Abstract
Long transcripts that do not encode protein have only rarely been the subject of experimental scrutiny. Presumably, this is owing to the current lack of evidence of their functionality, thereby leaving an impression that, instead, they represent “transcriptional noise.” Here, we describe an analysis of 3122 long and full-length, noncoding RNAs (“macroRNAs”) from the mouse, and compare their sequences and their promoters with orthologous sequence from human and from rat. We considered three independent signatures of purifying selection related to substitutions, sequence insertions and deletions, and splicing. We find that the evolution of the set of noncoding RNAs is not consistent with neutralist explanations. Rather, our results indicate that purifying selection has acted on the macroRNAs’ promoters, primary sequence, and consensus splice site motifs. Promoters have experienced the greatest elimination of nucleotide substitutions, insertions, and deletions. The proportion of conserved sequence (4.1%–5.5%) in these macroRNAs is comparable to the density of exons within protein-coding transcripts (5.2%). These macroRNAs, taken together, thus possess the imprint of purifying selection, thereby indicating their functionality. Our findings should now provide an incentive for the experimental investigation of these macroRNAs’ functions.
Whether it is 2.5% (Lunter et al. 2006) or 5% (Waterston et al. 2002) of the human genome that has been purified of deleterious mutations within functional sequence, this proportion is much greater than the 1.2% of the genome that encodes proteins (International Human Genome Sequencing Consortium 2004). Certainly, a small amount of this additional noncoding sequence represents small regulatory RNAs (Pang et al. 2006), validated binding sites for transcription factors, and other regulatory sites. Yet, the bulk of this “dark matter” (Yamada et al. 2003) represents sequence whose type and principal functions remain ill-determined.
Evidence from both large-scale studies of extensive full-length mouse cDNA libraries (Okazaki et al. 2002; Carninci et al. 2005) and high-density genome tiling arrays (Kapranov et al. 2002; Bertone et al. 2004; Cawley et al. 2004; Cheng et al. 2005) reveals that much of this dark matter is transcribed, both within the introns and untranslated regions of protein-coding genes, and within a set of long, apparently noncoding, transcripts. Only a small number of long ncRNAs have been functionally well-characterized, including prominent examples such as Xist and Air (Brockdorff et al. 1992; Sleutels et al. 2002), and most exhibit poor conservation when their sequences are compared between diverse mammals (Pang et al. 2006).
If long ncRNAs have preserved their functions over long time spans, then the imprint of purifying selection should be apparent within their sequences when sampled from diverse mammalian species. However, initial surveys have been discouraging and provide scant evidence of purifying selection (Wang et al. 2004; Lau et al. 2006; Pang et al. 2006). Wang et al. (2004) reported that the ncRNAs identified in Okazaki et al. (2002) are, in general, as poorly conserved as intergenic sequence, and thus concluded that most of these transcripts are unlikely to be functional. Others have argued that an apparent lack of sequence conservation need not imply an absence of function if positive, rather than negative, selection has prevailed (Pang et al. 2006). The occurrence of ncRNAs in some, but not all, related species is consistent with the rapid emergence, by adaptive evolution and/or decline, of a subset of ncRNAs (Hyashizaki 2004).
The issues that remain to be clarified are whether most long ncRNAs are biologically relevant and, if so, whether they have persisted because of the benefit accrued from their functions over long time intervals, such as since the last common ancestor of primates and rodents ∼90 million years ago (Mya) (Springer et al. 2003). If, instead, they are not biologically relevant, might these ncRNAs represent “transcriptional noise,” having been transcribed from illegitimate promoters? Studies have demonstrated distinct spatiotemporal expression patterns for ncRNAs, implying that the phenomenon of transcriptional noise, although plausible, is rare (Blake et al. 2003; Ravasi et al. 2006). Another possibility is that long ncRNA sequences do not themselves convey function, but their transcription promotes, by inducing a more “open” chromatin structure, the transcription of neighboring protein-coding genes (Gribnau et al. 2000; Schmitt and Paro 2004). Such transcripts are expected to be constrained in their promoters, but not in their transcribed sequences.
In this study, we sought to investigate whether long ncRNAs exhibit signatures of purifying selection that would provide indications of their functionality. To provide evidence for selection requires reliable estimates of neutral evolution. As virtually all ancestral repeats (ARs), defined as transposable elements present in the last common ancestor of, for instance, mouse and human, appear to have evolved neutrally (Lunter et al. 2006), their evolutionary rates provide appropriate proxies for mutational rates in selectively neutral sequence. These rates vary above the megabase scale (Gaffney and Keightley 2005) and thus need to be estimated locally. Although most studies consider nucleotide sequence conservation when inferring past selection, a complementary approach has been developed that considers the patterns of insertion and deletion (indel) events in nucleotide alignments (Lunter et al. 2006). Extended regions devoid of indels (“indel-purified segments,” or IPSs) are likely to be functional, and a significant association of long ncRNAs with these regions is thus an indicator of purifying selection. As both indel rates and the density of indel-purified regions vary strongly with G+C content (Lunter et al. 2006), it is imperative to account for spurious associations between them that arise simply through G+C content. To exclude such confounding effects, we assessed the significance of association between IPSs and ncRNAs using a sampling procedure that controls for G+C content.
We took advantage of a well-defined large set of 3122 long putative ncRNAs of unknown function obtained from the FANTOM 2 and 3 Consortia (Okazaki et al. 2002; Carninci et al. 2005) from which we have discarded sequences with evidence of protein-coding capacity. These have been termed macro-noncoding RNAs (macro-ncRNAs) (Furuno et al. 2006), but hereafter we refer to them as macroRNAs, in order to differentiate them from smaller microRNAs. We investigated signatures of purifying selection, in both the transcript and its predicted promoter region, of substitutions and transversions (relative to local ARs), and insertions and deletions (indels). In addition, we asked whether splice site donor and acceptor dinucleotides in mouse macroRNAs have been preferentially conserved within macroRNAs. If mouse macroRNAs are not functional, our null hypothesis is that they should accumulate substitutions, insertions, and deletions at the same rates as selectively neutral sequence, here taken to be ARs or else intergenic sequence.
Our studies show that the set of macroRNAs appears to exhibit suppressed rates of nucleotide substitution, insertion, and deletion, relative to proximal ARs and general intergenic sequence. Suppressed rates were observed for transcript sequences, promoters, and splice-site dinucleotide motifs. We interpret these suppressed rates as indicative of recurrent events of purifying selection that acted within functional sequence. Neutralist explanations of suppressed rates, such as varying mutational rates due to CpG substitutions, transcription-coupled repair, or nucleotide composition, were not consistent with our findings. We thus conclude that many of the macroRNAs we considered are functional, and thus deserve more intensive investigation of their evolution and functions.
Results
A validated set of macroRNAs
We investigated the evolutionary properties of a set of 3122 apparent ncRNAs (average length 4.2 kb) from which known protein-coding genes had previously been discarded. These transcripts were identified from mouse cDNA libraries collected by the FANTOM Consortium (Okazaki et al. 2002; Carninci et al. 2005). The FANTOM filtering procedure used to obtain this set comprised several steps. Using an automated annotation pipeline, the coding sequence and function of each cDNA were predicted, and its transcript was further described, manually curated, and finally reviewed by expert curators (Maeda et al. 2006). A set of CDS prediction algorithms such as CRITICA (Badger and Olsen 1999), mTRANS (M. Furuno, unpubl.), CombinerCDS (Allen et al. 2004), and rsCDS (Furuno et al. 2003) were used for the FANTOM3 project, for example; similar algorithms were used for FANTOM2. Depending on the prediction of CDS status and additional information of the transcript, the cDNAs were classified into protein- and non-protein-coding transcripts. In this study, we used the most stringent sets of noncoding transcripts identified by the FANTOM2 and FANTOM3 projects. For instance, the FANTOM3 most stringent noncoding set contains macroRNAs whose transcript start and termination sites are experimentally supported by ESTs, CAGE tags, or other cDNAs, and thus, as full-length cDNAs, are not partial sequences of, for example, longer protein-coding transcripts. These sets do not contain members of known functional and structural classes of ncRNAs, such as microRNAs and small nucleolar RNAs.
To exclude the possibility that evolutionary constraints we observed within these putative macroRNAs arise from overlaps with protein-coding exons not annotated by FANTOM2 or FANTOM3 or with regulatory intronic regions, we conservatively applied two additional filtering steps in order to create our own candidate noncoding set. We excluded macroRNAs that overlap with Ensembl-annotated protein-coding genes (including introns), and others exhibiting significant alignments with well-established protein-coding genes (see Methods). We believe that all remaining candidate macroRNAs are thus located within intergenic regions.
Suppressed substitution and transversion rates
We first sought evidence for the elimination of deleterious point mutations, in both evolutionary lineages, since the last common ancestor of mouse and human. For each sequence in our macroRNA set, we compared its estimated rate of nucleotide substitution (dRNA) to the equivalent rate (dAR) within neighboring ARs that we infer to have been present in this ancestor. The ratio of these two rates, dRNA/dAR, is expected to be 1 if selection has not distinguished substitutions within macroRNAs and substitutions within nearby ARs. (This ratio is analogous to dN/dS, the ratio of nonsynonymous to synonymous substitution rates in protein-coding sequence.) If this ratio, however, is significantly less than 1, then this would be an indication either that purifying selection on substitutions has been more prevalent in macroRNAs than in neighboring ARs, or that underlying mutation rates are lower in the former than in the latter. To ensure a sufficiently accurate estimation of the substitution rates, this analysis was performed only for those transcripts for which at least 1 kb of mouse sequence could be aligned to either human or rat sequence (1552 of 3122 and 2016 of 3122 transcripts, respectively).
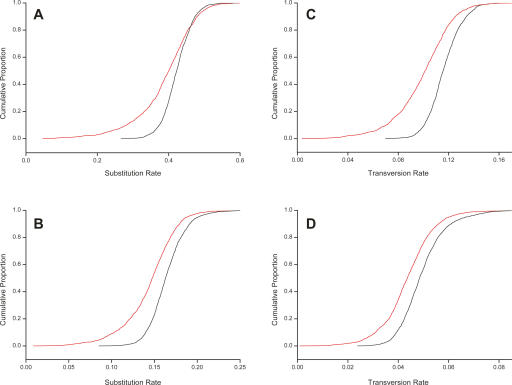
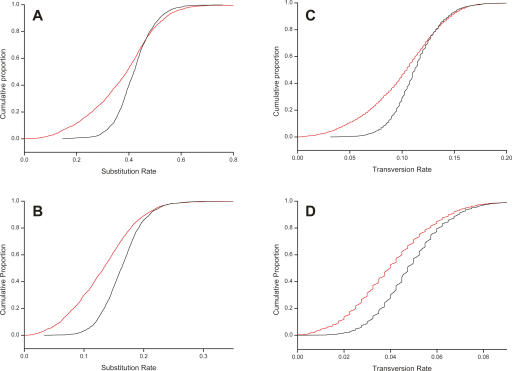
Our initial finding was that nucleotide substitutions have been fixed at a significantly reduced rate in macroRNAs compared to in neighboring ARs. The distributions of dRNA estimated between the mouse putative macroRNA sequences aligned to their rat or human orthologous sequence were both found to be significantly lower than those of dAR for these species pairs (P < 10−15; two-sided Kolmogorov-Smirnov test) (see Fig. 1A,B). Median dRNA/dAR values for macroRNAs were 0.899 (mouse–rat) and 0.948 (mouse–human) (Table 1). For these species pairs, substitution rates on macroRNAs are thus suppressed by, approximately, 10% and 5%.
Figure 1.
Nucleotide substitution and transversion rates are suppressed within macroRNA transcripts. Panels show the cumulative distributions of substitution (A,B) and transversion rates (C,D) as measured on macroRNA transcripts (red curves), and the same rates measured on nearby nonoverlapping AR sequence of matched length (black curves). (A) Mouse–human substitution rates; (B) mouse–rat substitution rates; (C,D) mouse–human and mouse–rat transversion rates. All macroRNA rates are significantly different from, and lower than, the putatively neutral AR rates (Kolmogorov-Smirnov test, P < 10−15 for all panels). The suppression of transversion rates in macroRNAs compared to AR sequence demonstrates that these observations are not a consequence of a higher density of highly mutable CpG sites within ARs, since the associated mutations are mainly transitions rather than transversions.
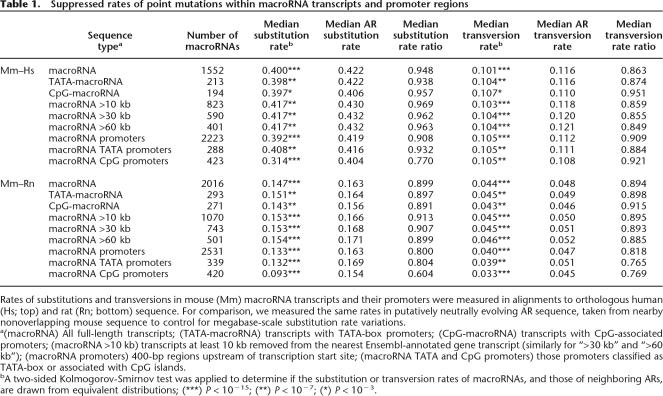
Table 1.
Suppressed rates of point mutations within macroRNA transcripts and promoter regions
Rates of substitutions and transversions in mouse (Mm) macroRNA transcripts and their promoters were measured in alignments to orthologous human (Hs; top) and rat (Rn; bottom) sequence. For comparison, we measured the same rates in putatively neutrally evolving AR sequence, taken from nearby nonoverlapping mouse sequence to control for megabase-scale substitution rate variations.
a(macroRNA) All full-length transcripts; (TATA-macroRNA) transcripts with TATA-box promoters; (CpG-macroRNA) transcripts with CpG-associated promoters; (macroRNA >10 kb) transcripts at least 10 kb removed from the nearest Ensembl-annotated gene transcript (similarly for “>30 kb” and “>60 kb”); (macroRNA promoters) 400-bp regions upstream of transcription start site; (macroRNA TATA and CpG promoters) those promoters classified as TATA-box or associated with CpG islands.
bA two-sided Kolmogorov-Smirnov test was applied to determine if the substitution or transversion rates of macroRNAs, and those of neighboring ARs, are drawn from equivalent distributions; (***) P < 10−15; (**) P < 10−7; (*) P < 10−3.
We considered whether these departures of dRNA/dAR from unity might be causally related to the known high rate of substitutions in CpG dinucleotides (Cooper and Youssoufian 1988; Sved and Bird 1990) if, for example, CpG dinucleotides are, on average, more frequent in ARs than they are in macroRNAs. As CpG substitutions are, in the main, transitions (Ebersberger et al. 2002), we compared the parsimony number of transversion events per base (tRNA) for aligned macroRNAs against the equivalent counts (tAR) for aligned ARs. Again, these distributions were significantly different (P < 10−15; two-sided Kolmogorov-Smirnov test), with median values of tRNA/tAR ratios for macroRNAs of 0.894 (mouse–rat) and 0.863 (mouse–human) (Table 1; Fig. 1C,D).
Although differential CpG content does not appear to explain the observed higher divergence within putatively neutral AR sequence compared to macroRNAs, we remained concerned that other AR-specific sequence features might underlie this difference. We therefore constructed a second, independent set of putative neutral sequence. For this, we considered all intergenic and nonrepetitive sequence, not overlapping with, but in the vicinity of, macroRNAs. To remove the majority of functional sequence, we discarded from this set regions that exhibit the signature of purifying selection upon indels (see Methods). Comparisons of macroRNAs with this second set of putative neutral sequence also demonstrated significant suppression of substitution and transversion rates within macroRNAs (Supplemental Fig. S1).
We were also concerned that this signature of purifying selection might be associated less with macroRNAs, and more with cis-regulatory elements, unannotated alternative first exons, or other elements of protein-coding genes. To investigate this, we repeated these analyses, now including only those macroRNAs located at least a well-defined distance away from protein-coding genes. For all substitution and transversion rate analyses, macroRNA sequences located >60 kb (or >10 kb, or >30 kb) from Ensembl protein-coding genes were seen to exhibit evolutionary rate distributions similar to those of the complete candidate macroRNA set (Table 1; Supplemental Figs. S2 and S3). These results re-emphasize the suppression of substitution rates in macroRNAs and further suggest that protein-associated regulatory regions do not contribute the only signature of substitution rate suppression from our putative macroRNA data set.
Although the vast majority of ARs appear to have evolved neutrally, it was possible that ARs harbored within macroRNAs might have been under greater constraint than neighboring ARs lying outside. However, we determined that rates of substitutions, or of transversions, within ARs inside and outside of macroRNAs were not significantly different at the 5% level (Supplemental Figs. S4 and S5). This held true for LINEs, LTRs, SINEs, or DNA transposons, whether considered together or in separate repeat classes. While substitution or transversion rates inside macroRNAs are reduced in general, such reductions thus do not appear to have occurred uniformly throughout each transcript.
Finally, we extended our pairwise sequence comparison and examined whether multispecies conserved sequences (MCSs) (Siepel et al. 2005) are enriched within our macroRNA set. These MCSs are mouse sequences that are well conserved with four multiple amniote species (rat, human, dog, and chicken). After accounting for biases in G+C composition, we find that these MCSs are significantly over-represented in our macroRNA set compared to their average density in intergenic sequence (2.18-fold increase, P < 10−4).
Suppressed rates of insertion/deletion (indel) mutations
We next considered a second mutational process that might provide an additional signature of purifying selection complementary to that from point mutations. We analyzed a 90-Mb set of human, mouse, and dog alignments that are uninterrupted by insertions or deletions over relatively long (approximately >80–100 bp) stretches. These, which we term “indel-purified segments” (IPSs), were identified previously at a false-discovery rate of 10% by comparing predictions of a neutral indel model with observations on real data (Lunter et al. 2006). Whereas false-positive segments within this set are expected to be uniformly distributed across the mammalian genome, any significant enrichment of IPSs within our macroRNA set is proposed to indicate the past action of purifying selection on deleterious indels.
Indeed, we find that IPSs are strongly and significantly over-represented within macroRNAs compared with their density in intergenic sequence (1.78-fold increase, P < 10−4). In these analyses, we took care to account for relevant nucleotide composition (G+C) biases (see Methods). In order, once more, to exclude the possibility of protein-coding genes contributing to our findings, we also restricted both the macroRNAs and the intergenic space to regions at a minimum distance of 10 kb, 30 kb, and 60 kb away from the nearest Ensembl protein-coding genes. For these sets, the significant over-representations remained and, indeed, progressively increased in magnitude (1.95-fold, 2.15-fold, and 2.32-fold, respectively; all P < 10−4).
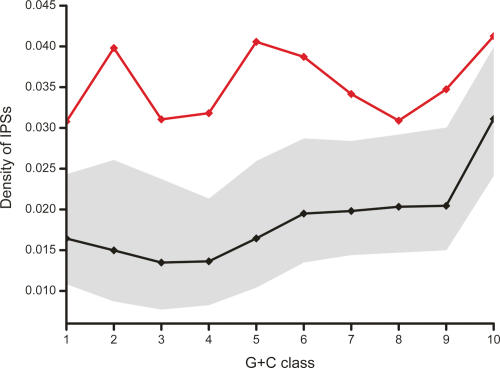
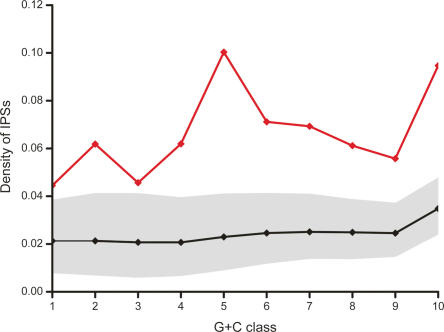
We next wished to investigate whether the observed associations with purifying selection exhibited any G+C biases. Consequently, we returned to considering all intergenic sequence and separately analyzed the density of IPSs for 10 sequence classes each with approximately equal G+C content. These classes were designed to partition 10-kb windows, from the intergenic portion of the mouse genome, into 10 equally populated isochores (see Methods). Across all 10 G+C classes we found significant over-representations of IPSs within our macroRNA data set, ranging from a 1.33-fold increase for the highest G+C class, to a 1.87-fold increase for the most A+T-rich sequence (Fig. 2).
Figure 2.
Density of indel-purified segments in macroRNA transcripts. Shown are the IPS densities within macroRNA transcripts (red line) for 10 G+C content bins (horizontal axis), and the expected density based on the intergenic distribution of IPSs (black line; the gray band indicates 95% confidence intervals obtained by randomization; see Methods for details). The IPS densities within macroRNAs exceed significantly the levels expected in G+C-matched intergenic sequence, indicating the past action of purifying selection.
These results should not be taken to imply that A+T-rich transcripts contain more functional sequence than G+C-rich ones. IPSs, and all functional segments, are considerably more abundant within high G+C sequence (Lunter et al. 2006), so that the relatively modest over-representation of 1.33-fold in the highest G+C category represents a large overall increase in IPS density. The density of IPSs in high G+C macroRNAs is 4.1%, whereas in A+T-rich transcripts it is 3.1%. Since ∼75% of conserved functional sequence is expected to be found within IPS segments (Lunter et al. 2006), this suggests that our candidate set of putative macroRNAs contains, on average, a considerable fraction of functional material (4.1%–5.5%,), with the highest density present in G+C-rich sequence. This is similar to the proportion (5.2%; 56.9 of 1083 Mb; Ensembl annotations) of coding sequence in protein-coding transcripts.
macroRNAs often possess conserved splice sites
The splicing of introns from pre-mRNAs of protein-coding genes requires 5′-donor and 3′-acceptor site motifs, which most often are GT and AG intronic terminal dinucleotides (Sheth et al. 2006). We next examined whether the 33% (1042/3122) of all mouse macroRNAs that possess introns exhibit higher than expected conservation of the GT-AG splice site dinucleotides in orthologous human and rat sequence. If so, this might indicate functional constraint, over tens of millions of years, on the maturation of pre-mRNAs. Of 1985 mouse macroRNA intron annotations, 87% were found to possess the canonical GT-AG splice site consensus sequence motifs at their 5′-donor and 3′-acceptor sites.
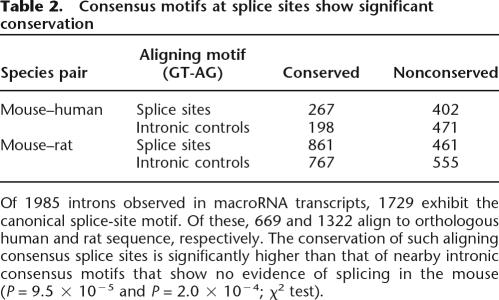
The association of pre-mRNA splicing with these consensus dinucleotides need not imply function, because the splicing machinery might have been recruited inconsequentially to consensus sites within otherwise nonsensical transcripts. To assess the functional significance of the consensus splice sites, we investigated their conservation in orthologous human or rat sequence. Against this, we compared the level of conservation of proximal and intronic GT and AG dinucleotides that are not known to be splice-site signals. We observed that 40% and 65% of mouse macroRNA GT-AG splice sites are conserved in human and rat, respectively, significantly more than for intronic GT and AG dinucleotides not involved in splicing (30% and 58%, respectively; P = 9.5 × 10−5 and P = 2.0 × 10−4; χ2 test; see Methods; Table 2).
Table 2.
Consensus motifs at splice sites show significant conservation
Of 1985 introns observed in macroRNA transcripts, 1729 exhibit the canonical splice-site motif. Of these, 669 and 1322 align to orthologous human and rat sequence, respectively. The conservation of such aligning consensus splice sites is significantly higher than that of nearby intronic consensus motifs that show no evidence of splicing in the mouse (P = 9.5 × 10−5 and P = 2.0 × 10−4; χ2 test).
To determine whether spliced and unspliced macroRNAs exhibit different signatures of purifying selection, we split the set into 1208 multi-exon and 1914 single-exon macroRNAs. Both subsets are significantly enriched in IPS sequence, and to similar degrees (1.85-fold and 1.80-fold, respectively; both P < 10−4). Single-exon macroRNAs exhibit a greater suppression of substitution rates when compared to the corresponding human and rat counterparts (9% vs. 3% in single-exon vs. multi-exon macroRNAs for human; 13% vs. 8% for rat), as expected if macroRNA exons show a higher average conservation than their introns, as is the case for protein-coding transcripts.
Conservation within macroRNA promoters
As functional elements, promoters would be expected to have been subject to purifying selection, and thus to have evolved more slowly than neutral sequence. To investigate constraint within promoters, we surveyed the evolutionary trends of macroRNA core promoter sequences, taken to be the 400 bp upstream (Cooper et al. 2006) of experimentally determined transcription start sites.
We tested first whether these putative core promoter sequences appeared to be evolutionarily conserved with respect to substitutions, by comparing the mouse promoter sequences with their orthologous human and rat counterparts. In both comparisons, the substitution rate within promoter sequences (dpro) was found to be significantly lower than dAR (P < 10−15; two-sided Kolmogorov-Smirnov test) (Fig. 3A,B). To account for potential CpG effects, we next considered the rate of transversions. For both the mouse–human and mouse– rat comparisons, we again observed transversion rate (tpro) distributions that are significantly different and below those of tAR (P < 10−15; two-sided Kolmogorov-Smirnov test) (Fig. 3C,D).
Figure 3.
Strong conservation of macroRNA promoters. Panels show the cumulative distributions of substitution (A,B) and transversion rates (C,D), as measured on the core putative promoter regions of macroRNA transcripts (red curves; 0–400 bp upstream of transcription start site), and the same rates on nearby AR sequence of the same length (black curves). Mouse macroRNA putative core promoter regions exhibit significantly suppressed substitution and transversion rates compared to selectively neutral ARs (P < 10−15 for all panels). This is true both for mouse–human (A,C) and mouse–rat (B,D) comparisons.
We also observed a clear signature of purifying selection on indels within promoters. IPSs were strongly over-represented within promoters (2.70-fold increase; P < 10−4), with 7.0% of the core promoter regions being contained within IPSs (compared with an expected IPS density of 2.6% within all intergenic G+C-matched sequence). Similar over-representations were seen when analyzing each G+C class separately (2.09-fold to 4.37-fold enrichments; P < 0.014 for all classes; one-sided test), indicating that, just as for the transcripts themselves, promoter regions show evidence of purifying selection across the G+C spectrum. The density of IPSs within promoters did vary considerably with G+C content, with promoters in G+C-rich regions showing very high densities of IPSs (9.5%; 3.5% expected), whereas IPS enrichments within promoters of A+T-rich regions were more modest (4.4%; 2.1% expected) (Fig. 4).
Figure 4.
Density of indel-purified segments in macroRNA promoters. Shown are the IPS densities within 400-bp regions upstream of macroRNA transcripts (red line) for 10 G+C content bins (horizontal axis), and the expected density based on the intergenic distribution of IPSs (black line; the gray band indicates 95% confidence intervals). IPS densities are substantially and significantly higher within putative core promoter regions.
Next, we identified within the promoter set (1) 450 TATA-driven promoters and (2) 448 CpG-associated promoters (including 28 promoters classified as both TATA-driven and CpG-associated). Putative TATA-boxes were identified, as previously (Ponjavic et al. 2006), using position weight matrices (Bucher 1990); CpG-associated promoters were classified based on the overlap to predicted locations of CpG islands obtained from the UCSC Genome Database (Hinrichs et al. 2006) (see Methods). For each promoter type, we analyzed the patterns of indel-purifying selection on the promoter and on their downstream transcripts as before. We observed significant over-representations of IPSs within both TATA-driven and CpG-associated promoters (2.24-fold, P = 2.1 × 10−3 and 7.19-fold, P < 10−4, respectively), and, to a lesser extent, within their associated transcripts (1.93-fold, P = 2.7 × 10−3 and 1.49-fold, P = 1.3 × 10−2, respectively). As might now be expected, substitution and transversion rates (dpro, tpro) were also significantly (P < 2.5 × 10−8) exceeded by rates (dAR, tAR) in local ARs, for each promoter class individually (Table 1; Supplemental Figs. S6 and S7).
Discussion
We have provided evidence for the suppression of substitution and transversion rates, by between 3% and 40% (Table 1), within a large set of macroRNA transcripts and their promoters. The same sequences also have experienced fewer indel mutations and fewer splice site consensus dinucleotide changes than expected by our neutral models. We interpret these results as indicating that the macroRNAs we investigated are enriched in sequence that has been subject to purifying selection to conserve the functional integrity of three main aspects of a functional transcript: its primary sequence, its promoter sequence, and its pattern of splicing.
We considered, but then discounted, the possibility that these observations arise from decreased rates of mutation, as opposed to purifying selection, within these transcripts. First, we considered whether substitution, insertion, and deletion rates would be decreased because of preferential repair of sequence within macroRNAs (“transcription-coupled repair”) (Svejstrup 2002), relative to repair within neighboring ARs that are not necessarily transcribed. We found no evidence for transcription-coupled repair because the rates of substitution or transversion for ARs either within, or else those neighboring, macroRNAs were not significantly different. It must be pointed out, however, that even if we had observed evidence for transcription-coupled repair, then this would, of itself, represent a signature of purifying selection. This is because for the signature of repair to have become apparent, transcription would have needed to have occurred over a time interval extended beyond that for inconsequential transcripts.
Second, we considered whether mutational biases, arising from single and dinucleotide (specifically, CpG) sequence composition, were associated with the suppression of substitution rates observed within macroRNAs. To account for the higher mutability of methylated CpG dinucleotides we considered transversions, rather than substitutions, and once more observed significantly suppressed rates in macroRNAs. Again, however, we note that even if CpG-associated mutations were to be, in general, higher in ARs than in macroRNAs, then this might indicate sustained functionality of macroRNAs, since CpG methylation is known to be incompatible with transcriptional activity (Ng and Bird 1999).
We also ensured that we controlled for nucleotide composition biases and large-scale mutation rate variations in our analyses (see Methods) by only comparing macroRNAs against putatively neutral sequence in the vicinity of the macroRNA. Previous analyses, which had not found differences in conservation levels between noncoding RNA, and other, sequences (Wang et al. 2004; Lau et al. 2006; Pang et al. 2006), did not consider G+C content as a possible confounding variable, despite the well-known relationship between G+C content and neutral substitution rates (Hardison et al. 2003). It is likely that this accounts for our observation of substitution rate suppression in macroRNAs, not seen by these other investigators. This is because noncoding transcripts are enriched in high G+C sequences, and such sequences possess elevated neutral rates, whereas intergenic, or other putatively neutral sequence, on average, exhibits lower G+C content and thus lower neutral rates.
For these reasons, we believe that purifying selection, rather than mutational biases, underlie the observed suppression of substitution, transversion, and indel rates in macroRNA sequence. We do not mean to imply that our evidence necessarily indicates that all macroRNAs in our set have been subject to evolutionary constraint throughout the ∼90 Myr separating humans and mice from their last common ancestor. It is possible that some macroRNAs are, indeed, wholly or partly “transcriptional noise,” and others may have been more ephemeral, having been subject to selection only in much shorter time periods (Ponting and Lunter 2006). Mouse macroRNAs that arose more recently, after the divergences of human or rat lineages from the mouse lineage, would present increasingly less evidence for purifying selection than would more ancient macroRNAs. As some ncRNAs, for example, Air and Xist (Oudejans et al. 2001; Duret et al. 2006), are well-known as being lineage-specific, this remains a strong possibility.
While our filtering procedure ensures that no known gene or any of its close homologs has any overlap with our macroRNAs, unannotated short peptides might still have passed our coding filters. To consider whether protein-coding contaminants explain our results, we created a conservative secondary test set of macroRNAs (2303 transcripts), by excluding all those that show any overlap with GenScan-predicted gene transcripts (Burge and Karlin 1997). This test set showed similar signatures of purifying selection as in the complete set (substitution rates suppressed by 5% and 11% in mouse–human and mouse–rat comparisons, respectively; 1.65-fold enrichment of IPSs; P < 10−4). We conclude that the macroRNA set contains a large number of functional noncoding transcripts and few, if any, protein-coding sequences.
We do not mean to imply that the entire lengths of macroRNAs represent functional sequence, even after accounting for transcription run-through. In particular, those transposable elements present within macroRNAs, appear, on average, not to have been subject to selection. A general picture emerges of macroRNAs harboring a density of functional sequence (4.1%–5.5%), similar to the density of coding exons within protein-coding genes (5.2%). This low amount of functional material may explain, in part, why these macroRNA transcripts were considered previously to be nonfunctional.
As observed previously (Carninci et al. 2005), macroRNA promoters, in particular, are often better conserved than their transcript sequences. Of the set we considered, CpG-associated promoters are the best conserved, with a 40% suppression of substitution rate between mouse and rat sequences, and an impressive 7.2-fold enrichment in IPSs.
If, as now appears likely, many macroRNAs have been subject to purifying selection, then what might be their functions? The greater constraint we observed within promoters than within transcript sequences is consistent with some, but not all, of the macroRNA transcripts possessing functions that are independent of their sequences. Transcription of such macroRNAs might induce a more open chromatin state that would be more amenable for the transcription of neighboring genes (Gribnau et al. 2000; Schmitt and Paro 2004). We note that individual macroRNAs showing evidence of selection are not specific to any one tissue or organ, neither are they evenly placed on the genome, with many of these full-length transcripts adjacent to the untranslated regions of protein-coding genes. These macroRNAs’ functions might thus be diverse, with possible contributions to the development and function of the nervous system (Mehler and Mattick 2006) and of spermatozoa (Miller et al. 1999), for example. That we know little of them has clearly been because of the scarcity of evidence demonstrating their importance (Huttenhofer et al. 2005; Mendes Soares and Valcarcel 2006). We hope that our findings now provide an incentive for detailed investigations of these enigmatic molecules.
Methods
Experimental data sources
We used the stringent sets of putative ncRNAs from the mouse (Mus musculus) FANTOM2 (4280 transcripts) (Okazaki et al. 2002; Numata et al. 2003; Pang et al. 2005) and FANTOM3 projects (2886 transcripts) (Carninci et al. 2005; ftp://fantom.gsc.riken.jp/FANTOM3/noncoding/README.txt). These were identified using several filtering procedures (for details, see Okazaki et al. 2002; Carninci et al. 2005) and therefore represent the strongest candidates for noncoding sequences from among the full-length FANTOM cDNA collection. Nevertheless, to exclude the possibility that signatures of purifying selection on transcripts are caused by the occurrence of unannotated protein-coding exons within them, we applied three additional conservative filtering steps. We removed a macroRNA if it fulfilled any of the following criteria: (1) Its nonrepetitive sequence yields a significant BLASTX (Altschul et al. 1990) hit to a known protein in the National Center for Biotechnology Information’s nonredundant (nr) protein database (E-value < 10−3). For this, we did not consider significant sequence similarity to “hypothetical,” “unknown,” or unnamed sequences as being sufficient evidence for protein-coding sequence, since short open-reading frames are often predicted in transcribed sequence that are not supported by evolutionary conservation in other mammals. (2) It overlaps on the same strand with a transcript of any Ensembl-annotated mouse protein-coding gene (Birney et al. 2006) (assembly mm5, obtained from Hinrichs et al. 2006). (3) It overlaps on its complementary strand with >20% of its length with the closest transcriptional start or end position of any Ensembl-annotated protein-coding transcript (in the remaining macroRNA set 97% [3030/3122] do not overlap at all). The data set used for the subsequent analyses consisted of 3122 putative macroRNAs.
To create a secondary test set to exclude the possibility of small peptide contaminants, we excluded those macroRNAs from the candidate set that overlap with the predicted transcripts of GenScan exons (Burge and Karlin 1997) obtained from the UCSC Browser (Hinrichs et al. 2006) and are transcribed from the same strand. Note that it suffices to remove potential peptides on the sense strand, since any random association with antisense peptides will occur, to the same degree, within randomized samples, whereas any nonrandom overlap suggests a biological role of the antisense transcript and thus legitimately contributes to any association.
Partition of intergenic space
All macroRNAs in our set are located within the intergenic space of Ensembl-annotated protein-coding genes (see above). To distinguish between macroRNAs that are closely or distantly located to the protein-coding genes, we created four overlapping partitions of this intergenic space based on the minimum distance l to the nearest transcriptional start or end base of Ensembl-annotated transcripts: (1) l > 0 kb, the complete intergenic space, (2) l > 10 kb, (3) l > 30 kb, and (4) l > 60 kb. The choices of l were informed by the median physical distance between Ensembl protein-coding genes in the same orientation, which is 60.9 kb. If not further indicated in the following section, we refer to analyses considering the complete intergenic space (i).
Definition of macroRNA core promoters and further classification
We defined the core promoter region for macroRNAs as the region extending from 400 bp upstream up until an associated transcription start site (Carninci et al. 2006; Cooper et al. 2006). Additionally, we classified these promoters into (1) CpG-island-associated promoters, if the majority of the region overlaps with a predicted CpG-island (annotation taken from the UCSC Genome Browser Database [assembly mm5] [Hinrichs et al. 2006]) and (2) TATA-box driven promoters, as previously (Ponjavic et al. 2006). Using the TFBS Perl module (Lenhard and Wasserman 2002), we scanned for TATA-boxes 40–20 bp upstream of the transcription’s first base with the TATA model constructed by Bucher (1990) deposited in the JASPAR database (Vlieghe et al. 2006). We accepted site predictions on the same strand as the transcript that exceeded a relative score threshold of 75%.
Nucleotide substitution and transversion rates in noncoding genomic DNA
We independently extracted both the mouse macroRNA (without distinguishing exonic from intronic sequence) and its core promoter genomic sequence (from here on referred to as “noncoding segments”) and identified putatively orthologous genomic regions in human (Homo sapiens) and rat (Rattus norvegicus), using the mouse–human and mouse–rat BlastZ NET alignments from the UCSC Genome Browser Database (Schwartz et al. 2003; Hinrichs et al. 2006) (assembly mm5, hg17, and rn3), and the AT Perl libraries for genome analysis (P. Engström, M. Andersen, A. Sandelin, D. Fredman, and B. Lenhard, in prep.). We discarded alignments for those cases in which a mouse noncoding segment mapped to multiple locations in either human or rat genomes.
We estimated the nucleotide substitution rates between orthologous mouse–human and mouse–rat aligned sequences using baseml with the REV substitution model (Yang 1994). All alignments were masked for transposable elements using RepeatMasker annotations (Smit 1999) obtained from the UCSC Genome Browser Database (Hinrichs et al. 2006). To ensure the accuracy of these rate estimates, we only considered alignments of minimal length 1 kb for macroRNAs and 150 bp for promoter sequences, after removing transposable elements and gaps. We used mouse–human and mouse–rat ancestral repeats (ARs) as a proxy for neutrally evolving sequence. To obtain an estimate of the local neutral rate whose variance is matched to the substitution rate estimate for the noncoding segment, we selected local ARs with a total ungapped alignment length matching that of the noncoding segment. In this process, the local ARs must fulfill each of two criteria: (1) no overlap with its local noncoding segment and (2) a length of at least 100 bp.
To test whether the substitution rate estimates are not biased by the higher mutability of CpG dinucleotides, we additionally determined the transversion rate of each noncoding segment, since CpG-associated substitutions are largely transitions (Ebersberger et al. 2002). To implement this, we calculated for each noncoding segment the parsimonious number of transversion mutations between the identified mouse–human and mouse–rat aligned and nonrepetitive sequences, respectively, and normalized this count by the total number of nonrepetitive and ungapped alignment positions (minimum length for macroRNA, 1 kb; for promoter sequence, 150 bp). Local neutral transversion rates were obtained with the same procedure, but instead using ARs as identified above. Because the transversion rate is low and our interest is in relative rather than absolute rate estimates, Jukes-Cantor-type corrections for repeated substitutions were not necessary.
All analyses were performed independently for the four different intergenic spaces defined for (1) l > 0, (2) l > 10 kb, (3) l > 30 kb, and (4) l > 60 kb.
We further determined whether the substitution or transversion rates for ARs inside and outside of macroRNAs are different. We applied the same cross-species comparison procedure between aligned mouse–human and mouse–rat sequences, as described above, and required the minimum AR length within macroRNAs to be 100 bp. We analyzed each repeat class (LINE, SINE, LTR, and DNA transposon) individually, as well as pooled together.
In a next step, we created a second control of putatively neutral sequence that is independent from the neutral AR sequence defined above. We considered the intergenic sequence of Ensembl-annotated protein-coding genes from which we discarded mouse repetitive sequence (assembly mm5) obtained from Hinrichs et al. (2006) and segments of indel-purifying selection as defined elsewhere (Lunter et al. 2006), using the 10% false-discovery rate set (see also below). When performing the substitution and transversion analyses, we applied the same criteria as above and in addition required the minimum distance of the local alignable neutral intergenic segment to the macroRNA to be 1 kb.
Multispecies conserved sequences (MCSs) (Siepel et al. 2005) for assembly mm5 were obtained from the UCSC Browser Database (Hinrichs et al. 2006).
Indel-purifying selection in noncoding genomic DNA
We performed a second, independent test to see whether noncoding segments show a significant association with purifying selection, using a genome-wide set of mouse DNA segments that have been subject to indel-purifying selection (referred to as IPSs), which were previously identified at a false-discovery rate of 10% (Lunter et al. 2006). These segments were identified using a “neutral indel model” that utilizes the evolutionary impact of insertions and deletions (indels) to identify functional DNA sequence under purifying selection. We investigated whether the noncoding segments show an over-representation of these IPS segments, when compared to the expected coverage if the IPS segments were to be uniformly distributed within the intergenic space. In this test, we accounted for any G+C-content biases (for details, see below).
This analysis for the noncoding transcripts was performed independently for the four different intergenic spaces defined above (l > 0 to l > 60 kb), whereas for their core promoter sequences, it was carried out using the complete intergenic space (l > 0). To investigate the association of these noncoding segments with IPSs depending on G+C class, we performed the association study within each of the 10 G+C classes separately.
Genome-wide partition based on G+C-content
We divided the mouse genome into nonoverlapping 10-kb windows and partitioned these into 10 equally populated categories by G+C content using the defined 10th percentiles: 0, 0.213, 0.365 0.379, 0.389, 0.400, 0.411, 0.423, 0.437, 0.454, 0.478, 0.651, 1.
Genome-wise association procedure controlling for G+C-content biases
When determining the association of noncoding segments S with other genomic elements E (such as IPSs or MCSs) within an intergenic space I, it is essential to control for G+C-content biases, since both noncoding segments and the genomic elements we consider show nonuniform distributions with respect to G+C content.
The basis for the procedure is a randomization test, which compares the intersection S ∩ E ∩ I with randomized intersections, S′∩ E ∩ I, where S′ is a randomized set of segments whose length distribution is matched with that of S overlapping with I, and whose locations are chosen uniformly across I. To account for G+C biases, the genome is partitioned into 10 G+C classes C1 . . . C10 as described above, and the same procedure is applied with S, E, and I replaced by S ∩ Ci, E ∩ Ci, and I ∩ Ci, for i = 1, . . . , 10, resulting in 10 randomized sets Si′. This process was performed independently for each chromosome, to account for any chromosome-specific effect, resulting in 210 randomized sets (10 for each mouse chromosome). To obtain P-values for any observed over- or under-representation, this procedure was repeated 10,000 times, and the number of nucleotides in the original intersection S ∩ E ∩ I was compared with the distribution of nucleotides in the combined intersection (S′1 ∪ . . . ∪ S′210) ∩ E ∩ I; the proportion of times this number exceeded (fell short of) the observed number was reported as the one-sided P-value for under-representation (over-representation). We added one pseudo-count to the randomized number to avoid reporting spuriously low P-values. The ratio of the expected number to the observed number was used to calculate the percentage under- or over-representation.
Determining splice-site consensus in orthologous mouse–human and mouse–rat introns
We retrieved the intron–exon boundaries for the mouse macroRNA sequences from the UCSC Genome Browser Database (Hinrichs et al. 2006) (1985 introns in total). Although we chose a minimum intron length of 4 bp, to accommodate the two dinucleotide motifs, only 4% of identified introns were of length 50 bp or less. We then searched for the splice site consensus sequence motif having a GT at the 5′-donor splice site (positions +1, +2 of the 5′-intron) and a AG at the 3′-acceptor splice site (−1, −2 of the 3′-intron) in the mouse macroRNA introns. For those introns possessing this splice-site consensus, we examined the orthologous regions in the human and rat genomes using their respective alignments (see above), and counted (1) how many were alignable to all four nucleotides, and (2) how many showed a fully conserved GT-AG consensus site at these locations.
To test for conservation, we scanned along the intron locating the first 5′-GT and 3′-AG dinucleotides that did not overlap with the splice site and that could be aligned to human or rat sequence. We counted the number of times both putatively neutral GT and AG sites were conserved. These two counts were compared using a χ2 test.
Statistical methodology
We used the R language (Ihaka and Gentleman 1996) and Python for statistical analysis and visualization.
Acknowledgments
We thank Andreas Heger and Caleb Webber for generously providing their toolsets, and members of the C.P.P. research group for advice and helpful discussions. We thank the UK Medical Research Council (MRC) for financial assistance. J.P. gratefully acknowledges a graduate Clarendon Award, Oxford Balliol College Domus Award, and a graduate scholarship by the Studienstiftung des deutschen Volkes. G.L. is a MRC Bioinformatics Research Fellow.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6036807
References
- Allen J.E., Pertea M., Salzberg S.L., Pertea M., Salzberg S.L., Salzberg S.L. Computational gene prediction using multiple sources of evidence. Genome Res. 2004;14:142–148. doi: 10.1101/gr.1562804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Badger J.H., Olsen G.J., Olsen G.J. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Tongprasit W., Samanta M., Weissman S., Samanta M., Weissman S., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., Cox T., Cunningham F., Curwen V., Cutts T., Cunningham F., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake W.J., Kaern M., Cantor C.R., Collins J.J., Kaern M., Cantor C.R., Collins J.J., Cantor C.R., Collins J.J., Collins J.J. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S., Ashworth A., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S., Kay G.F., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S., McCabe V.M., Norris D.P., Cooper P.J., Swift S., Rastan S., Norris D.P., Cooper P.J., Swift S., Rastan S., Cooper P.J., Swift S., Rastan S., Swift S., Rastan S., Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Burge C., Karlin S., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Oyama R., Ravasi T., Lenhard B., Wells C., Ravasi T., Lenhard B., Wells C., Lenhard B., Wells C., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Taylor M.S., Engstrom P.G., Frith M.C., Engstrom P.G., Frith M.C., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Cooper D.N., Youssoufian H., Youssoufian H. The CpG dinucleotide and human genetic disease. Hum. Genet. 1988;78:151–155. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Cooper S.J., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Trinklein N.D., Anton E.D., Nguyen L., Myers R.M., Anton E.D., Nguyen L., Myers R.M., Nguyen L., Myers R.M., Myers R.M. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Chureau C., Samain S., Weissenbach J., Avner P., Chureau C., Samain S., Weissenbach J., Avner P., Samain S., Weissenbach J., Avner P., Weissenbach J., Avner P., Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Ebersberger I., Metzler D., Schwarz C., Paabo S., Metzler D., Schwarz C., Paabo S., Schwarz C., Paabo S., Paabo S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno M., Kasukawa T., Saito R., Adachi J., Suzuki H., Baldarelli R., Hayashizaki Y., Okazaki Y., Kasukawa T., Saito R., Adachi J., Suzuki H., Baldarelli R., Hayashizaki Y., Okazaki Y., Saito R., Adachi J., Suzuki H., Baldarelli R., Hayashizaki Y., Okazaki Y., Adachi J., Suzuki H., Baldarelli R., Hayashizaki Y., Okazaki Y., Suzuki H., Baldarelli R., Hayashizaki Y., Okazaki Y., Baldarelli R., Hayashizaki Y., Okazaki Y., Hayashizaki Y., Okazaki Y., Okazaki Y. CDS annotation in full-length cDNA sequence. Genome Res. 2003;13:1478–1487. doi: 10.1101/gr.1060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno M., Pang K.C., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Pang K.C., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Kai C., Kawai J., Carninci P., Hayashizaki Y., Kawai J., Carninci P., Hayashizaki Y., Carninci P., Hayashizaki Y., Hayashizaki Y., et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney D.J., Keightley P.D., Keightley P.D. The scale of mutational variation in the murid genome. Genome Res. 2005;15:1086–1094. doi: 10.1101/gr.3895005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J., Diderich K., Pruzina S., Calzolari R., Fraser P., Diderich K., Pruzina S., Calzolari R., Fraser P., Pruzina S., Calzolari R., Fraser P., Calzolari R., Fraser P., Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- Hardison R.C., Roskin K.M., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Roskin K.M., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Yang S., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Diekhans M., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Kent W.J., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Weber R., Elnitski L., Li J., O’Connor M., Kolbe D., Elnitski L., Li J., O’Connor M., Kolbe D., Li J., O’Connor M., Kolbe D., O’Connor M., Kolbe D., Kolbe D., et al. Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 2003;13:13–26. doi: 10.1101/gr.844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., Diekhans M., Furey T.S., Harte R.A., Hsu F., Furey T.S., Harte R.A., Hsu F., Harte R.A., Hsu F., Hsu F., et al. The UCSC Genome Browser database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A., Schattner P., Polacek N., Schattner P., Polacek N., Polacek N. Non-coding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Hyashizaki Y. Mouse transcriptome: Neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431:757. [PubMed] [Google Scholar]
- Ihaka R., Gentleman R., Gentleman R. R: A language for data analysis and graphics. J. Comput. Graph. Statist. 1996;5:299–314. [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E., Nakano T., Bartel D.P., Kingston R.E., Bartel D.P., Kingston R.E., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lenhard B., Wasserman W.W., Wasserman W.W. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- Lunter G., Ponting C.P., Hein J., Ponting C.P., Hein J., Hein J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput. Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N., Kasukawa T., Oyama R., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Kasukawa T., Oyama R., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Oyama R., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., Aturaliya R.N., Batalov S., Beisel K.W., Batalov S., Beisel K.W., Beisel K.W., et al. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler M.F., Mattick J.S., Mattick J.S. Non-coding RNAs in the nervous system. J. Physiol. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes Soares L.M., Valcarcel J., Valcarcel J. The expanding transcriptome: The genome as the ‘Book of Sand.’. EMBO J. 2006;25:923–931. doi: 10.1038/sj.emboj.7601023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D., Briggs D., Snowden H., Hamlington J., Rollinson S., Lilford R., Krawetz S.A., Briggs D., Snowden H., Hamlington J., Rollinson S., Lilford R., Krawetz S.A., Snowden H., Hamlington J., Rollinson S., Lilford R., Krawetz S.A., Hamlington J., Rollinson S., Lilford R., Krawetz S.A., Rollinson S., Lilford R., Krawetz S.A., Lilford R., Krawetz S.A., Krawetz S.A. A complex population of RNAs exists in human ejaculate spermatozoa: Implications for understanding molecular aspects of spermiogenesis. Gene. 1999;237:385–392. doi: 10.1016/s0378-1119(99)00324-8. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Bird A., Bird A. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Numata K., Kanai A., Saito R., Kondo S., Adachi J., Wilming L.G., Hume D.A., Hayashizaki Y., Tomita M., Kanai A., Saito R., Kondo S., Adachi J., Wilming L.G., Hume D.A., Hayashizaki Y., Tomita M., Saito R., Kondo S., Adachi J., Wilming L.G., Hume D.A., Hayashizaki Y., Tomita M., Kondo S., Adachi J., Wilming L.G., Hume D.A., Hayashizaki Y., Tomita M., Adachi J., Wilming L.G., Hume D.A., Hayashizaki Y., Tomita M., Wilming L.G., Hume D.A., Hayashizaki Y., Tomita M., Hume D.A., Hayashizaki Y., Tomita M., Hayashizaki Y., Tomita M., Tomita M. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 2003;13:1301–1306. doi: 10.1101/gr.1011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Nikaido I., Osato N., Saito R., Suzuki H., Osato N., Saito R., Suzuki H., Saito R., Suzuki H., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Oudejans C.B., Westerman B., Wouters D., Gooyer S., Leegwater P.A., van Wijk I.J., Sleutels F., Westerman B., Wouters D., Gooyer S., Leegwater P.A., van Wijk I.J., Sleutels F., Wouters D., Gooyer S., Leegwater P.A., van Wijk I.J., Sleutels F., Gooyer S., Leegwater P.A., van Wijk I.J., Sleutels F., Leegwater P.A., van Wijk I.J., Sleutels F., van Wijk I.J., Sleutels F., Sleutels F. Allelic IGF2R repression does not correlate with expression of antisense RNA in human extraembryonic tissues. Genomics. 2001;73:331–337. doi: 10.1006/geno.2001.6522. [DOI] [PubMed] [Google Scholar]
- Pang K.C., Stephen S., Engstrom P.G., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Stephen S., Engstrom P.G., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Engstrom P.G., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S., Lenhard B., Hayashizaki Y., Mattick J.S., Hayashizaki Y., Mattick J.S., Mattick J.S. RNAdb—A comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K.C., Frith M.C., Mattick J.S., Frith M.C., Mattick J.S., Mattick J.S. Rapid evolution of noncoding RNAs: Lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Lenhard B., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Lenhard B., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Carninci P., Hayashizaki Y., Sandelin A., Hayashizaki Y., Sandelin A., Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Lunter G., Lunter G. Signatures of adaptive evolution within human non-coding sequence. Hum. Mol. Genet. 2006;15(Suppl 2):R170–R175. doi: 10.1093/hmg/ddl182. [DOI] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., Fukuda S., Ru K., Frith M.C., Gongora M.M., Ru K., Frith M.C., Gongora M.M., Frith M.C., Gongora M.M., Gongora M.M., et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S., Paro R., Paro R. Gene regulation: A reason for reading nonsense. Nature. 2004;429:510–511. doi: 10.1038/429510a. [DOI] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Baertsch R., Hardison R.C., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Haussler D., Miller W., Miller W. Human–mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R., Roeder T., Krainer A.R., Sachidanandam R., Krainer A.R., Sachidanandam R., Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F., Zwart R., Barlow D.P., Zwart R., Barlow D.P., Barlow D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Springer M.S., Murphy W.J., Eizirik E., O’Brien S.J., Murphy W.J., Eizirik E., O’Brien S.J., Eizirik E., O’Brien S.J., O’Brien S.J. Placental mammal diversification and the Cretaceous–Tertiary boundary. Proc. Natl. Acad. Sci. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J., Bird A., Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup J.Q. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 2002;3:21–29. doi: 10.1038/nrm703. [DOI] [PubMed] [Google Scholar]
- Vlieghe D., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B., Wasserman W.W., van Roy F., Lenhard B., van Roy F., Lenhard B., Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang J., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K., Zhang J., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K., Zheng H., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K., Li J., Liu D., Li H., Samudrala R., Yu J., Wong G.K., Liu D., Li H., Samudrala R., Yu J., Wong G.K., Li H., Samudrala R., Yu J., Wong G.K., Samudrala R., Yu J., Wong G.K., Yu J., Wong G.K., Wong G.K. Mouse transcriptome: Neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431:757. Comment on Okazaki et al. 2002. [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., Mouse Genome Sequencing Consortium. et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yamada K., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Southwick A.M., Wu H.C., Kim C., Nguyen M., Wu H.C., Kim C., Nguyen M., Kim C., Nguyen M., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yang Z. Estimating the pattern of nucleotide substitution. J. Mol. Evol. 1994;39:105–111. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]