Abstract
Reverse genetic or gene-driven knockout approaches have contributed significantly to the success of model organisms for fundamental and biomedical research. Although various technologies are available for C. elegans, none of them scale very well for genome-wide application. To address this, we implemented a target-selected knockout approach that is based on random chemical mutagenesis and detection of single nucleotide mutations in genes of interest using high-throughput resequencing. A clonal library of 6144 EMS-mutagenized worms was established and screened, resulting in the identification of 1044 induced mutations in 109 Mbp, which translates into an average spacing between exonic mutations in the library of only 17 bp. We covered 25% of the open reading frames of 32 genes and identified one or more inactivating mutations (nonsense or splice site) in 84% of them. Extrapolation of our results indicates that nonsense mutations for >90% of all C. elegans genes are present in the library. To identify all of these mutations, one only needs to inspect those positions that—given the known specificity of the mutagen—can result in the introduction of a stop codon. We define these positions as nonsense introducing mutations (NIMs). The genome-wide collection of possible NIMs can be calculated for any organism with a sequenced genome and reduces the screening complexity by 200- to 2000-fold, depending on the organism and mutagen. For EMS-mutagenized C. elegans, there are only ∼500,000 NIMs. We show that a NIM genotyping approach employing high-density microarrays can, in principle, be used for the genome-wide identification of C. elegans knockouts.
The use of Caenorhabditis elegans as a model system has contributed increasingly to biological research over the past decades. Initially, C. elegans was introduced as a model organism allowing phenotype-driven (forward) genetic research (Brenner 1974). At present, this has resulted in the identification and elucidation of many novel components and pathways that were found to be involved in equivalent biological processes in other animal species, including mammals. The success of C. elegans as a model system was boosted by the development of powerful experimental technologies (Wood 1998), including reverse (gene-driven) genetics tools (for review, see Barstead 1999). With the completion and annotation of the whole genome sequence for C. elegans (C. elegans Sequencing Consortium 1998), gene-driven genetics has become very important for assigning function to protein-coding genes, as only ∼10% of the almost 20,000 encoded genes have been subject to systematic study. Efficient technology for disrupting gene function is therefore essential.
Currently, several methods are available for the genetic inactivation of genes in C. elegans. The first method developed is based on mutator strains that show a high rate of spontaneous mutations as a result of novel insertions from active transposable elements. Using such strains, animals can be recovered that carry Tc1 transposable elements in target loci of choice (Rushforth et al. 1993; Zwaal et al. 1993). However, transposon insertions do not always result in inactivation of the target gene, as the transposable element may be inserted in intronic regions or deleted from the transcript because of post-transcriptional splicing (Rushforth and Anderson 1996). Nevertheless, transposon insertions can be used to generate deletion derivatives that may result from imprecise excision after activation of transposon activity (Zwaal et al. 1993). As this approach is laborious and success is not guaranteed, chemical mutagens are preferred to introduce deletions in the C. elegans genome. Both ethylmethanesulfonate (EMS) and UV/trimethylpsoralen (TMP) have been used successfully to generate mutant libraries for PCR-based identification of deletions in genes of interest (Jansen et al. 1997; Liu et al. 1999), and modifications (Edgley et al. 2002) of the original PCR-based deletion detection protocol have increased the odds of finding the desired deletion as it improves the retrieval efficiency of smaller deletions that occur most frequently. Recently, gene disruption by homologous recombination has been demonstrated (Berezikov et al. 2004), but this method is laborious and not very amenable for scaling. Besides genetic approaches, RNAi-based approaches have proven their value for fast genome-wide screens for effects in a wide variety of phenotypic tests (Fraser et al. 2000; Gonczy et al. 2000). However, gene inactivation by RNAi may be spatially or temporally restricted or for some genes result in only partial (knockdown) or no gene inactivation at all. On the basis of our own experience, we find that RNAi-based and genetic knockout approaches are complementary to each other, each with their own specific experimental and technological advantages and disadvantages. Furthermore, we and others recently showed that systematic RNAi screens on genetic knockout backgrounds are very powerful for the mapping of genetic interaction networks (van Haaften et al. 2004; Lehner et al. 2006).
Here we describe the establishment of a cryopreserved mutant clonal library containing >12,000 EMS-mutagenized genomes. Although EMS induces genomic deletions at some level, the most frequently induced mutations are single base pair changes (Anderson 1995). We established a screening method for the identification of such point mutations by high-throughput dideoxy resequencing and found that the average spacing between induced mutations in the library is only 17 nucleotides. Our results suggest that there are nonsense or splice site mutations present in the library for >90% of all C. elegans genes. Although the resequencing approach presented here can be scaled relatively well, it is not suited for a genome-wide approach. We discuss an approach that could be used for larger scale or even genome-wide identification of gene knockouts by reducing the genomic screening complexity by three orders of magnitude.
Results and Discussion
Library construction
Although EMS is used in C. elegans reverse genetic approaches to introduce genomic deletions (Jansen et al. 1997), the most common type of DNA damage introduced by this mutagen are point mutations, primarily G/C to A/T transitions. The molecular mutation frequency at the standard 0.05 M dose is estimated to be ∼7 × 10−6 per mutagenized G/C base pair (Anderson 1995). Corrected for the genomic GC content of C. elegans (36%), this translates into a molecular mutation frequency of one induced mutation every 400,000 nucleotides per mutagenized genome. A clonal library of 6144 EMS mutagenized C. elegans was constructed as outlined in Figure 1A. P0 worms were EMS-mutagenized and F1 progeny were singled out and allowed to grow for two to three generations (10,000–50,000 worms). The resulting population was split in three parts, where two parts were used to cryopreserve the mutant stocks in duplicate in 96-well format and the third part was used for DNA extraction. In contrast to libraries generated for the identification of deletions, this library was not multiplexed, which, in combination with cryopreservation in duplicate, should facilitate the retrieval of mutants from the archive. Assuming that EMS induces only transitions from G/C to A/T at a frequency of 7 × 10−6 per G/C base pair per haploid genome and taking into account the C. elegans codon usage (Nakamura et al. 2000), one can calculate that for an average gene of 1000 coding bp, the expected frequency of silent, missense, and nonsense mutations will be 8.6 × 10−4, 1.8 × 10−3, and 1.7 × 10−4, respectively. For a library of 6144 animals, corresponding to 12,228 mutagenized haploid genomes, this translates to an expected induction of 10 silent, 21 missense, and two nonsense mutations per 1-kb gene. These numbers may be slight underestimates, because the mutation frequency per base is slightly higher in coding regions compared with the whole genome sequence as the GC content is higher in coding regions (43%) than in the whole genome (36%).
Figure 1.
Schematic outline of the mutant C.elegans library generation and screening. (A) Library generation: P0 worms were treated with EMS and F1 progeny were singled out and expanded clonally until the F2 to F3 generation. Cultures were split in two, where the first half was used for cryopreservation in duplicate and the other half for DNA isolation and subsequent mutation detection in genes of interest. (B) Library screening: Genic regions of interest were amplified by a nested PCR and screened for induced mutations by dideoxy resequencing. Mutations were scored by PolyPhred 5.0 (Nickerson et al. 1997) and stored and annotated in LIMSTILL (http://limstill.niob.knaw.nl; V. Guryev and E. Cullen, unpubl.). When an interesting mutation was identified, it was reconfirmed in an independent assay and the mutant was retrieved from the frozen archive, outcrossed, and analyzed for phenotypic consequences. (C) Nested PCR setup: In the first PCR multiple regions of interest are amplified in a multiplex of various oligo 1 and 4 combinations. The second PCR was performed with a single pair of M13-tailed oligos 2 and 3. The resulting products were sequenced using universal M13F and/or M13R oligonucleotides.
Library screening
Although several methods have been developed for single nucleotide mutation discovery and target-selected mutagenesis in various organisms (Bentley et al. 2000; McCallum et al. 2000; Perry et al. 2003; Wienholds et al. 2003; Quwailid et al. 2004; Winkler et al. 2005), we employed dideoxy resequencing, which directly provides sequence context information and is very well suited for automation, at both the experimental and the data interpretation levels. We implemented a robust and economical procedure for semiautomated high-throughput analysis of C. elegans mutant DNA (Fig. 1B), based on our work in zebrafish (Wienholds et al. 2002) and rat (Smits et al. 2006). First, DNA was isolated in 96-well format from half of the worms from a full-grown single 6-cm plate. Stock genomic DNA was diluted, providing sufficient starting material for 2000 PCR reactions. For mutation screening, we used a nested PCR approach in 384-well format to amplify coding regions of genes of interest (Fig. 1C). This approach allowed us to multiplex the first PCR (PCR1) at least up to 10-fold, increasing the total number of target sequences to be screened with the limited amount of DNA template. This gave the most uniform PCR product yields in the second PCR and allowed us to titrate down the oligo and dNTP concentrations in the second PCR (PCR2), making it possible to sequence diluted PCR2 products directly without the need for any expensive and laborious purification. Furthermore, oligos for the second PCR were tailed with universal M13 adapters allowing for standardized sequencing reactions under universal conditions. Sequencing results were semiautomatically processed using the PolyPhred packages (Stephens et al. 2006), custom scripts, and web-based front-ends (http://limstill.niob.knaw.nl; V. Guryev and E. Cuppen, unpubl.). We have optimized experimental and logistic procedures, resulting in a direct supply cost per sample (from genomic DNA to a sequencing read of max. 500 bp) of only 15 eurocents (including all chemicals and disposable involved but excluding investments in equipment and personnel costs). Direct supply costs for screening the complete library for a single amplicon would currently add up to ∼1000 Euro.
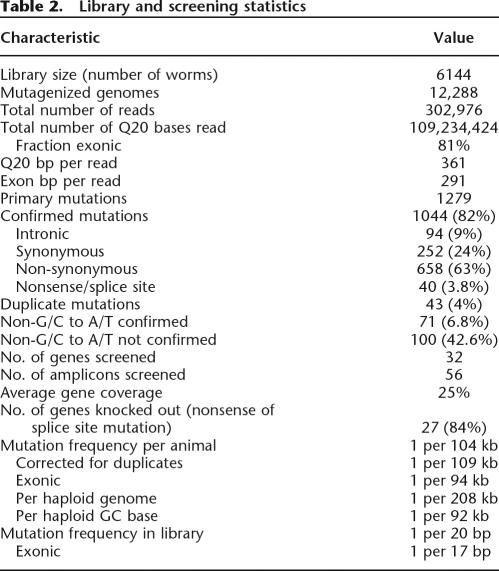
We have screened 56 amplicons in 32 different genes and identified 1279 candidate mutations (Tables 1, 2). Although we used relatively loose criteria for mutation calling to prevent false negatives, we reconfirmed 1044 mutations (82%) by an independent PCR and sequencing reaction (see Supplemental Material for details). Most false positives were non-G/C to A/T transitions (43%, 100 out of 235), although a significant number of non-G/C to A/T mutations (66, 6.3% of the total number of confirmed mutations) could be confirmed as well (Table 2).
Table 1.
Library screening overview
aWhen alternative splice forms were annotated, the longest open reading frame is shown.
bSome amplicons cover multiple exons.
cThe number of 384-well plates that were actually screened for a gene. Although the library consists of 16 384-well plates, we did not screen the complete library for all amplicons.
dOnly bases with a Phred score of ≥20 were included in the analysis.
ePercent of library screened is calculated by dividing the total number of exonic bases read by the multiplication of the open reading frame length of a gene with the total number of mutants (6144) in the library.
fThe number of non-G/C to A/T mutations is shown in parentheses.
gMutations that could not be reconfirmed in an independent PCR and resequencing assay. The number of non-G/C to A/T mutations is shown in parentheses.
Table 2.
Library and screening statistics
Mutation characteristics
An example of a gene (C01G5.2) that was screened very intensively is shown in Figure 2. As the oligos used for PCR amplification were mostly designed in the exon-flanking intronic segments and because various amplicons included multiple exons because of the compact organization of the C. elegans genome, some intronic mutations (94) were identified in the screen (Tables 1, 2). Although some of these could have an effect on the efficiency of mRNA splicing, such effects are extremely difficult to predict. Twelve mutations were found to affect a splice donor or acceptor site. Such mutations are frequent (up to 1% of all mutations) as the canonical splice donor/acceptor sites are relatively GC-rich. Most of these mutations are expected to result in a loss-of-function phenotype because of the introduction of a frameshift resulting from the use of alternative splice donor/acceptor sites or exon skipping. Even when the open reading frame remains intact, these mechanisms could result in the introduction of additional amino acids or deletion of parts of the mature protein, thereby affecting the function of the protein (domain) or resulting in an improperly folded, unstable mature protein.
Figure 2.
Schematic overview of the screening results for C01G5.2. The coding exons are shown in red and the five amplicons used to screen the majority of the gene are shown in black (first PCR product) and gray (second PCR product). All mutations (118 in total) are annotated on the bottom: intronic (8, black); silent (39, green); missense conserved (34, blue); missense nonconserved (33, pink); stop (3, red); and splice site (1, orange).
Most induced mutations reside in exonic segments and either are silent (252) or result in an amino acid change (658) or introduction of a premature stop codon (28) (Tables 1, 2). Although the ratio between silent and missense mutations is in the expected range, the percentage of identified premature stop codons is ∼3% of the exonic mutations, where theoretically one would expect 6%. Examination of potential premature stop codons that could be introduced in the amplicons that were screened did not reveal a sequence bias (i.e., mutation of 6.1% of the GCs would result in the introduction of a stop codon), indicating that there is a significant selection in the current library against premature stop codons, which may be caused by selection against lethals during clonal growth expansion. It should be mentioned that 6 out of 26 retrieved mutants have a sterile or lethal phenotype (Supplemental Table 2), indicating that this type of mutant can be retrieved from the library. On the other hand, loss of recessive lethal homozygotes during growth would decrease the amount of mutant alleles in the worm pool used for DNA isolation to 20% and 11% in the F3 and F4 generation, respectively, which could compromise PolyPhred-based mutation discovery and result in an increased false-negative rate.
The large category of nonsynonymous mutations could be of interest for studying protein subdomain function as they may have an effect on, for example, protein–protein interaction properties or enzymatic characteristics. As a result, both (partial) gain- or loss-of-function phenotypes can be expected. Especially when full knockout alleles result in lethality or sterility, more subtle protein changes may be very useful.
For 21 out of the 32 genes, one or multiple independent nonsense mutations were identified (Table 1). When assuming that all splice site and nonsense mutations result in a loss of function of the gene, 27 out of the 32 genes (84%) were knocked out by one or more mutations in the current screen. Knockout mutations may be present in the library for the remaining five genes as on average only 40% of the coding capacity of these genes was screened (Table 1).
Mutant retrieval
Although characterization of mutants is beyond the scope of this article, we found that all attempts for retrieval of the mutant alleles from the cryopreserved resource were successful, without the need to use the duplicate backup archive (Supplemental Table 2). One of the major advantages of the strategy used is that every sample is derived from a single mutagenized worm. Hence, every EMS-induced mutation was heterozygous in the F1 animal that was clonally expanded by self-fertilization. Without any selection pressure on the population, this would result in 75%, 62.5%, and 56% of the F2, F3, and F4 progeny, respectively, carrying at least one mutant allele. In the case of a recessive lethal mutation, homozygous mutants are removed from the population and these numbers would be 67%, 40%, and 22%, respectively. Although the lower levels of these latter numbers are not expected to interfere with mutant retrieval, a mutant-to-wild type allele ratio of 1:4 may become difficult to detect with the dideoxy sequencing-based screening setup and PolyPhred-based mutation discovery, resulting in an increased false-negative rate. This may explain the lower than expected observed frequency of premature stop codons, as discussed above. Upon thawing a sample from the cryopreserved mutant archive, we found that the number of viable worms varied between 10 and several hundred and that the percentage of worms carrying the mutant allele was between 10% and 80%—well within the range of the expected values. Several of the identified and retrieved mutants were found to be lethal when homozygous, illustrating that the library construction and screening procedure does not completely select against this type of mutation.
A complication that could occur because of the relatively high mutation load in the library is lethality due to a linked mutation, which would prevent the retrieval of the homozygous mutant. In only one instance where we expected to recover homozygous viable progeny (based on RNAi results) did we fail to do so (Supplemental Table 2). Perhaps this resulted from a closely linked lethal mutation. It should be mentioned that one more mutant, which eventually had no obvious phenotype, required substantially more effort to find homozygotes, suggesting the presence of a weakly linked mutation. Finally, we also retrieved a mutant that was almost exclusively homozygous in the initial thawed sample, most likely because of a lethal mutation on the other chromosome.
Taken together, in practice, retrieval of (homozygous) mutants does not seem to pose a significant problem. This observation can be substantiated by a theoretical calculation. With the observed mutation frequency of one per ±100 kb per animal, there are ∼1000 induced point mutations in the diploid genome and 500 per haploid genome. Given that ∼25% of the C. elegans genome is protein-coding, the estimate that ∼10% of the exonic mutations results in a loss of function of the gene (Wienholds and Plasterk 2004) and that approximately one-third of the genes in C. elegans are essential (Johnsen et al. 2000), approximately eight lethal mutations are expected per haploid genome, which equals <1.5 lethal per chromosome.
EMS-induced mutation spectrum in C. elegans
Our results provide the first large-scale and completely unbiased systematic analysis of the single base pair mutation frequency and spectrum for EMS in C. elegans. About 6.8% of the mutations are not G/C to A/T, which is well within range with forward (phenotype-driven) genetic data in C. elegans, for which it was found that 13 out of 238 EMS-induced single base pair mutations (5.5%) are not G/C to A/T (Anderson 1995), but much higher than the 1% found in Arabidopsis in a large-scale EMS-driven TILLING screen (Greene et al. 2003) and lower than the 16% observed in a similar but smaller-scale screen in Drosophila (Winkler et al. 2005).
We identified 43 identical mutations and although we cannot fully exclude clonal effects caused by mutagenesis of germline precursor cells, statistically up to 53 G/C positions can be expected to be mutated twice or more, indicating that no mutational hotspots exist. Nevertheless, a more subtle bias may still exist depending on the local sequence context. To investigate this, we calculated the ratio between the observed base frequencies for the positions flanking the mutated G/C base pairs from our screen and the expected base frequencies based on the sequence context of all G/C base pairs in the regions that were screened (Fig. 3). The most prominent biases are seen at positions −1 and +2, where a G is strongly over-represented, and at positions −2, −1, and +1, where As are clearly under-represented. Additional, but smaller, effects are seen at various positions between −4 and +4, and no significant biases are seen for positions that are further away. These observations show that there are local, sequence-dependent effects that are similar, but not identical, in size and spectrum as those observed in Arabidopsis (Greene et al. 2003). This may either be the result of chemical properties of the molecules involved or the presence of a common repair mechanism with a sequence context-dependent activity. The latter could also explain the intriguing asymmetric pattern.
Figure 3.
Sequence context bias around the mutated G position. The ratio was calculated between the observed nucleotide occurrence around the G/C to A/T mutations in our screen (n = 973) and the expected frequency for a random G/C base pair in the complete coding part of the 32 genes that were screened (n = 11,740). Sequence data was analyzed with the G base centered on the 0 position of the positive DNA strand. *P < 0.001, #P < 0.05.
EMS-induced mutation frequency in C. elegans
The average molecular mutation frequency per animal varied per locus from one mutation per 61–192 kb with an average of one per 104 kb (Table 2), to, corrected for duplicates, one per 109 kb. As both the male and the female germ lines were mutagenized in the founder animals, the mutation frequency per haploid genome is one per 208 kb and translates into a mutation frequency per haploid sequenced G/C base of one per 92,000 or 1.1 × 10−5, which is ∼1.5-fold higher than calculated previously (Anderson 1995). The intronic mutation frequency is significantly lower (one per 440 kb), but this is most likely due to technical issues because the intronic sequences occurred largely at the beginnings and ends of the reads where base accuracy is less, compromising mutation scoring. When considering the complete library, the average spacing between two adjacent mutations is only 20 bp and for coding sequences only 17 bp. Taking into account that ∼70% of the exonic mutations results in a nonsynonymous change of an amino acid, this means that at the amino acid level there is a mutation present in the library for every eighth amino acid of every C. elegans protein.
From gene-based to genome-wide screening
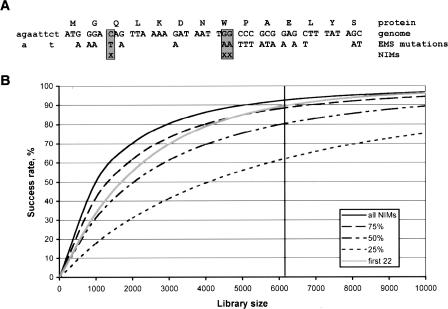
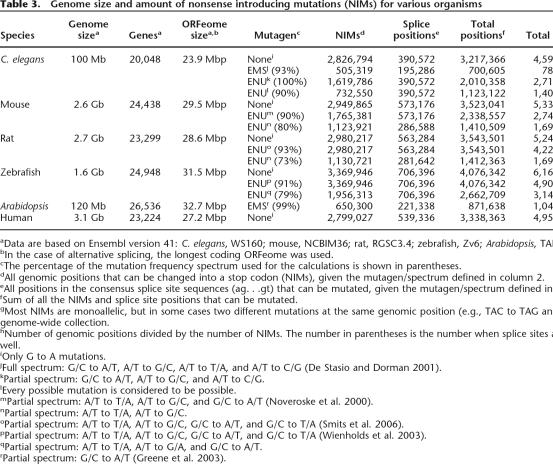
Extrapolation of the observed mutation frequency indicates that knockouts for the vast majority of the genes are present in the current library. However, screening the complete library with the current gene-based technology would not be feasible. In practice, stop and splice site mutations are the preferred type of knockout mutations, as these are most likely to result in a full loss of function of the gene. It should be noted that not every premature stop, especially those that are close to the 5′ and 3′ ends of a gene, results in a loss-of-function phenotype and that nonsense mutations for certain genes, such as small genes and those with multiple transcriptional isoforms, may be more difficult to recover. When the complete and well-annotated genomic sequence of an organism in combination with the mutation spectrum of the mutagen applied is known, one can simply deduce those positions in the genome that can result in a premature stop codon upon mutation (Fig. 4A). We define these positions as nonsense introducing mutations (NIMs). Although the C. elegans genome is ∼100 Mbp and the complete coding part of the transcriptome (ORFeome) is ∼24 Mbp, the total number of NIMs based on EMS mutagenesis is only slightly more than 500,000 positions, reducing the genome complexity by ∼200-fold (Table 3). For this calculation, we took into account only the most frequently EMS-induced point mutation G to A (93% of all mutations). When splice site mutations are taken into account as well, the total number of genomic positions increases to 700,000. To identify all knockout mutations in an animal, one only has to inspect the genome-wide collection of NIMs, for example by genotyping. The genome-wide NIM collection can be calculated for any species of interest and, interestingly, its size does not scale linearly with genome size or animal complexity but is similar for Arabidopsis and increases only slightly for vertebrates (up to sixfold, depending on the mutagen and species) (Table 3). As a result, the genome-wide screening complexity is reduced >2000-fold.
Figure 4.
Nonsense introducing mutations (NIMs) and genome-wide NIM-based knockout screening. (A) A NIM is defined by those positions in the genome that introduce a premature stop codon in the open reading frame of a gene upon mutation. In this example, only a small fraction of the positions that can be mutated by EMS results in the introduction of a stop codon (boxed positions). (B) Success rate for genome-wide knockout retrieval. The percentage of all C. elegans genes with an expected nonsense mutation is plotted as a function of the library size screened. Rates are plotted for all NIMs (all), the first 25%, 50%, and 75% of the NIMs per gene, and the first maximum of 22 NIMs per gene (first 22). Only G/C-to-A/T mutations (mutation frequency one per 46,000 GC-base) are taken into account for this calculation. The vertical line indicates the size of the current library.
Table 3.
Genome size and amount of nonsense introducing mutations (NIMs) for various organisms
aData are based on Ensembl version 41: C. elegans, WS160; mouse, NCBIM36; rat, RGSC3.4; zebrafish, Zv6; Arabidopsis, TAIR6.0; human, NCBI36.
bIn the case of alternative splicing, the longest coding ORFeome was used.
cThe percentage of the mutation frequency spectrum used for the calculations is shown in parentheses.
dAll genomic positions that can be changed into a stop codon (NIMs), given the mutagen/spectrum defined in column 2.
eAll positions in the consensus splice site sequences (ag. . .gt) that can be mutated, given the mutagen/spectrum defined in column 2.
fSum of all the NIMs and splice site positions that can be mutated.
gMost NIMs are monoallelic, but in some cases two different mutations at the same genomic position (e.g., TAC to TAG and TAA) contribute to the genome-wide collection.
hNumber of genomic positions divided by the number of NIMs. The number in parentheses is the number when splice sites are taken into account as well.
iOnly G to A mutations.
jFull spectrum: G/C to A/T, A/T to G/C, A/T to T/A, and A/T to C/G (De Stasio and Dorman 2001).
kPartial spectrum: G/C to A/T, A/T to G/C, and A/T to C/G.
lEvery possible mutation is considered to be possible.
mPartial spectrum: A/T to T/A, A/T to G/C, and G/C to A/T (Noveroske et al. 2000).
nPartial spectrum: A/T to T/A, A/T to G/C.
oPartial spectrum: A/T to T/A, A/T to G/C, G/C to A/T, and G/C to T/A (Smits et al. 2006).
pPartial spectrum: A/T to T/A, A/T to G/C, G/C to A/T, and G/C to T/A (Wienholds et al. 2003).
qPartial spectrum: A/T to T/A, A/T to G/A, and G/C to A/T.
rPartial spectrum: G/C to A/T (Greene et al. 2003).
Although genotyping technologies with increasing capacity and sensitivity are continuously emerging, current techniques are limited to several hundreds of thousands of genotypes per experiment. When all NIMs (stop mutations only, no splice site mutations) of C. elegans would be screened in our library, we expect to find at least one premature stop codon for 93% of all genes (Fig. 4B). The remaining parts of the genes are difficult to inactivate because of their small size or low NIM content. Even when only 25% of all NIMs are screened, nonsense mutations for >60% of the genes are expected to be retrieved. Furthermore, screening only half of the NIMs, or a maximum of the first 22 NIMs per gene (which equals about half of all NIMs), the success rate will still be ∼90% (Fig. 4B).
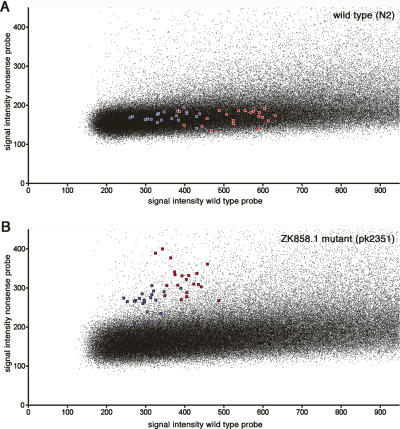
To test whether current technologies could be used for NIM genotyping, we designed a high-density array (Nimblegen, 370,000 features) for genotyping based on differential hybridization to allele-specific oligonucleotides and assayed a wild-type strain (N2) and an EMS-induced knockout strain (pk2351) that was mixed 1:1 with N2 to mimic the situation as present in a mutant library (Fig. 5). The array covered 97,500 NIMs and included 29-mer probes for the wild-type and nonsense allele for both genomic strands, resulting in four probes per NIM. The known knockout mutation present in pk2351 was represented 10 times on the array. Using this approach, we were able to distinguish the control knockout mutation clearly when comparing the mutant sample with the wild-type sample (Fig. 5). Although one could expect ∼11 NIMs per mutant and the detection of two NIMs on the current array, we were not able to retrieve additional candidate mutated positions that are expected as background in the unbackcrossed pk2351 strain from this experiment. This could be because (1) only part of the NIMs were present on this array (∼20%); (2) differential hybridization to probes with a single mismatch is not sensitive enough for all NIMs, which may also explain the high background signal observed our experiments; and (3) every NIM is represented only once (with four probes) per array. When analyzing the control NIM data, we find that with the current technology a NIM should preferably be represented three- to fivefold per experiment to reliably call the mutation, or the analysis should be performed in duplicate. Although the current custom-designed platforms do not yet allow for densities to accommodate sufficiently large numbers of features to meet these criteria, making the suggested genome-wide approach currently impractical, the expectation is that densities will grow to up to 10 million features, which would be sufficient to accommodate every C. elegans NIM in multifold. Alternatively, other high-throughput genotyping assay, such as methods based on match/mismatch strategy (Matsuzaki et al. 2004), single-base extension (Steemers et al. 2006), or molecular inversion probes (Hardenbol et al. 2005), or novel emerging techniques, could be considered to assay (genome-wide collections of) NIMs.
Figure 5.
Array-based NIM analysis. The signal of the nonsense (vertical) versus the wild-type (horizontal) allele is plotted for wild-type worms (N2) (A) and pk2351, containing an EMS-induced stopcodon in ZK858.1 (B). For nonmutated positions, the nonsense signal (vertical axis) is expected to be background, whereas the wild-type signal spreads over the X-axis, most likely reflecting GC content differences. A heterozygous mutation would have similar intensities for both the nonsense and wild-type allele, and these are thus expected to be located close to the diagonal in the figure. Microarrays with 390,000 features were used to assay part of the C. elegans genome-wide NIM collection. Every NIM was represented by four probes that were temperature-normalized (Tm = 72°C, probe length between 25 and 40 nt), representing the wild-type and mutant alleles for both DNA strands. The stop codon in ZK858.1 was represented 10 times (red, plus strand probes; blue, minus strand probes).
Conclusions
We showed that a target-selected mutagenesis approach based on chemically induced point mutations is highly effective for the identification of knockouts in C. elegans. Using high-throughput dideoxy resequencing, we identified the largest unbiased collection of EMS-induced point mutations after mutagenesis of an animal. Extrapolation of our results indicates that knockouts for >90% of all C. elegans genes are present in the current resource. We calculated that the screening complexity for genome-wide identification of knockouts can be decreased by two to three orders of magnitude when focusing on NIMs and speculate that genome-wide NIM genotyping could be used to retrieve all knockouts from the mutant library.
The NIM-based target-select mutagenesis strategy is not limited to C. elegans. Reverse genetics using chemical mutagenesis is already very popular in a variety of species, including zebrafish, rat, and several plant species, partially because no other methods are available for the generation of knockouts and partially because the resulting models are not transgenic. The latter is relevant because of legal, ethical, and safety issues that are associated with the introduction of foreign DNA into organisms. Especially for crop species, where one would like to inactivate “adverse” characteristics, such as a gene encoding an allergen, there is no need to generate a genetically modified organism. The approach described here can, in principle, be used for any organism of interest that can be mutagenized and bred, and for which a genome sequence is available. Although the genome-wide NIM collection increases several-fold from C. elegans to vertebrates, it does not scale linearly with genome size or complexity.
NIMs can also be calculated without a specific mutation spectrum assuming that every mutation is possible, thereby providing an equivalent for spontaneous mutations. Screening the human NIM collection (3.3 × 106 positions including all stop and splice mutations), potentially in combination with all relevant disease-related mutations as, for example, present in the OMIM (Online Mendelian Inheritance in Man, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM) database, could be a powerful diagnostic tool. This would allow the immediate identification of all stop mutations in the genome of an individual.
Methods
Nematode culturing and mutagenesis
C. elegans (strain Bristol N2) were cultured (Lewis and Fleming 1995) and mutagenized (Anderson 1995) as described. Animals were grown on NGM plates seeded with E. coli OP50 and cultured at room temperature. Stage-synchronized L4 worms were mutagenized with 50 mM EMS (Sigma) in M9 buffer (44 mM Na2HPO4, 22 mM KH2PO4, 86 mM NaCl, 1 mM MgSO4) for 6 h at room temperature. After overnight recovery on NGM/OP50 plates, 200 P0 animals were singled and grown for a few days. Subsequently, 9000 F1 animals were singled from these plates onto 6-cm plates. Worms were grown until plates were just starved and enriched for L1 stage larvae and subsequently washed off in M9-buffer into 96-well deep-well blocks (2.5 mL capacity, Riplate). All subsequent pipetting steps were performed using a 96-channel pipettor (Hydra, Matrix). Worms were allowed to settle by putting the sample block on ice for 15 min, and supernatant was removed until 200 μL was left in the block. Half of the worm suspension was used for cryopreservation of the worms in duplicate in 96-well plates and the other half for DNA isolation. Worms were resuspended by three times pipetting 150 μL up and down, and 100 μL of worm suspension was transferred to a 96-well plate (strips of 8 wells, Costar), prefilled with 100 μL of 2× concentrated freezing medium (50 mM phosphate buffer at pH 6.0, 100 mM NaCl, 30% glycerol, 0.3 mM MgSO4). After mixing by three times pipetting 150 μL up and down, 100 μL was transferred to an empty 96-well plate (duplicate frozen stock). Both plates were sealed using adhesive foil, wrapped in cotton wool and towels, and slowly frozen in a −80°C freezer.
DNA isolation and gridding
Genomic DNA was isolated from the remaining 100 μL of worm suspension in 96-well deep-well blocks by adding 50 μL of 3× concentrated worm lysis buffer (150 mM KCl, 30 mM Tris-HCl at pH 8.0, 7.5 mM MgCl2, 1.35% NP-40, 1.35% Tween-20, 300 mg/L gelatin, 0.3 mg/mL ProtK) and incubating at 60°C for 45 min. ProtK was inactivated by heating for 15 min at 95°C. Samples were further purified using protein precipitation by adding 50 μL of Protein Precipitation Solution (Promega) and centrifuging for 20 min at 5000g. Supernatant was transferred to a new plate, and DNA was precipitated by adding 150 μL of isopropanol, centrifuging for 30 min at 5000g, and washing the pellet with 150 μL of 70% ethanol. The DNA pellet was dissolved in 100 μL of TE (10 mM Tris-HCl at pH 8.0, 0.1 mM EDTA) by gently shaking for a few hours at room temperature and stored at –20°C. For further automated robotic handling, DNA was diluted 100 times with water and gridded into 384-well plates (5 μL per well).
Mutation discovery
Genomic regions of interest were amplified by PCR. Nested sets of oligonucleotides (1 and 4 for the first PCR and 2 and 3 for the second PCR) were designed using a customized version of Primer3 (Rozen and Skaletsky 2000), which is integrated in the LIMSTILL (Laboratory Information Management System for the Identification of Mutations by Sequencing and TILLing) interface (http://limstill.niob.knaw.nl; V. Guryev and E. Cuppen, unpubl.). Universal M13 adapters were added to oligos 2 and 3 (forward oligo adapter: M13F, TGTAAAACGACGGCCAGT; reverse oligo adapter: M13R, AGGAAACAGCTATGACCAT). The first PCR contained 5 μL of genomic template DNA (pregridded), 2 μL of 5× PCR buffer (0.125 M Tricine, 40% glycerol, 10% DMSO, 0.43 M NH4Ac, 11.2 mM MgCl2, pH adjusted to 8.7 using 25% ammonia), 0.2 μL of 10 mM dNTP, 0.2 μL of oligo 1 (10 mM), 0.2 μL of oligo 4 (10 mM), and 0.08 μL of Taq polymerase (5 U/μL) in a total volume of 10 μL. Thermocycling conditions were: 94°C for 60 sec, followed by 30 cycles of 92°C for 20 sec, 58°C for 20 sec, and 72°C for 60 sec. PCR 1 was diluted with 25 μL of H2O and 1 μL was used as template for the second PCR reaction that, in addition, contained 1 μL of 5× PCR buffer, 0.1 μL of 10 mM dNTP, 0.04 μL of M13F-tailed oligo 2 (10 mM), 0.02 M13R-tailed oligo 3 (10 mM), and 0.04 μL of Taq polymerase (5 U/μL) in a total volume of 5 μL. Thermocycling conditions were the same as for the first PCR.
Subsequently, PCR2s were diluted with 25 μL of water and 1 μL was used as template in a 5-μL dideoxy sequencing reaction, containing 0.01 μL of universal primer (100 μM stock, either M13F or M13R), 1.9 μL of 2.5× sequencing dilution buffer (Applied Biosystems), and 0.1 μL of BigDye terminator v3.1 (Applied Biosystems). Samples were processed under the following conditions: 35 cycles of 92°C for 10 sec, 50°C for 5 sec, and 60°C for 120 sec. The resulting reactions were purified by ethanol precipitation in the presence of 40 mM sodium-acetate as recommended by the manufacturer (Applied Biosystems) and analyzed on AB3730XL 96-capillary sequencers using the standard RapidSeq fast run protocol. All PCR and sequencing reactions were set up using high-throughput robotic automation (Tecan Genesis Workstation RSP200 with integrated TeMo96 and Velocity11 VPrep96/384 with BenchCell) in 384-well format. The resulting sequencing reads were automatically analyzed using in-house developed scripts running the PolyPhred 5.0 package (Nickerson et al. 1997) in the background for mutation discovery and imported into the management system (http://limstill.niob.knaw.nl) for storage and automated annotation. Every individual mutation was reconfirmed by an independent nested PCR and sequencing reaction.
Duplicate mutation calculation
The number of mutations that are expected to be observed twice or more can be calculated by: N2+ = N − N0 − N1, where N is the number of effective GC sites per animal, which is calculated by N = GC/A, where GC is the total number of GC positions screened (44,805,188) and A is the effective number of animals screened (5453, which is lower than the total number of animals in the library because of sample failures and incomplete library screening for some amplicons); N0 is the number of monomorphic positions (not mutated sites) and can be calculated by N0 = N(1 − p)A, where p is the GC mutation frequency (2.17 × 10−5); and N1 is the number of positions that are expected to be mutated once, which is calculated by N1 = NAp1(1 − p)(A−1).
NIM calculation
Ensembl release 41 sequences and annotations of C. elegans, human, rat, mouse, and zebrafish genomes and Arabidopsis thaliana chromosome annotations (NCBI entries NC_003070.5, NC_003071.3, NC_003074.4, NC_003075.3, NC_003076.4) were used for calculations of their NIMs.
Microarray design and analysis
Probes for genotyping of NIMs were fixed length 29-mer oligonucleotides with the NIM position fixed at base 15. N2 and pk2351 (containing an EMS-induced stop codon in ZK858.1) strains were grown on 9-cm NGM plates, worms were collected, and genomic DNA was isolated using minicolumns (DNeasy Tissue kit, Qiagen). Microarray hybridization and processing was done under standard conditions for resequencing by Nimblegen. Raw hybridization intensities were used for plotting wild-type versus nonsense values.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6080607
References
- Anderson P. Mutagenesis. Methods Cell Biol. 1995;48:31–58. [PubMed] [Google Scholar]
- Barstead R.J. Reverse genetics. In: Hope I.A., editor. C. elegans: —A Practical Approach. Oxford University Press; New York: 1999. pp. 97–118. [Google Scholar]
- Bentley A., MacLennan B., Calvo J., Dearolf C.R., MacLennan B., Calvo J., Dearolf C.R., Calvo J., Dearolf C.R., Dearolf C.R. Targeted recovery of mutations in Drosophila. Genetics. 2000;156:1169–1173. doi: 10.1093/genetics/156.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Bargmann C.I., Plasterk R.H., Bargmann C.I., Plasterk R.H., Plasterk R.H. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 2004;32:e40. doi: 10.1093/nar/gnh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- De Stasio E.A., Dorman S., Dorman S. Optimization of ENU mutagenesis of Caenorhabditis elegans. Mutat. Res. 2001;495:81–88. doi: 10.1016/s1383-5718(01)00198-x. [DOI] [PubMed] [Google Scholar]
- Edgley M., D'Souza A., Moulder G., McKay S., Shen B., Gilchrist E., Moerman D., Barstead R., D'Souza A., Moulder G., McKay S., Shen B., Gilchrist E., Moerman D., Barstead R., Moulder G., McKay S., Shen B., Gilchrist E., Moerman D., Barstead R., McKay S., Shen B., Gilchrist E., Moerman D., Barstead R., Shen B., Gilchrist E., Moerman D., Barstead R., Gilchrist E., Moerman D., Barstead R., Moerman D., Barstead R., Barstead R. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 2002;30:e52. doi: 10.1093/nar/gnf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Martinez-Campos M., Sohrmann M., Ahringer J., Sohrmann M., Ahringer J., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gonczy P., Echeverri C., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., Echeverri C., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., Duperon J., Oegema J., Brehm M., Cassin E., Oegema J., Brehm M., Cassin E., Brehm M., Cassin E., Cassin E., et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Enns L.C., Burtner C., Johnson J.E., Odden A.R., Burtner C., Johnson J.E., Odden A.R., Johnson J.E., Odden A.R., Odden A.R., et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003;164:731–740. doi: 10.1093/genetics/164.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardenbol P., Yu F., Belmont J., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Yu F., Belmont J., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Belmont J., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Mackenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Boudreau A., Chow S., Eberle J., Erbilgin A., Chow S., Eberle J., Erbilgin A., Eberle J., Erbilgin A., Erbilgin A., et al. Highly multiplexed molecular inversion probe genotyping: Over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Hazendonk E., Thijssen K.L., Plasterk R.H., Hazendonk E., Thijssen K.L., Plasterk R.H., Thijssen K.L., Plasterk R.H., Plasterk R.H. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- Johnsen R.C., Jones S.J., Rose A.M., Jones S.J., Rose A.M., Rose A.M. Mutational accessibility of essential genes on chromosome I(left) in Caenorhabditis elegans. Mol. Gen. Genet. 2000;263:239–252. doi: 10.1007/s004380051165. [DOI] [PubMed] [Google Scholar]
- Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A.G., Crombie C., Tischler J., Fortunato A., Fraser A.G., Tischler J., Fortunato A., Fraser A.G., Fortunato A., Fraser A.G., Fraser A.G. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Lewis J.A., Fleming J.T., Fleming J.T. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- Liu L.X., Spoerke J.M., Mulligan E.L., Chen J., Reardon B., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., Spoerke J.M., Mulligan E.L., Chen J., Reardon B., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., Mulligan E.L., Chen J., Reardon B., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., Chen J., Reardon B., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., Reardon B., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., Sun L., Abel K., Armstrong B., Hardiman G., Abel K., Armstrong B., Hardiman G., Armstrong B., Hardiman G., Hardiman G., et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 1999;9:859–867. doi: 10.1101/gr.9.9.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H., Dong S., Halina L., Di X., Liu G., Hubbel E., Law J., Berntsen T., Chadha M., Hui H., Dong S., Halina L., Di X., Liu G., Hubbel E., Law J., Berntsen T., Chadha M., Hui H., Halina L., Di X., Liu G., Hubbel E., Law J., Berntsen T., Chadha M., Hui H., Di X., Liu G., Hubbel E., Law J., Berntsen T., Chadha M., Hui H., Liu G., Hubbel E., Law J., Berntsen T., Chadha M., Hui H., Hubbel E., Law J., Berntsen T., Chadha M., Hui H., Law J., Berntsen T., Chadha M., Hui H., Berntsen T., Chadha M., Hui H., Chadha M., Hui H., Hui H., et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- McCallum C.M., Comai L., Greene E.A., Henikoff S., Comai L., Greene E.A., Henikoff S., Greene E.A., Henikoff S., Henikoff S. Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 2000;123:439–442. doi: 10.1104/pp.123.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Gojobori T., Ikemura T., Gojobori T., Ikemura T., Ikemura T. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D.A., Tobe V.O., Taylor S.L., Tobe V.O., Taylor S.L., Taylor S.L. PolyPhred: Automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noveroske J.K., Weber J.S., Justice M.J., Weber J.S., Justice M.J., Justice M.J. The mutagenic action of N-ethyl-N-nitrosourea in the mouse. Mamm. Genome. 2000;11:478–483. doi: 10.1007/s003350010093. [DOI] [PubMed] [Google Scholar]
- Perry J.A., Wang T.L., Welham T.J., Gardner S., Pike J.M., Yoshida S., Parniske M., Wang T.L., Welham T.J., Gardner S., Pike J.M., Yoshida S., Parniske M., Welham T.J., Gardner S., Pike J.M., Yoshida S., Parniske M., Gardner S., Pike J.M., Yoshida S., Parniske M., Pike J.M., Yoshida S., Parniske M., Yoshida S., Parniske M., Parniske M. A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 2003;131:866–871. doi: 10.1104/pp.102.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quwailid M.M., Hugill A., Dear N., Vizor L., Wells S., Horner E., Fuller S., Weedon J., McMath H., Woodman P., Hugill A., Dear N., Vizor L., Wells S., Horner E., Fuller S., Weedon J., McMath H., Woodman P., Dear N., Vizor L., Wells S., Horner E., Fuller S., Weedon J., McMath H., Woodman P., Vizor L., Wells S., Horner E., Fuller S., Weedon J., McMath H., Woodman P., Wells S., Horner E., Fuller S., Weedon J., McMath H., Woodman P., Horner E., Fuller S., Weedon J., McMath H., Woodman P., Fuller S., Weedon J., McMath H., Woodman P., Weedon J., McMath H., Woodman P., McMath H., Woodman P., Woodman P., et al. A gene-driven ENU-based approach to generating an allelic series in any gene. Mamm. Genome. 2004;15:585–591. doi: 10.1007/s00335-004-2379-z. [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Rushforth A.M., Anderson P., Anderson P. Splicing removes the Caenorhabditis elegans transposon Tc1 from most mutant pre-mRNAs. Mol. Cell. Biol. 1996;16:422–429. doi: 10.1128/mcb.16.1.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushforth A.M., Saari B., Anderson P., Saari B., Anderson P., Anderson P. Site-selected insertion of the transposon Tc1 into a Caenorhabditis elegans myosin light chain gene. Mol. Cell. Biol. 1993;13:902–910. doi: 10.1128/mcb.13.2.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits B.M., Mudde J.B., de van Belt J., Verheul M., Olivier J., Homberg J., Guryev V., Cools A.R., Ellenbroek B.A., Plasterk R.H., Mudde J.B., de van Belt J., Verheul M., Olivier J., Homberg J., Guryev V., Cools A.R., Ellenbroek B.A., Plasterk R.H., de van Belt J., Verheul M., Olivier J., Homberg J., Guryev V., Cools A.R., Ellenbroek B.A., Plasterk R.H., Verheul M., Olivier J., Homberg J., Guryev V., Cools A.R., Ellenbroek B.A., Plasterk R.H., Olivier J., Homberg J., Guryev V., Cools A.R., Ellenbroek B.A., Plasterk R.H., Homberg J., Guryev V., Cools A.R., Ellenbroek B.A., Plasterk R.H., Guryev V., Cools A.R., Ellenbroek B.A., Plasterk R.H., Cools A.R., Ellenbroek B.A., Plasterk R.H., Ellenbroek B.A., Plasterk R.H., Plasterk R.H., et al. Generation of gene knockouts and mutant models in the laboratory rat by ENU-driven target-selected mutagenesis. Pharmacogenet. Genomics. 2006;16:159–169. doi: 10.1097/01.fpc.0000184960.82903.8f. [DOI] [PubMed] [Google Scholar]
- Steemers F.J., Chang W., Lee G., Barker D.L., Shen R., Gunderson K.L., Chang W., Lee G., Barker D.L., Shen R., Gunderson K.L., Lee G., Barker D.L., Shen R., Gunderson K.L., Barker D.L., Shen R., Gunderson K.L., Shen R., Gunderson K.L., Gunderson K.L. Whole-genome genotyping with the single-base extension assay. Nat. Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- Stephens M., Sloan J.S., Robertson P.D., Scheet P., Nickerson D.A., Sloan J.S., Robertson P.D., Scheet P., Nickerson D.A., Robertson P.D., Scheet P., Nickerson D.A., Scheet P., Nickerson D.A., Nickerson D.A. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat. Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- van Haaften G., Vastenhouw N.L., Nollen E.A., Plasterk R.H., Tijsterman M., Vastenhouw N.L., Nollen E.A., Plasterk R.H., Tijsterman M., Nollen E.A., Plasterk R.H., Tijsterman M., Plasterk R.H., Tijsterman M., Tijsterman M. Gene interactions in the DNA damage-response pathway identified by genome-wide RNA-interference analysis of synthetic lethality. Proc. Natl. Acad. Sci. 2004;101:12992–12996. doi: 10.1073/pnas.0403131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Plasterk R.H., Plasterk R.H. Target-selected gene inactivation in zebrafish. Methods Cell Biol. 2004;77:69–90. doi: 10.1016/s0091-679x(04)77004-1. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Schulte-Merker S., Walderich B., Plasterk R.H., Schulte-Merker S., Walderich B., Plasterk R.H., Walderich B., Plasterk R.H., Plasterk R.H. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- Wienholds E., van Eeden F., Kosters M., Mudde J., Plasterk R.H., Cuppen E., van Eeden F., Kosters M., Mudde J., Plasterk R.H., Cuppen E., Kosters M., Mudde J., Plasterk R.H., Cuppen E., Mudde J., Plasterk R.H., Cuppen E., Plasterk R.H., Cuppen E., Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler S., Schwabedissen A., Backasch D., Bokel C., Seidel C., Bonisch S., Furthauer M., Kuhrs A., Cobreros L., Brand M., Schwabedissen A., Backasch D., Bokel C., Seidel C., Bonisch S., Furthauer M., Kuhrs A., Cobreros L., Brand M., Backasch D., Bokel C., Seidel C., Bonisch S., Furthauer M., Kuhrs A., Cobreros L., Brand M., Bokel C., Seidel C., Bonisch S., Furthauer M., Kuhrs A., Cobreros L., Brand M., Seidel C., Bonisch S., Furthauer M., Kuhrs A., Cobreros L., Brand M., Bonisch S., Furthauer M., Kuhrs A., Cobreros L., Brand M., Furthauer M., Kuhrs A., Cobreros L., Brand M., Kuhrs A., Cobreros L., Brand M., Cobreros L., Brand M., Brand M., et al. Target-selected mutant screen by TILLING in Drosophila. Genome Res. 2005;15:718–723. doi: 10.1101/gr.3721805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W., editor. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. [Google Scholar]
- Zwaal R.R., Broeks A., van Meurs J., Groenen J.T., Plasterk R.H., Broeks A., van Meurs J., Groenen J.T., Plasterk R.H., van Meurs J., Groenen J.T., Plasterk R.H., Groenen J.T., Plasterk R.H., Plasterk R.H. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc. Natl. Acad. Sci. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]