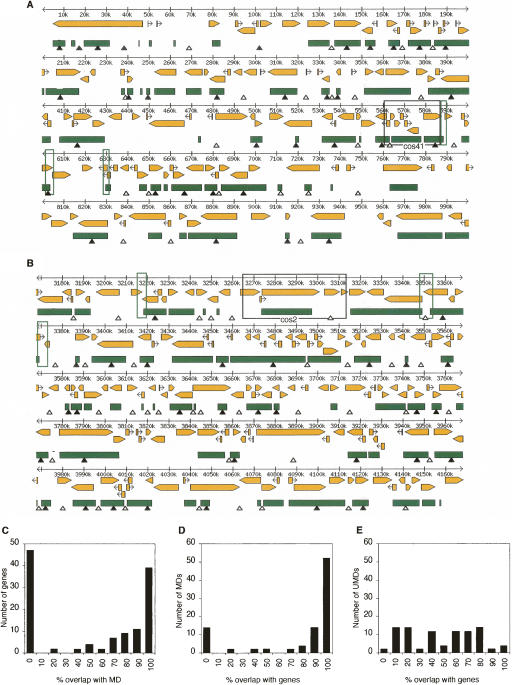
To further illuminate the relationship between MDs and genes, a global methylation pattern was deduced using a combination of genome sequence analysis and computation. Although whole-genome tiling microarrays have been shown as a powerful strategy to map DNA methylation (Weber et al. 2005; Zhang et al. 2006), high polymorphism in the C. intestinalis genome renders this an unreliable option in this species. Sequence analysis revealed 1%–2% allelic polymorphism between individuals, with local peaks as high as 10%–15% within a window of 100 nucleotides (Dehal et al. 2002; Suzuki et al. 2005). Fortunately, MDs and UMDs can be reliably predicted from plots of CpG frequency. This is due to the well-known mutability of 5-methylcytosine, which leads to depletion of CpG in consistently methylated DNA sequences over evolutionary time (Simmen et al. 1999). The MD in Figure 1D, for example, projects onto a region of the genome in which CpG is depleted compared with flanking DNA. On the basis of the observed relationship between DNA methylation and CpG frequency in our bisulfite sequence analysis, we designated regions in which CpG frequency (observed/expected) was greater or less than 0.8 as candidate UMDs or MDs, respectively. We screened two 1-Mb regions from chromosomes 7 and 4 that contain 123 and 156 genes, respectively, including cos41 and cos2 (see Fig. 1C,D), using a 1000-bp window and a 200-bp step (Fig. 2A,B). To test the reliability of these predictions, we carried out methylation-sensitive PCR for all of the inferred MDs and UMDs in both regions. Out of 128 primer pairs, 122 confirmed the predicted methylation status (95%; see data example in Supplemental Fig. 1). It is possible that the 5% rate of false positives is due to the relatively large window size, which was necessary to minimize random fluctuations in base composition and CpG frequency. We selected an additional six regions (a total of 25 kb) to test predicted UMD/MD boundaries by bisulfite sequencing (Supplemental Fig. 2). The observed methylation patterns agreed with expectation and boundaries were reliably within 2 kb of the predicted location.
Figure 2.
Methylated domains (MDs) within two 1-Mb genomic regions from chromosome 7q (A) and chromosome 4q (B) colocalize with transcription units. MDs (green bars) were inferred from CpG[o/e] ratios as described in the text. The inferred pattern was experimentally verified by methylation-sensitive PCR at sites marked by closed triangles (methylated) and open triangles (unmethylated). Yellow arrows show the size and orientation of predicted transcription units. The positions of bisulfite sequenced cosmid inserts cos41 and cos2 (see Fig. 1) are shown by black boxes. Green boxes indicate regions that were bisulfite sequenced in order to test the accuracy of predicted unmethylated domain (UMD)/MD boundaries (Supplemental Fig. 2). (C) Frequency of predicted genes that overlap to varying degrees with MDs. (D) Frequency of MDs that overlap to varying degrees with predicted genes. (E) Frequency of UMDs with gene overlap.
The profiles of mosaic DNA methylation in these regions of the C. intestinalis genome show a striking relationship between DNA methylation and predicted transcription units. Approximately 38% of genes in the sample (47/123) were entirely within UMDs, whereas 48% of genes (59/123) overlapped with an MD for at least 70% their length (Fig. 2C). Correspondingly, 84% of MDs (77/92) overlapped with predicted genes over at least 70% of their length (Fig. 2D). By contrast, UMDs showed no consistent relationship with genes (Fig. 2E). The tested regions have one predicted gene per 7.2 kb, which is typical of the C. intestinalis genome (one gene per 7.5 kb on average: Dehal et al. 2002). Analyses of additional relatively gene-poor and gene-dense regions gave equivalent results (data not shown). The data indicate that DNA methylation in C. intestinalis is targeted to a subset of transcription units.
Shared properties of methylated and unmethylated genes
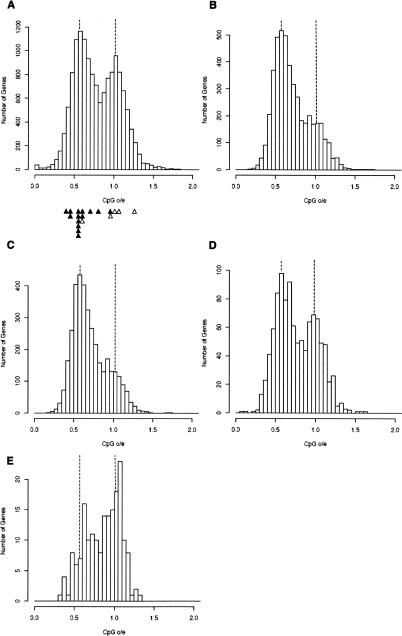
Methylated and unmethylated genes are evidently interspersed in the C. intestinalis genome. To identify all methylated and unmethylated genes, we plotted 14,515 C. intestinalis Ensembl gene models against CpG[o/e] frequency (Fig. 3A) and found a bimodal distribution indicating two distinct groups: (1) CpG-deficient genes with mean CpG[o/e] frequencies of ∼0.6, and (2) genes with the expected frequency of CpG (mean o/e = 1.1). Approximately 54% of all genes belonged to the CpG-deficient fraction (CpG[o/e] < 0.8) and were therefore predicted to be methylated at CpGs within their transcription units. Genes in which CpG was close to the predicted frequency, on the other hand, were expected to lie within UMDs. We tested these predictions for a set of 19 C. intestinalis genes by bisulfite sequencing (see Fig. 1 and Supplemental Fig. 2) and the results agreed with the expected DNA methylation status (see Fig. 3A, triangles).
Figure 3.
Features that distinguish C. intestinalis methylated and unmethylated genes. (A) Histogram showing the frequency of all genes with CpG[o/e] frequencies between 0 and 2. CpG[o/e] of genes whose methylation status was analyzed by bisulfite sequencing are shown in closed (methylated) and open (unmethylated) triangles below. (B) Frequencies of genes that are evolutionarily conserved between C. intestinalis, Homo sapiens, and D. melanogaster plotted against CpG[o/e]. (C) Frequencies of C. intestinalis orthologs of human CpG island genes plotted against CpG[o/e]. (D) Frequencies of C. intestinalis orthologs of human non-CpG island genes plotted against CpG[o/e]. (E) Frequencies of 188 genes that recorded the highest number of EST hits (>500) in the UniGene database plotted against CpG[o/e].
We next asked whether genes in each category shared specific properties. Neither gene orientation nor gene length nor gene ontology was obviously related to DNA methylation status (data not shown). Also, analysis of ESTs from nine developmental stages and seven adult tissues (Satou et al. 2005), including unfertilized eggs, gonad, and testis, showed that methylated and unmethylated genes were expressed at similar ratios in a variety of stages and tissues (data not shown). Strong enrichment within the methylated gene fraction was observed, however, when only evolutionarily conserved genes with identifiable orthologs in human and D. melanogaster genomes were selected (Fig. 3B; Supplemental Fig. 3A,B). Genes encoding proteins that perform core functions required by most or all cells tend to be evolutionarily more conserved than genes whose expression is restricted to one or a few cell types (Zhang and Li 2004). As these “housekeeping genes” often possess CpG island promoters in mammalian cells (Bird 1987; Larsen et al. 1992), we asked whether C. intestinalis orthologs of human CpG island genes were also enriched in the methylated group. When C. intestinalis orthologs of human genes were separately pooled in CpG island-positive and -negative subsets, significant enrichment of the CpG island positive subset was observed in the methylated fraction (CpG[o/e] < 0.8; Fig. 3C). In contrast, the CpG island negative subset (CpG[o/e] > 0.8; Fig. 3D) was relatively enriched in the unmethylated fraction. These two distributions are significantly different from one another, as measured by the Kolmogorov-Smirnov test (D = 0.1293, P < 0.0001). The finding that housekeeping genes, many of which are transcribed at low/moderate levels, are highly represented within the methylated gene fraction raised the possibility that genes with low transcription rates are preferentially methylated. If so, the most highly transcribed genes would be predicted to belong to the unmethylated gene category. To test this, we determined the CpG frequency of 188 genes from the UniGene database (Wheeler et al. 2006) that had the highest number of EST reports. The results showed that these most highly expressed genes are significantly enriched in the unmethylated gene fraction (Fig. 3E).
DNA methylation and intragenic transcriptional initiation
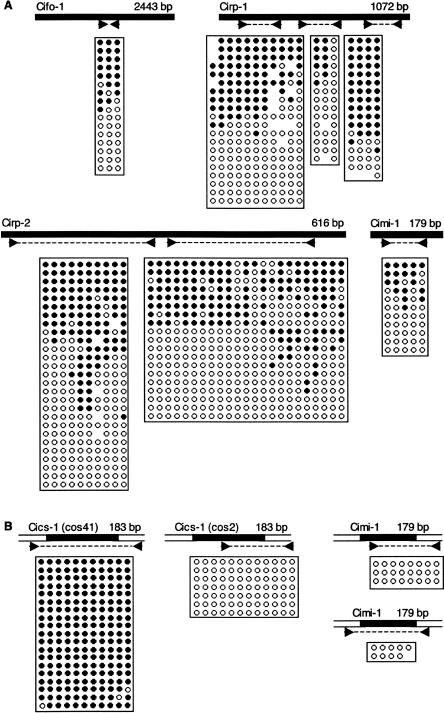
The determinants that trigger methylation of a sharply defined DNA sequence are unknown, but our data show that neither DNA sequence per se nor repetitiveness are sufficient as multicopy DNA elements occur in either methylated or unmethylated states. Previous work has shown that several transposons, including a foldback element (Cifo-1) and the Cigr-1 transposon, are predominantly unmethylated (Simmen et al. 1999). We surveyed all copies of these elements by bisulfite sequencing using internal primers that amplify most copies of a specific transposon. Cifo-1, Cirp-1, Cirp-2, and Cimi-1 were partially or fully methylated in ∼50% of copies, whereas Cigr-1 was methylated in ∼30% of copies (Fig. 4A; Supplemental Fig. 4). As ∼50% of genomic DNA is methylated in this organism, it is evident that transposons are not preferred targets for this modification. When we examined the methylation status of a single transposon copy using location-specific primers, all clones were either homogeneously methylated or unmethylated (Fig. 4B). Specifically, two different copies of Cics-1 had opposite modification patterns regardless of their almost identical sequences (98%). In addition, the methylation pattern of the single transposon copies agreed with that of the surrounding DNA sequence. These results suggest that the methylation status of a transposon is determined by its insertion site. As in the case of mammals (Rabinowicz et al. 2003), there is no evidence that transposons are direct targets of the CpG methylation system in this organism.
Figure 4.
Variable DNA methylation of repetitive elements in C. intestinalis matches flanking DNA. Elements as named here or by Simmen and Bird (2000) are diagrammed as solid bars above each bisulfite profile showing primer pair location. A horizontal row within each box corresponds to one sequenced clone in which specific CpGs were methylated (solid) or unmethylated (open). Mutated CpG sites are shown as blanks within a row. (A) Results using primer sets within the element showed a mixture of methylated and unmethylated clones. (B) Locus-specific primers showed that the methylation patterns of two Cics-1 elements and two Cimi-1 elements at specific genomic locations are homogeneous and match the methylation status of flanking genomic DNA (open bars above). The locations of the elements analyzed in B are shown in Fig. 1.
CpG methylation is recognized as an important transcriptional silencing mechanism, yet, paradoxically, actively transcribed genes in animals and plants have high levels of this modification. The hypothesis that a function of CpG methylation might be to dampen spurious transcriptional initiation (Bird 1995) received earlier support from the observation that many expressed genes are specifically methylated at CpG in mosaically methylated invertebrate genomes. This led to the proposal that CpG methylation within transcription units serves specifically to quell transcriptional initiation within infrequently transcribed genes (Simmen et al. 1999). Mapping of DNA methylation in the plant Arabidopsis thaliana strengthened this interpretation (Tran et al. 2005; Zhang et al. 2006; Zilberman et al. 2007). The present findings establish that transcription units of housekeeping genes are major targets for DNA methylation, perhaps because they are more likely to suffer interference from spurious transcriptional noise. In contrast, the most abundant transcripts, which presumably originate from strong promoters, are preferentially derived from unmethylated genes.
The relationship between DNA methylation and transcription units fits with recent reports that repressive histone modifications are associated with actively transcribed genes. The histone H3 lysine 36 methyltransferase Set2 travels with RNA polymerase and promotes deacetylation of chromatin during transcription. Failure of this process causes spurious transcriptional initiation within genes (Carrozza et al. 2005). It has also been reported that methylation of histone H3 lysine 9, which is linked with the formation of constitutive heterochromatin, occurs within actively transcribed genes and recruits HP1γ (Vakoc et al. 2005). It is possible that intragenic CpG methylation is an additional mechanism to minimize transcriptional interference.
Does intragenic CpG methylation represent an evolutionarily conserved function of this DNA modification within the animal kingdom? In addition to the present data, bisulfite sequencing in two insect species shows comparable intragenic CpG methylation. Analysis of the amplified esterase E4 gene of the aphid Myzus persicae showed CpG methylation within the gene but not at the 5′ and 3′ regions of the transcription unit (Field 2000). Recently, a similar analysis of several honey bee genes again showed CpG methylation within the transcription units but not at their extremities (Wang et al. 2006). DNA methylation is also found within most vertebrate genes. Although it is often assumed that DNA methylation in vertebrates is preferentially targeted to repetitive DNA sequences, the probability that a CpG is methylated appears similar in genes, transposons, and tandemly repeated sequences. This means that vertebrates resemble invertebrates with respect to intragenic DNA methylation. The similarity raises the possibility that the functional role played by intragenic CpG methylation is conserved between plants and both invertebrate and vertebrate animals. To test this, it will be important to unravel the relationship between DNA methylation and transcription in organisms across the phylogenetic spectrum.
Methods
DNA sequence data
C. intestinalis genomic sequence was obtained from the Ensembl release 40 (http://www.ensembl.org). British cosmid sequences cos41 and cos2 were determined by Simmen et al. (1999). For C. intestinalis gene annotation, Kyotograil2005, which predicts gene models based on ∼680,000 ESTs and ∼6,500 full-length insert cDNAs, was used (Satou et al. 2005). Some of the neighboring gene models which share 5′ and 3′ ESTs from the same transcript were merged manually as one transcriptional unit. The whole list of the gene models in two 1-Mb regions from chromosomes 7 and 4 is shown in Supplemental Table 1. UniGene was downloaded from NCBI. Repetitive sequence Cirp-1 and Cirp-2 are from cos41: 29330–30402 and cos2: 24848–25464, respectively.
Genomic DNA extraction
Specimens were obtained from Oban, Scotland, UK and Santa Barbara, CA. Genomic DNA was extracted from sperm with TRIzol reagent (GIBCO-BRL).
Bisulfite sequencing
Genomic DNA was subjected to bisulfite treatment according to Feil et al. (1994). Approximately 2 μg of genomic DNA digested by EcoRI or EcoRV was used for the sodium bisulfite reaction. The DNA was denatured in 0.3 M NaOH for 20 min at 37°C. Sodium bisulfite (Sigma-Aldrich) was then added to a final concentration of 4.3 M, and hydroquinone (Sigma) to a final concentration of 0.12 M. The reaction was adjusted to pH 5.0 and incubated at 55°C for 5 h. The sample was desulfonated by 0.3 M NaOH and cleaned by QIAquick PCR purification kit (QIAGEN). Bisulfite-treated DNA was used as template in subsequent PCR assays. A series of PCR primer sets targeting around 350-bp regions were designed by MethPrimer (Li and Dahiya 2002) to cover genomic regions of interest. The primer sequences are available on request. PCR with bisulfite-treated DNA was carried out using FastStart Taq DNA polymerase (Roche) for a 4-min denaturation step at 95°C; followed by 45 cycles of 30 sec at 95°C, 30 sec at 45°C, and 45 sec at 72°C; with a final extension at 72°C for 7 min. PCR products were cloned using TOPO TA Cloning Kit (Invitrogen) and at least 10 individual clones containing PCR amplicons from each primer set were sequenced. We determined the methylation status of each CpG dinucleotide within a region by comparing the sequences obtained from the cloned PCR products of the bisulfite-treated DNA with the untreated DNA sequences. Only data from bisulfite PCR clones showing complete C-to-T conversion by bisulfite treatment at Cs not located in CpG dinucleotides throughout a region were used.
DNA methylation status prediction.
Candidate MDs were identified by plotting a moving average of CpG[o/e] frequency. On the basis of empirical bisulfite sequencing results, MDs had a CpG[o/e] frequency of <0.8 (average = 0.6), whereas UMDs had a CpG[o/e] of >0.8 (average = 1.1). We eliminated very short or low-scoring regions that may constitute noise by discounting domains with an area above or below the 0.8 threshold of <0.46. Repetitive sequences were excluded from this analysis by examining the Ensembl repeat masked genome data set. The methylation status of C. intestinalis genes was determined by calculating CpG[o/e] in primary transcription region of Ensembl gene models including those of human and D. melanogaster orthologs. Human CpG island genes were defined as genes with a 500-bp region with CpG[o/e] > 0.6 and G + C > 0.5 within 1 kb upstream of 5′ UTR (Ponger et al. 2001). The best-hit Ensembl gene models of UniGene were extracted by BLAST search.
Methylation-sensitive PCR
Genomic DNA (10 μg) was digested by an excess of HhaI or MspI (New England Biolabs). Semi-quantitative PCR was conducted with 20 ng of digested or undigested genomic DNA as templates, using primers designed for the area that include at least one HpaII/MspI recognition sites (primers available on request). The reaction was carried out for 25 cycles with GoTaq DNA polymerase (Promega) in accordance with the manufacturer’s instructions. PCR products were visualized on 2% agarose gels.
Acknowledgments
We thank Shota Chiba and William Smith (University of California, Santa Barbara), and members of Craobh Haven (Oban, Scotland) for kindly providing animals. We thank members of the Bird laboratory for comments on the manuscript and Aileen Greig for technical assistance. This work was supported by a grant (no. 074948) from the Wellcome Trust.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6163007
References
- Bird A.P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1594. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A.P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- Bird A.P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird A.P., Taggart M.H., Smith B.A., Taggart M.H., Smith B.A., Smith B.A. Methylated and unmethylated DNA compartments in the Sea Urchin genome. Cell. 1979;17:889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., Shia W.J., Anderson S., Yates J., Washburn M.P., Anderson S., Yates J., Washburn M.P., Yates J., Washburn M.P., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Dehal P., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Satou Y., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Campbell R.K., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Chapman J., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Degnan B., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., De Tomaso A., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Davidson B., Di Gregorio A., Gelpke M., Goodstein D.M., Di Gregorio A., Gelpke M., Goodstein D.M., Gelpke M., Goodstein D.M., Goodstein D.M., et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Duncan B.K., Miller J.H., Miller J.H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Feil R., Charlton J., Bird A.P., Walter J., Reik W., Charlton J., Bird A.P., Walter J., Reik W., Bird A.P., Walter J., Reik W., Walter J., Reik W., Reik W. Methylation analysis on individual chromosomes: Improved protocol for bisulphite genomic sequencing. Nucleic Acids Res. 1994;22:695–696. doi: 10.1093/nar/22.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field L.M. Methylation and expression of amplified esterase genes in the aphid Myzus persicae (Sulzer) Biochem. J. 2000;349:863–868. doi: 10.1042/bj3490863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C., Hindermann W., Rosenthal A., Hindermann W., Rosenthal A., Rosenthal A. Large-scale methylation analysis of human genomic DNA reveals tissue-specific differences between the methylation profiles of genes and pseudogenes. Hum. Mol. Genet. 2000;9:2651–2663. doi: 10.1093/hmg/9.18.2651. [DOI] [PubMed] [Google Scholar]
- Larsen F., Gundersen G., Lopez R., Prydz H., Gundersen G., Lopez R., Prydz H., Lopez R., Prydz H., Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Li L.C., Dahiya R., Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Ponger L., Duret L., Mouchiroud D., Duret L., Mouchiroud D., Mouchiroud D. Determinants of CpG islands: expression in early embryo and isochore structure. Genome Res. 2001;11:1854–1860. doi: 10.1101/gr.174501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz P.D., Palmer L.E., May B.P., Hemann M.T., Lowe S.W., McCombie W.R., Martienssen R.A., Palmer L.E., May B.P., Hemann M.T., Lowe S.W., McCombie W.R., Martienssen R.A., May B.P., Hemann M.T., Lowe S.W., McCombie W.R., Martienssen R.A., Hemann M.T., Lowe S.W., McCombie W.R., Martienssen R.A., Lowe S.W., McCombie W.R., Martienssen R.A., McCombie W.R., Martienssen R.A., Martienssen R.A. Genes and transposons are differentially methylated in plants, but not in mammals. Genome Res. 2003;13:2658–2664. doi: 10.1101/gr.1784803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan V.K., Hildmann T., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Hildmann T., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., Andrews T.D., Howe K.L., Otto T., Olek A., Howe K.L., Otto T., Olek A., Otto T., Olek A., Olek A., et al. DNA methylation profiling of the human major histocompatibility complex: A pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y., Kawashima T., Shoguchi E., Nakayama A., Satoh N., Kawashima T., Shoguchi E., Nakayama A., Satoh N., Shoguchi E., Nakayama A., Satoh N., Nakayama A., Satoh N., Satoh N. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zoolog. Sci. 2005;22:837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- Simmen M.W., Bird A.P., Bird A.P. Sequence analysis of transposable elements in the sea squirt, Ciona intestinalis. Mol. Biol. Evol. 2000;17:1685–1693. doi: 10.1093/oxfordjournals.molbev.a026267. [DOI] [PubMed] [Google Scholar]
- Simmen M.W., Leitgeb S., Charlton J., Jones S.J.M., Harris B.R., Clark V.H., Bird A., Leitgeb S., Charlton J., Jones S.J.M., Harris B.R., Clark V.H., Bird A., Charlton J., Jones S.J.M., Harris B.R., Clark V.H., Bird A., Jones S.J.M., Harris B.R., Clark V.H., Bird A., Harris B.R., Clark V.H., Bird A., Clark V.H., Bird A., Bird A. Nonmethylated transposable elements and methylated genes in a chordate genome. Science. 1999;283:1164–1167. doi: 10.1126/science.283.5405.1164. [DOI] [PubMed] [Google Scholar]
- Suzuki M.M., Nishikawa T., Bird A., Nishikawa T., Bird A., Bird A. Genomic approaches reveal unexpected genetic divergence within Ciona intestinalis. J. Mol. Evol. 2005;61:627–635. doi: 10.1007/s00239-005-0009-3. [DOI] [PubMed] [Google Scholar]
- Tran R.K., Henikoff J.G., Zilberman D., Ditt R.F., Jacobsen S.E., Henikoff S., Henikoff J.G., Zilberman D., Ditt R.F., Jacobsen S.E., Henikoff S., Zilberman D., Ditt R.F., Jacobsen S.E., Henikoff S., Ditt R.F., Jacobsen S.E., Henikoff S., Jacobsen S.E., Henikoff S., Henikoff S. DNA methylation profiling identifies CG methylation clusters in Arabidopsisgenes. Curr. Biol. 2005;15:154–159. doi: 10.1016/j.cub.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Tweedie S., Charlton J., Clark V., Bird A., Charlton J., Clark V., Bird A., Clark V., Bird A., Bird A. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol. Cell. Biol. 1997;17:1469–1475. doi: 10.1128/mcb.17.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc C.R., Mandat S.A., Olenchock B.A., Blobel G.A., Mandat S.A., Olenchock B.A., Blobel G.A., Olenchock B.A., Blobel G.A., Blobel G.A. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Wang Y., Jorda M., Jones P.L., Maleszka R., Ling X., Robertson H.M., Mizzen C.A., Peinado M.A., Robinson G.E., Jorda M., Jones P.L., Maleszka R., Ling X., Robertson H.M., Mizzen C.A., Peinado M.A., Robinson G.E., Jones P.L., Maleszka R., Ling X., Robertson H.M., Mizzen C.A., Peinado M.A., Robinson G.E., Maleszka R., Ling X., Robertson H.M., Mizzen C.A., Peinado M.A., Robinson G.E., Ling X., Robertson H.M., Mizzen C.A., Peinado M.A., Robinson G.E., Robertson H.M., Mizzen C.A., Peinado M.A., Robinson G.E., Mizzen C.A., Peinado M.A., Robinson G.E., Peinado M.A., Robinson G.E., Robinson G.E. Functional CpG methylation system in a social insect. Science. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- Weber M., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Davies J.J., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Wittig D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Oakeley E.J., Haase M., Lam W.L., Schubeler D., Haase M., Lam W.L., Schubeler D., Lam W.L., Schubeler D., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., Church D.M., DiCuccio M., Edgar R., Federhen S., DiCuccio M., Edgar R., Federhen S., Edgar R., Federhen S., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Li W.H., Li W.H. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol. Biol. Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Shinn P., Pellegrini M., Jacobsen S.E., Pellegrini M., Jacobsen S.E., Jacobsen S.E., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S., Gehring M., Tran R.K., Ballinger T., Henikoff S., Tran R.K., Ballinger T., Henikoff S., Ballinger T., Henikoff S., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]