Abstract
Long Interspersed Element-1 (LINE-1 or L1) sequences comprise ∼17% of human DNA and ongoing L1 retrotransposition continues to impact genome evolution. The L1-encoded proteins also can mobilize other cellular RNAs (e.g., Alu retrotransposons, SVA retrotransposons, and U6 snRNAs), which comprise ∼13% of human DNA. Here, we demonstrate that the trans-mediated mobilization of non-L1 RNAs can occur by either template choice or template-switching mechanisms. Remarkably, these mechanisms are not mutually exclusive, as both processes can operate sequentially on the same RNA template. Finally, we provide evidence that efficient U6 snRNA retrotransposition requires both ORF1p and ORF2p, providing indirect evidence for the action of ORF1p in U6 snRNA retrotransposition. Thus, we propose that the LINE-1-encoded reverse transcriptase can mediate the retrotransposition of non-L1 RNAs by distinct mechanisms.
The average human genome is estimated to contain ∼80–100 retrotransposition competent L1s (Sassaman et al. 1997; Brouha et al. 2003). These elements are ∼6 kb in length and contain two open reading frames (ORF1 and ORF2) (Scott et al. 1987; Dombroski et al. 1991). ORF1 encodes a ∼40-kDa RNA-binding protein (p40 or ORF1p) that possesses nucleic acid chaperone activity (Holmes et al. 1992; Hohjoh and Singer 1996; Martin and Bushman 2001; Martin et al. 2005). ORF2 encodes an ∼150-kDa protein with demonstrated endonuclease (EN) and reverse transcriptase (RT) activities (Mathias et al. 1991; Feng et al. 1996; Ergun et al. 2004). Both proteins are responsible for retrotransposition in cis (Moran et al. 1996; Wei et al. 2001; Kulpa and Moran 2005), which likely occurs via target-site primed reverse transcription (TPRT) (Luan et al. 1993; Feng et al. 1996; Kazazian and Moran 1998; Cost et al. 2002; Kulpa and Moran 2006).
The LINE-encoded proteins can mobilize other cellular RNAs, and a number of pathways have been proposed to account for this phenomenon (Denison et al. 1981; Van Arsdell et al. 1981; Brosius 1991, 1999; Schmitz et al. 2004; Ohshima and Okada 2005; Hulme et al. 2006). For example, in humans, Alu retrotransposition requires the enzymatic activities encoded by ORF2p (Dewannieux et al. 2003), whereas processed pseudogene formation appears to require activities encoded by both ORF1p and ORF2p (Esnault et al. 2000; Wei et al. 2001). Previous studies indicate that the L1-encoded proteins also mobilize SINE-R/VNTR/ALU (SVA) elements and certain uracil-rich small nuclear RNAs (snRNAs) (Buzdin et al. 2002, 2003; Ostertag et al. 2003; Bennett et al. 2004; Gilbert et al. 2005; Gogvadze et al. 2005; Wang et al. 2005; Weber 2006). Here, we experimentally demonstrate that the L1-encoded reverse transcriptase can retrotranspose cellular RNAs by discrete mechanisms.
Results
In silico analysis of small noncoding RNA sequences in the human genome
Previous in silico and in vitro data suggest that the L1 reverse transcriptase can “switch” from its own mRNA to U6 snRNA during TPRT, resulting in the formation of chimeric U6/L1 pseudogenes (Buzdin et al. 2002, 2003; Gilbert et al. 2005; Gogvadze et al. 2005). Similarly, in silico analyses have suggested that the L1 retrotransposition machinery also can mobilize other uracil-rich small nuclear RNAs, small nucleolar RNAs, and hY RNAs, which are components of the Ro/SS-A autoantigen by a fundamentally different template choice mechanism (Buzdin et al. 2003; Perreault et al. 2005; Weber 2006). To gain greater insight about how these sequences have impacted the genome, we conducted a BLAST search of the human genome working draft sequence (HGWD) using separate small RNA sequences as queries (see Methods). We restricted the analysis to sequences that presented an E-value of less than 5.0 × 10−27, as they are <10% divergent from the sequence of the functional gene.
One-hundred-ninety-seven U6 snRNA sequences met these criteria. These candidates then were inspected manually to characterize the sequences flanking the paralogous U6 copies. Three sequences were discarded because they either were part of a genomic duplication (two instances) or were contained within an unassigned cosmid (one instance). Approximately 90% of U6 sequences (173 instances) were flanked at their 3′ end by a retrotrotransposon. Of these, 78% (135 instances) had sequence characteristics suggesting that they were interspersed by retrotransposition (i.e., the presence of variable-sized target site duplications [TSDs] and a 3′ poly[A] tail). Consistent with previous analyses, we identified 74 U6/L1 and 17 U6/processed pseudogene chimeras (Buzdin et al. 2002, 2003). We also identified 76 U6 sequences that terminated in a poly(A) tail that could have been generated by template switching from the poly(A) tail of a cellular RNA to U6 snRNA or by the retrotransposition of an unusual, polyadenylated U6 snRNA (Table 1). Interestingly, we only identified 15 U6/Alu pseudogenes, despite the fact that ∼1.5 million Alu elements are dispersed throughout the genome (see below). We also identified U6/L1 chimeras in the genomes of other placental mammals and the marsupial Monodelphis domestica (opossum; A.J. Doucet and N. Gilbert, unpubl.). Thus, the above data extend previous analyses (Buzdin et al. 2002, 2003), and suggest that the majority (i.e., >90%) of U6 snRNA sequences in the human genome have been dispersed by retrotransposition.
Table 1.
U6 snRNA pseudogenes in the human genome
Columns 1 and 2 indicate the human chromosomes and their respective sizes. Column 3 indicates the number of U6 sequences identified on each chromosome using a BLAST search E-value <5.0 × 10−27. Columns 4 through 7 indicate the number of U6/L1, U6/Alu, U6/processed pseudogene, or U6/poly(A) sequences present on each chromosome. Column 8 (Alone) indicates U6 sequences that were not associated with repetitive sequences (18 instances). Column 9 (Others) indicates U6 sequences associated with other repetitive sequences (e.g., DNA transposon or LTR retrotransposon; three instances). The total number of each class of pseudogene, the number that are flanked by target site duplications (TSD), and the percentage of U6 sequences that the pseudogenes represent are indicated at the bottom of the table. The single asterisk (*) denotes two U6 sequences that were identified in distinct duplicated sequences. Only one representative U6 sequence within the duplicon was analyzed. The double asterisk (**) denotes a U6 sequence found in an unassigned contig that was not used in the analyses. Thus, 194 U6 sequences were analyzed.
We next extended our analysis to a cohort of other snRNAs that are used in the splicing of conventional GT-AG (i.e., U1, U2, U4, and U5) or minor AT-AC introns (U11, U12, U4atac, and U6atac), as well as U3 snoRNA, which is involved in ribosome biogenesis. Using the criteria employed in the in silico U6 analysis, we identified numerous paralogs of the snRNA sequences (Table 2). For example, some U1, U2, U4, and U4atac sequences ended in a poly(A) tail and are flanked by TSDs, consistent with the idea that they were mobilized to new genomic locations by the L1-encoded reverse transcriptase (Table 2). Akin to the situation observed for U6/L1 chimeric pseudogenes, it is possible that the retrotransposed U1, U2, U4, and U4atac sequences could have been generated by template switching from the L1 mRNA poly(A) to the snRNA. However, the lack of other L1 sequences 3′ of these retrotransposed copies leads us to speculate that unusual polyadenylated snRNAs (Houseley et al. 2006) occasionally can be used as reverse-transcription templates by the L1 retrotransposition machinery.
Table 2.
Small RNA pseudogenes in the human genome
Column 1 indicates the name of the small RNA. Column 2 indicates the number of sequences identified in the human genome using a BLAST search E-value <5.0 × 10−27. Columns 3 through 6 indicate the number of Ux/L1, Ux/Alu, Ux/processed pseudogene, or Ux/poly(A) sequences identified in our analyses. Column 7 (Alone) indicates Ux sequences identified in our screen that are not associated with repetitive sequences in their 3′ end. Column 8 (Others) indicates sequences that are associated with other repetitive sequences. The frequency of each category of pseudogene is indicated.
Interestingly, 7/16 U6atac snRNAs were followed by L1 sequences and exhibited structural similarities to the U6/L1 processed pseudogenes identified above (Table 2). We also detected a single U3/L1 chimera using the above criteria. However, when we lowered the cutoff value (1 × 10−9) we identified 11 U3/L1 chimeras of 72 sequences examined. Similarly, the same cutoff values detected two U5/L1 chimeras of 50 sequences examined. In both cases, the U3 and U5 sequences were 3′ truncated and then were followed by a 5′-truncated L1. Together, these data suggest that U6 snRNA, and to a lesser extent U6atac snRNA, can serve as templates for the L1-encoded reverse transcriptase more frequently than other snRNAs, but that a template-switching event from L1 mRNA to U3 snoRNA or U5 snRNA can occur during the course of genome evolution, albeit rarely.
An experimental system to detect snRNA/L1 processed pseudogenes
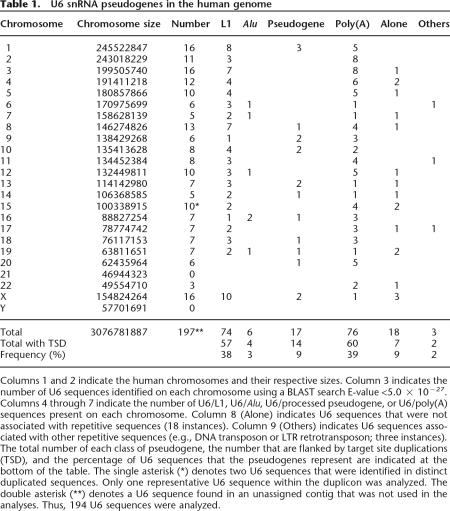
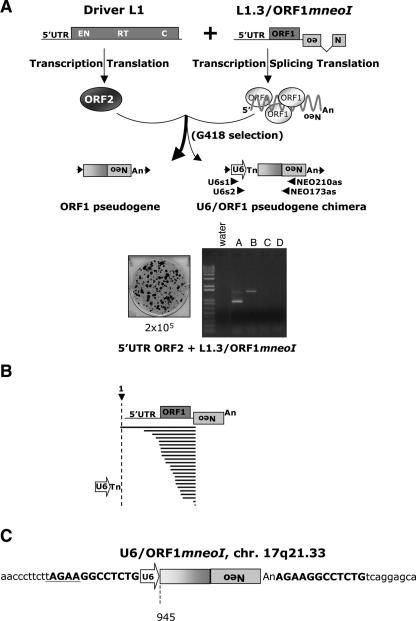
To experimentally recapitulate U6/L1 pseudogene formation, we used an established cultured cell retrotransposition assay. We transfected HeLa cells with a retrotransposition-competent L1 that was tagged with a retrotransposition indicator cassette in its 3′-untranslated region (pJM101/L1.3mneoI) (Dombroski et al. 1993; Sassaman et al. 1997). The retrotransposition cassette consists of a backward copy of the neomycin phosphotransferase gene containing its own promoter and polyadenylation signal (Jensen and Heidmann 1991; Freeman et al. 1994; Moran et al. 1996). The neo gene also is interrupted by an intron in the same transcriptional orientation as the L1. This arrangement ensures that G418-resistant foci will arise only if a spliced L1 mRNA undergoes a successful round of retrotransposition (Fig. 1A).
Figure 1.
A cultured cell assay to detect U6/L1 pseudogenes. (A) Rationale of the assay. The 3′ UTR of a retrotransposition-competent L1 (RC-L1) was tagged with a retrotransposition indicator cassette (light gray box labeled with a backward Neo). ORF1 and ORF2 are indicated by the dark-gray rectangles and the relative positions of the endonuclease (EN), reverse transcriptase (RT), and cysteine-rich domains (C) of ORF2 are indicated. Cartoons depicting the structures of the resultant retrotransposition events that confer G418 resistance (G418R) to HeLa cells are shown at the bottom of the diagram. A 5′-truncated L1 insertion is shown at the left and a U6/L1 pseudogene is shown at the right. Target site duplications flanking the elements are represented as horizontal arrows. PCR primers used to detect U6/L1 pseudogenes are indicated below the U6/L1 schematic. (B) Results of the assay: Shown are schematic representations of two RC-L1s (pJM101/L1.3 and pL1.3neoIII). pJM101/L1.3 contains our standard retrotransposition indicator cassette (Moran et al. 1996). pL1.3neoIII contains a retrotransposition indicator cassette disrupted by a self-splicing group I intron (Esnault et al. 2002; Dewannieux et al. 2003). Retrotransposition assays conducted by transfecting 2 × 104 HeLa cells revealed that both L1s retrotranspose at similar efficiencies (Wei et al. 2000). Genomic DNAs derived from six independent pools of G418-resistant foci then were used as templates in nested PCR reactions with the primers noted in A to detect the U6/L1 pseudogenes (lanes A–F). Molecular size standards are indicated on the gels (1-kb ladder plus from Invitrogen). (Water) PCR reactions conducted without the genomic DNA template. (C) Structures of the U6/L1 pseudogenes. Sequence analysis of the PCR products amplified in B confirmed the existence of the U6/L1 pseudogenes. All sequences contain the 3′ terminus of U6 (depicted as an horizontal open arrow) and a variable 5′−truncated L1. A full-length RC-L1 with the spliced reporter cassette is represented at the top. The U6 sequence ends in 4–8 thymidine residues and is followed by a 5′-truncated L1. The horizontal bold lines indicate the amount of the L1 sequence included in the pseudogene. The longest sequence extends to nucleotide 3503 of the L1. The numbering is based on the L1.2 reference sequence (Dombroski et al. 1991).
G418-resistant foci harboring L1 retrotransposition events were grouped into pools that ranged in size from 13 to 65 foci and aliquots of genomic DNA from each pool were used as templates in a series of nested PCRs using primers specific for U6 snRNA (U6s1 and U6s2) and the retrotransposition indicator cassette (210NEOas and 173NEOas; Fig. 1A). The data indicate that approximately one in 15 G418-resistant foci contain a U6/L1 pseudogene (Fig. 1B). Sequence analysis of 43 independent PCR products confirmed the identity of the U6/L1 pseudogenes (Fig. 1C; Supplemental Fig. 1A). The U6 sequence ended in four to eight thymidine residues, which is consistent with the fact that U6 snRNA is transcribed by RNA polymerase III; it was then followed by a variably 5′-truncated L1 (Fig. 1C; Supplemental Fig. 1A). The data further revealed that there is not a particular consensus sequence/motif in L1 RNA that may facilitate the template switch (Supplemental Fig. 1).
Similar data was observed when the above experiment was conducted with synthetic (L1SM) (Han and Boeke 2004) and natural (TGf21) (Goodier et al. 2001) mouse retrotransposition-competent L1s, consistent with the notion that there is not a particular consensus sequence/motif in L1 RNA that may facilitate the template switch (Supplemental Fig. 2). U6/L1 pseudogenes also could be detected at similar efficiencies (approximately one in 25 G418-resistant foci) using an L1 tagged with a retrotransposition indicator cassette, where the neo gene is interrupted with a self-splicing group I intron (Fig. 1B, L1.3neoIII) (Esnault et al. 2002; Dewannieux et al. 2003), indicating that U6/L1 pseudogene formation probably does not depend on the association of L1 mRNA with the spliceosome. Although individual G418-resistant foci may contain multiple retrotransposition events, the efficiency of U6/L1 pseudogene formation seems to be elevated by at least an order of magnitude when compared with the number of U6/L1 pseudogenes observed in the HGWD. That said, we conclude that this experimental system faithfully reproduces retrotransposition events that occurred during the course of human genome evolution.
We next tested whether other small RNAs could form chimeric pseudogenes with L1. To accomplish this task, we designed two sets of primers for each snRNA and then performed PCR on DNA isolated from the same pools of G418-resistant foci used above (Methods). We identified a single U3/L1 pseudogene (Supplemental Fig. 3A; Supplemental Table 1), and the U3 sequence was 3′ truncated at the same position identified in our in silico analysis. We also detected a single U6atac/L1 pseudogene (Supplemental Fig. 3B; Supplemental Table 1). In contrast, and consistent with our in silico analyses, we did not identify any U1/L1, U2/L1, U4/L1, U5/L1, hY1/L1, hY3/L1, or hY4/L1 pseudogenes in our assay (Supplemental Table 1). Thus, U6 snRNA, and to a much lesser extent U6atac and U3 small RNAs, can serve as template-switch substrates for the L1-encoded reverse transcriptase.
Processed pseudogene formation can occur by template choice
The L1-encoded proteins can act in trans to promote the retrotransposition of other cellular RNAs (Esnault et al. 2000; Wei et al. 2001; Dewannieux et al. 2003; Ostertag et al. 2003; Bennett et al. 2004). We previously showed that RNA derived from an artificial construct (L1.3/ORF1mneoI), which consists of the L1 5′UTR, ORF1, and that the mneoI retrotransposition indicator cassette could be used as a template for retrotransposition (Wei et al. 2001). Interestingly, the structure of L1.3/ORF1mneoI is similar to a repeated sequence present in all mammalian genomes (HAL1 [Half an L1], consensus sequence accessible from Repbase; Jurka et al. 2005). Thus, we examined the trans-mobilization of L1.3/ORF1mneoI RNA as a model to investigate the retrotransposition mechanism of HAL1-like sequences.
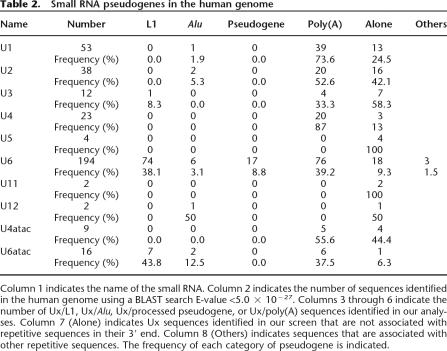
To monitor ORF1mneoI retrotransposition, we exploited a trans-complementation assay that monitors the ability of a retrotransposition-competent “driver” L1 to trans-mobilize a “reporter” construct (Fig. 2A). Although the trans-complementation assay is relatively inefficient when compared with the cis-based retrotransposition assay, the assay yields hundreds of G418-resistant foci and allows an experimental measure of pseudogene generation (Wei et al. 2001; Alisch et al. 2006). We demonstrated that efficient trans-complementation requires a functional ORF1p expressed from the reporter plasmid (Fig. 2A), and RNase protection experiments confirmed that wild-type ORF1mneoI RNA and its mutant derivatives were expressed in transfected cells at similar steady-state levels (Supplemental Fig. 4). These results indicate that ORF1mneoI retrotransposition requires the generation of a functional ribonucleoprotein (RNP) particle, which is consistent with our previous results using the cis-based retrotransposition assay (Kulpa and Moran 2005).
Figure 2.
ORF1p-dependent processed pseudogene formation occurs by template choice. (A) Rationale of the trans-complementation assay. A wild-type RC-L1 lacking the retrotransposition indicator cassette (pJM101/L1.3 No neo) or an ORF2 expression construct (ORF2 No neo) were cotransfected into HeLa cells with a construct consisting of the L1 5′UTR, ORF1, and the mneoI retrotransposition indicator cassette (ORF1mneoI or mutant derivatives), which is a preferential substrate for trans-complementation (Wei et al. 2001). The CMV immediate early promoter augments the expression of both the driver and reporter constructs. G418-resistant foci will arise only if ORF1mneoI RNA (i.e., the reporter mRNA) is trans-mobilized by ORF2p provided by the L1 lacking the indicator cassette (i.e., the driver L1). The inset Table indicates the names of the various reporter constructs (column 1), the status of ORF1 (column 2), the number of times the assay was conducted (column 3, N), and the number of G418-resistant foci obtained using either driver L1 (column 4). (B,C) Rationale and results of the fluorescent primer extension (FluPE) assay. The 3′ ends of the driver and reporter mRNAs were engineered so that the driver contains two extra nucleotides (indicated by the black star in the illustration of the driver construct and the inverted triangle in the depicted sequence). The driver and reporter constructs were cotransfected into HeLa cells and we isolated pools that contained ∼125–450 independent G418-resistant foci. Genomic DNA was extracted from the resultant pools and was subjected to PCR analysis using an oligonucleotide complementary to sequences in the indicator cassette (437NEOs) and another present just upstream of the poly(A) addition site in the vectors (Cep237as). Fluorescent Primer Extension (FluPE) then was conducted on the resultant PCR products using the Cy5 primer (black horizontal arrow). The arrows at bottom indicate the expected sizes of the Cy5 primer, the expected signal from the reporter construct (elongation of five nucleotides), and the expected signal from the driver construct (elongation of seven nucleotides). Retrotransposition events generated by template choice would be five nucleotides in size, whereas those formed via template switching would be seven nucleotides in size. The signals present in each pool (1–12) were five nucleotides in length, consistent with a template-choice mechanism for ORF1mneoI pseudogene formation. Traces t1, t2, and t3 represent positive control FluPE reactions performed on genomic DNA from cells transfected with an RC-L1 containing both the 2-bp insertion and the retrotransposition indicator cassette (pJM101/L1RP SDM). All experiments were repeated at least three times with the same results. The X-axis represents the running time of the reaction (minutes).
To determine whether ORF1mneoI pseudogene formation occurs by a template choice or template-switching mechanism, we introduced two nucleotides upstream of the poly(A) signal in our driver L1 (Fig. 2B, indicated by the star). If ORF1mneoI pseudogene formation occurs by template switching, the resultant retrotransposition events should contain the engineered SNPs at their respective 3′ ends. In contrast, if ORF1mneoI pseudogene formation occurs by template choice, the resultant retrotransposition events should lack the engineered SNPs.
Twelve pools that contained ∼125 (pools 1–9) or 450 (pools 10–12) G418-resistant foci were generated from HeLa cells that had been cotransfected with a driver L1 (pJM101/L1RP SDM No neo) and reporter (L1.3/ORF1mneoI) constructs (Fig. 2), and DNAs from each pool were then subjected to PCR to amplify the resultant retrotransposition events. The presence and/or absence of the engineered SNPs was then quantified using fluorescent primer-extension (FluPE) analysis (Fahy et al. 1997; Bennett-Baker et al. 2003). Serial dilution control experiments indicated that we could discriminate roughly one molecule in 250 (see Methods). However, despite examining genomic DNA from roughly 2500 G418-resistant foci, we never observed transfer of the diagnostic SNPs from the driver L1 to the resultant ORF1mneoI retrotransposition events (Fig. 2C). Thus, ORF1mneoI pseudogene formation predominantly occurs by a template choice mechanism. Although our experiments cannot formally exclude the possibility that template switching from the poly(A) tail of the driver L1 mRNA to the reporter mRNA generates some ORF1mneoI processed pseudogenes, our aforementioned studies of U6/L1 pseudogene formation make it very unlikely that all template-switching events would be confined to the poly(A) tails of the driver and reporter mRNAs.
Template choice and template switching can operate sequentially on the same RNA template
To determine whether the template choice and template-switching mechanisms could occur sequentially on the same RNA template, we combined the experimental strategies outlined in Figures 1 and 2 to test whether any L1.3/ORF1mneoI-processed pseudogenes are conjoined to U6 snRNA-derived sequences (Fig. 3A). Consistent with data presented above, we found that approximately one in 20 ORF1mneoI pseudogenes contain U6 snRNA sequences at their 5′ ends (Fig. 3A). Sequence analysis of 23 independent PCR products indicated that the U6 sequences end in four to six thymidine residues, and then are followed by sequences derived from the ORF1mneoI expression construct (Fig. 3B; Supplemental Fig. 1). We further demonstrated that a synthetic mouse L1 (L1SM/ORF1mneoI) can generate U6/pseudogenes (Supplemental Fig. 5), and that a U6/ORF1mneoI pseudogene exhibited hallmarks typical of an endonuclease-dependent L1 retrotransposition event (Fig. 3C), which is in agreement with our previously published data (Gilbert et al. 2005). Finally, our in silico analyses revealed that the HGWD contains at least 17 U6/processed pseudogene chimeras, and we propose that these events were formed by template-choice/template-switching mechanisms acting sequentially on the same RNA template in vivo.
Figure 3.
Template choice and template switching are not mutually exclusive. (A) Rationale and results of the assay. The trans-complementation assay is described in Figure 2. Retrotransposition assays were conducted by transfecting 2 × 105 HeLa cells with the driver (5′ UTR ORF2 No neo) and reporter (L1.3/ORF1mneoI) plasmids. Genomic DNAs from pools of G418-resistant foci were subjected to individual nested PCR reactions using the indicated primers (U6s1-NEO210as and U6s2-NEO173as) to detect U6/ORF1mneoI pseudogenes. (B) Results of the assay. Sequence analysis of 23 PCR products confirmed the existence of the U6/ORF1mneoI pseudogenes. A full-length ORF1mneoI mRNA is represented at top. The U6 sequence ends in four to six thymidine residues and is followed by ORF1mneoI RNA. The horizontal bold lines indicated the amount of the L1 included in the pseudogene. The longest U6/ORF1mneoI extends beyond the first nucleotide of ORF1mneoI RNA, indicating that the transcript was initiated from the CMV promoter upstream of the reporter gene. (C) The structure of a U6/ORF1mneoI pseudogene. The characterization of a single U6/ORF1mneoI pseudogene by inverse PCR revealed the hallmarks of L1 retrotransposition. Top bold characters on each side of the pseudogene represent the target site duplication. Lowercase characters represent the flanking genomic sequence. The complement of the L1 endonuclease cleavage recognition site (5′-TTCT/A) is underlined. The numbering is based on the L1.2 reference sequence (Dombroski et al. 1991).
U6/Alu pseudogenes are not formed efficiently in cultured cells
We next asked whether U6/Alu pseudogenes could be generated in HeLa cells. To accomplish this task, we cotransfected an Alu sequence tagged with a retrotransposition indicator cassette (pAluNF1neoIII) (Dewannieux et al. 2003) with either a retrotransposition-competent L1 lacking an indicator cassette (pJM101/L1RPNo neo) or an ORF2p expression construct (5′UTR ORF2 No neo; Supplemental Fig. 6A). The resultant G418-resistant foci were pooled in groups that ranged in size from 15 to 55 foci, and aliquots of genomic DNA from each pool were used as templates in nested PCR reactions using primers specific for U6 snRNA (U6s1 and U6s2) and the retrotransposition indicator cassette (210NEOas and 173NEOas). Although we could detect amplification products (Supplemental Fig. 6B), sequence analysis of 27 independent PCR amplicons indicated that they did not represent de novo U6/Alu pseudogenes, but instead were PCR artifacts (Supplemental Fig. 6C). Consistently, we determined that the majority (9/15) of U6/Alu pseudogenes identified in silico contained an A-rich linker between U6 and Alu and likely represent Alu insertions that occurred in the poly(A) tail of an existing U6 pseudogene. By comparison, at least four of the remaining six events might represent true U6/Alu-processed pseudogenes because they are flanked by target-site duplications that range from 13 to 20 bp in length.
Discussion
We have provided evidence that trans-mediated mobilization of cellular RNAs can occur via template-switching and/or template-choice mechanisms. U6 snRNA (as well as U3 snoRNA and U6atac snRNA to a much lesser extent) likely use the L1 RT in the nucleus by a template-switching mechanism after the initiation of target-site primed reverse transcription (Fig. 4). However, it remains unclear whether these small RNAs are occasionally copackaged into the L1 ribonucleoprotein complex in the cytoplasm or whether they gain access to the L1 retrotransposition machinery in the nucleus. We further propose that ORF1mneoI (and likely other cellular RNAs) associate with the L1 reverse transcriptase in the cytoplasm by a template-choice mechanism and then complete the process of retrotransposition by target-site primed reverse transcription (Fig. 4). Indeed, the experimental demonstration of U6/ORF1mneoI pseudogenes and the identification of U6/processed pseudogene chimeras in the HGWD further suggests that the mechanisms of template choice and template switching are not necessarily mutually exclusive, and that both processes can act sequentially on the same RNA template.
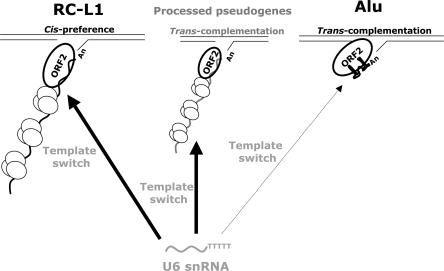
Figure 4.
Trans-mobilization of non-L1 mRNAs by the L1 retrotransposition machinery. The L1-encoded proteins preferentially bind to the RNA that encoded them in cis to promote their retrotransposition by target-site primed reverse transcription (left). Cellular RNAs (e.g., ORF1mneoI, middle; or Alu RNA, right) can occasionally use the L1 RT to mediate their mobility in trans. During target-site primed reverse transcription a template switch from L1 mRNA to U6 snRNA can lead to the formation U6/L1 pseudogenes. The data indicate that U6/ORF1mneoI pseudogenes can be generated by sequential template choice and template-switching mechanisms. However, U6/Alu pseudogenes are not efficiently generated in our experimental system, leading us to propose that ORF1p promotes efficient U6 snRNA retrotransposition. ORF1p molecules are depicted as small ovals associated in trimers (Martin et al. 2003). ORF2p is drawn as a large bolded oval. L1, ORF1mneoI, and Alu mRNA that are being reverse transcribed by target-site primed reverse transcription are indicated. U6 snRNA is indicated in gray toward the bottom of the figure. Whether U6 snRNA gains access to the L1 retrotransposition machinery in the cytoplasm or nucleus remains unknown. As proposed, the Alu RNP complex is shown without ORF1p (Dewannieux et al. 2003). However, it remains possible that some Alu RNA is associated with ORF1p in vivo.
Interestingly, Dewannieux et al. (2003) have reported that Alu RNA also likely associates with the L1 reverse transcriptase in the cytoplasm and that its retrotransposition does not require the ORF1-encoded protein. However, we did not observe a high efficiency of U6/Alu pseudogene formation in cultured cells. Thus, these data suggest that ORF1p enhances the retrotransposition of U6 snRNA. Our results also indicate that U6 snRNA is more efficiently dispersed throughout the genome when compared with other abundant snRNAs. Why U6 snRNA seems to serve as a preferential substrate for the L1 reverse transcriptase requires further study. However, because we detect efficient U6/L1 chimeric pseudogene formation with “driver” L1s that are equipped with a retrotransposition indicator cassette containing a self-splicing group I intron or a codon optimized “synthetic” mouse L1, we hypothesize that U6 snRNA does not necessarily gain access to the L1 retrotransposition machinery because of its association with the spliceosome.
In closing, our data highlight mechanisms by which the LINE-1 encoded proteins can mobilize non-L1 RNAs and strongly suggest that U6 snRNA can still undergo retrotransposition in the human genome. The copy number of U6/L1 pseudogenes and the finding that U6 snRNA serves as a preferential substrate for LINE-1-mediated retrotransposition when compared with other snRNAs would suggest that they represent a new SINE family. However, since a functional U6 snRNA gene contains a proximal sequence element located upstream of its transcription-initiation site (Kunkel and Pederson 1988; Lobo and Hernandez 1989), most retrotransposed U6/L1 chimeras should lack promoter sequences and will be unable to undergo subsequent rounds of retrotransposition and will be “dead on arrival.” It remains possible that some retrotransposed U6 snRNA sequences may integrate near a RNA pol III promoter, leading to the generation of a transcriptionally active “founder” locus. Indeed, such copies may represent “stealth” drivers that contribute to the ongoing interspersion of U6 elements in a manner analogous to that described previously for Alu elements (Han et al. 2005). Clearly, future experiments are needed to address this interesting possibility.
Methods
Oligonucleotides
173NEOs: 5′-CAAGAAGGCGATAGAAGGCGATG-3′; 437NEOs: 5′-GAGCCCCTGATGCTCTTCGTCC-3′; 664NEOs: 5′-CCC TTCCCGCTTCAGTGACAA-3′; 1808NEOs: 5′-GCGTGCAATCC ATCTTGTTCAATG-3′; 173NEOas: 5′-CATCGCCTTCTATCGCC TTCTTG-3′; 210NEOas: 5′-GACCGCTTCCTCGTGCTTTACG-3′; U1s1: 5′-TGTGGGAAACTCGACTGC-3′; U1s2: 5′-GTGGTAG TGGGGGACTGCG-3′; U2s1: 5′-GGAAATAGGAGATGGAATAG-3′; U2s2: 5′-GCTTGCTCCGTCCACTCC-3′; U3s1: 5′-CTATAC TTTCAGGGATCATTTC-3′; U3s2: 5′-GTTCGTTACTAGAGAAG TTTC-3′; U4s1: 5′-CAATACCCCGCCATGACGAC-3′; U4s2: 5′-GAAATATAGTCGGCATTGGC-3′; U5s1: 5′-CCGTGGAGAGGAA CAAC-3′; U5s2: 5′-CTGAGTCTTAACCCAATT-3′; U6s1: 5′-ACAGAGAAGATTAGCATGGC-3′; U6s2: 5′-CCCTGCGCAAG GATGAC-3′; U6atac_s1: 5′-TGACAAGGATGGAAGAGGCCCT-3′; U6atac_s2: 5′-CTGACAACACGCATACGGTTAAG-3′; hY1s1: 5′-GTCAGTTACAGATCGAAC-3′; hY1s2: 5′-CTTGTTCTACTC TTTCCCC-3′; hY3s1: 5′-CAACTAATTGATCACAAC-3′; hY3s2: 5′-GTTACAGATTTCTTTGTTCCTTC-3′; hY4s1: 5′-TATTAA CATTAGTGTCACTAAAG-3′; hY4s2: 5′-GGTATACAACCC CCCACTGCT-3′; U6as1: 5′-GCCATGCTAATCTTCTCTGT-3′; U6as2: 5′-GTCATCCTTGCGCAGGG-3′; JA10: 5′-GAT CTCGGCCGAAAAGCGCAATATTCGGGT-3′; JB3165: 5′-AATTAACCCTCACTAAAGGGCAGGTTGACGCAAATGGGCGG TAGGCGTGTACGG-3′; JB3168: 5′-TAATACGACTCACTATAGGG CAGCGGGCAGTTCGGTTTCAGGCAGGTCTTGC-3′; JB3167: 5′-AATAACCCTCACTAAAGGGCAGCCAGCGTCTTGTCAT TGGCGAATTCGAACACGC-3′; SDM295PacIs: (5′-GCTTTATT TGTAACCATTAATTAAGCTGCAATAAACAAGTTAAC-3′); SDM295PacIas: (5′-GTTAACTTGTTTATTGCAGCTTAATT AATGGTTACAAATAAAGC-3′); Cep273as: 5′-GTTAACTTGTTTAT TGCAGC-3′.
Recombinant DNA plasmids
The following plasmids contain the indicated fragments cloned in pCEP4 (Invitrogen), unless otherwise indicated. Cloning strategies are available upon request.
pJM101/L1.3 contains a 8.2-kb NotI–BamHI fragment containing a full-length copy of the L1.3 element and the mneoI indicator cassette (Dombroski et al. 1993; Sassaman et al. 1997).
pJM101/L1.3 No neo contains a 6.0-kb NotI–BamHI fragment containing a full-length copy of the L1.3 element (Wei et al. 2001).
pJM101/L1RP contains a 8.2-kb NotI–BamHI fragment containing a full-length copy of the L1RP element and the mneoI indicator cassette (Kimberland et al. 1999).
pJM101/L1RP No neo contains a 6.0-kb NotI–BamHI fragment containing a full-length copy of the L1RP element (Wei et al. 2001).
L1.3/ORF1mneoI contains a 3.8-kb NotI–BamHI fragment containing the L1.3 5′UTR, L1.3 ORF1, and the mneoI indicator cassette (Wei et al. 2001).
L1.3/ORF1mneoI-AAA was created by swapping the NotI–AgeI fragment from L1.3 ORF1mneoI with the corresponding fragment from pDK105 (Kulpa and Moran 2005). It contains two arginine-alanine mutations in ORF1 (RR261-262AA).
L1.3/ORF1mneoI-AKR was created by swapping the NotI–AgeI fragment from L1.3 ORF1mneoI with the corresponding fragment from pDK107 (Kulpa and Moran 2005). It contains an arginine-lysine mutation in ORF1 (R261K).
pJM101/L1RP No neo SDM is a derivative of pJM101/L1RP No neo that contains a two-base insertion (AT) 26 bp upstream the SV40 poly(A) site present in pCEP4. The insertion creates a PacI restriction site. Site-directed mutagenesis was performed using the QuickChange XL Site-Directed Mutagenesis Kit (Stratagene) using the SDM295PacIs and SDM295PacIas primers.
pJM101/L1RP SDM is a derivative of pJM101/L1RP No neo SDM that contains the mneoI indicator cassette.
pCEPTGf21 contains a 8.8-kb NotI–BamHI fragment containing a full-length mouse TGf21 L1 element and the mneoI indicator cassette (Goodier et al. 2001).
pCEPL1SM contains a 9.5-kb NotI–BamHI fragment containing a full-length synthetic mouse L1 (L1spa) and the mneoI indicator cassette. The sequence of both ORFs was codon optimized (Han and Boeke 2004).
pL1SM/ORF1mneoI contains a 4.1-kb NotI–BamHI fragment containing the L1SM 5′UTR, the L1SM ORF1, and the mneoI indicator cassette.
pCEP 5′UTR ORF2 No Neo contains a 5.1-kb NotI–BamHI fragment containing the L1.3 5′UTR and L1.3 ORF2 (Alisch et al. 2006).
pAluNF1-neoIII has been described previously (Dewannieux et al. 2003). It contains a 2.1-kb fragment containing a 7SL promoter, a copy of the NF1 Alu (Wallace et al. 1991), the neoIII self-splicing indicator cassette (Esnault et al. 2002), a 44-bp poly(A) tail, and a 7SL transcription termination sequence cloned in pUC19 (Dewannieux et al. 2003).
pL1.3neoIII contains a 8.0-kb NotI–BamHI fragment containing a full-length copy of the L1.3 element and the neoIII self-splicing indicator cassette (Dombroski et al. 1993; Sassaman et al. 1997; Esnault et al. 2002).
In silico analysis
Sequences of the small nuclear RNA genes used to perform BLAST search on Ensembl v40 and v41 (August and October 2006; http://www.ensembl.org/Homo_sapiens/blastview) are as follows: U1 accession no. J00318, U2 accession no. K02227, U3 accession no. AF020534, U4 accession no. M15957, U5 accession no. X04215, U6 accession no. M14486, U11 accession no. X58716, U12 accession no. J04119, U4atac accession no. U62822, and U6atac accession no. U62823. We limited our analysis to the sequences having an E value ≤5 × 10−27 corresponding to a U6 full-length sequence (107 bp) with <10% divergence from the query sequence.
DNA preparation
Plasmid DNAs were purified using QIAGEN midi-prep columns (QIAGEN). HeLa genomic DNA was isolated using either the Blood and Cell Midi-prep kit (QIAGEN) or the Cell and Tissue DNA isolation kit (Puregene, Gentra).
Cell culture
HeLa cells were grown at 37°C in an atmosphere containing 7% carbon dioxide and 100% humidity in Dulbecco’s modified Eagle medium (DMEM) lacking pyruvate (GIBCO BRL). DMEM was supplemented with 10% fetal bovine calf serum and 1× Penicillin-Streptomycin-L-glutamine (a 100× stock is sold by GIBCO BRL). Cell passage and cloning (by either limiting dilution or colony lifting) was performed using standard techniques.
Cis and trans retrotransposition assays
Both assays have been described previously (Moran et al. 1996; Wei et al. 2000, 2001; Bogerd et al. 2006). Briefly, in the cis-retrotransposition assay 2 × 104 and/or 2 × 103 HeLa cells were plated in duplicate 6-well tissue culture dishes. After 12–14 h, one set of 6-well plates was cotransfected with equal amounts of a reporter plasmid (humanized Renilla green fluorescent protein [hr-EGFP; Stratagene]) and a L1 tagged with the mneoI indicator cassette. The other set of 6-well plates was transfected with only the L1 construct. We routinely used 3 μL of Fugene 6 transfection reagent (Roche Molecular Biochemical) and 1.0 μg of DNA per well. Seventy-two hours post-transfection, the HeLa cells in one set of 6-well tissue culture plates was trypsinized and subjected to flow cytometry (Ostertag et al. 2000; Wei et al. 2000). The percentage of green fluorescent (GFP) cells was used to determine the transfection efficiency of each sample. The remaining set of 6-well plates was subjected to G418 selection (400 μg/mL). After 12 d, pools of G418-resistant cells were collected from three wells and genomic DNA was extracted. The remaining three wells were washed in 1× phosphate-buffered saline (PBS), fixed by incubation in FIX solution (2% formaldehyde [of a 37% stock solution in water], 0.2% glutaraldehyde, 1× PBS) at 4°C for 30 min. The fixed cells were stained with either 0.1% Brilliant Blue or 0.1% Crystal Violet at room temperature overnight, washed with water, and then manually counted. The retrotransposition efficiency is expressed as the number of G418-resistant foci/the number of transfected (EGFP-positive) cells. In the trans-retrotranspositon assay, 2 × 105 and/or 2 × 104 HeLa cells were plated in 6-well tissue culture dishes. Approximately 12–14 h after plating, cells were cotransfected with equal amounts of a “reporter plasmid” (e.g., L1.3/ORF1mneoI, L1SM/ORF1mneoI, or Alu-NF1 neoIII) and a “driver” L1 that lacked the mneoI indicator cassette (e.g., JM101/L1RP No neo or 5′UTR ORF2 No neo). Transfection conditions, transfection efficiency calculations, G418 selection, and pool harvesting was preformed as described above. When a mutant L1.3/ORF1mneoI construct was assayed in the trans-retrotranspositon assay, 6 × 106 HeLa cells were plated in a T-175 cm2 flask and transfected with equal amounts of a “driver” L1 and “reporter” plasmid using established protocols (Wei et al. 2000, 2001). Retrotransposition assays were performed as described above.
RNase protection assays
HeLa cells were plated in T-175 cm2 flasks at a density of 6 × 106 cells/flask and were transfected with 30 μg of the ORF1mneoI expression plasmids using 90 μL of the Fugene 6 transfection reagent (Roche Molecular Biochemicals) (Wei et al. 2000). Cells were harvested 72 h post-transfection and total RNA was prepared using the TRIzol reagent (Invitrogen). RNA samples were treated with 100 U of RNase-free DNase I (Roche Molecular Biochemicals) for 1 h at 37°C, extracted with acid-equilibrated phenolchloroform, and precipitated with 2 Vol of 100% ethanol. PCR products containing T7 promoter sequences were used as templates for in vitro transcription, which was carried out using T7 RNA polymerase in the presence of [α-32P]UTP using the Maxiscript in vitro transcription kit (Ambion). The primers JB3169 and JA10 were used to generate the L1 probes. JB3168 and JB3167 were used to generate the hyg probe. We used a Hybaid Thermocycler programmed as follows: One cycle of 95°C for 10 min, 55°C for 7 min, 72°C for 1 min, followed by 30 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 1 min, followed by a final extension step of 72°C for 10 min. RNase protection assays were performed using the RPA III kit (Ambion). Briefly, 30 μg of total RNA was hybridized to gel-purified probe (1 × 105 cpm/reaction) at 42°C overnight. The reactions were then digested in a mixture of RNase A (0.375 U) and RNase T1 (15 U) at 37°C for 1 h, precipitated, and resolved on a 6% denaturing polyacrylamide gel.
Fluorescent primer extension
Genomic DNAs from pools of G418-resistant colonies generated from the trans-complementation assay were used as templates in PCR reactions with oligonucleotides specific to the indicator cassette (437NEOs) and a sequence in the engineered L1 that was located upstream of the SV40 poly(A) site in the pCEP4 vector (cep273as). PCR was performed as follows: one cycle of 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. We used the PCR product to conduct Fluorescent Primer Extension (FluPE) reactions. Primer extension was performed in a volume of 8 μL containing 100 fmol of the PCR product, 100 fmol of the Cy5 primer, 25 μM each dNTP (we used ddCTP to limit the extension after the nucleotide polymorphism), 2 mM MgCl2, 10 mM Tris-HCl (pH 8.8), 10 mM KCl, 0,002% Tween-20, and 0.64 U of Thermo sequenase (Amersham Pharmacia Biotech). Extension reactions were cycled as follows: one cycle of 94°C for 2 min, followed by 25 cycles of 55°C for 40 sec and 94° for 30 sec. The products were resolved on a 19% denaturing polyacrylamide gel using the ALFexpress II (Amersham Pharmacia Biotech) automated sequencing system. The relative peak fluorescence signal was measured for each extension product using the AlleleLinks software package (v1.01) (Bennett-Baker et al. 2003). To calculate the sensitivity of the reaction, PCR was used to amplify DNA fragments containing or lacking the two-nucleotide polymorphism. The resultant DNA fragments were quantified and serially diluted to produce “reporter” (ORF1mneoI allele) to “driver” (JM101/L1RPSDM allele) ratio mixtures of 1:1, 1:5, 1:10, 1:50, 1:100, 1:250, 1:500, and 1:1000, respectively. Each mixture was used as a template in the primer-extension assay. We could reproducibly detect a signal in the 1:250 dilution sample.
Nested PCR amplification
Chimeric pseudogenes (snRNA/L1 and U6/Alu) were amplified using a nested PCR protocol on genomic DNAs derived from pools of G418-resistant colonies (see above). First-round PCR reactions were conducted in a volume of 50 μL using oligonucleotides specific to the snRNA (oligonucleotides s1) and NEO gene (210NEOas) using 200 ng of genomic DNA as a template. Second-round PCR reactions were performed using 4 μL of the first-round PCR as a template with the s2 primer and 173NEOas oligonucleotide primers. All PCR reactions were performed using High Fidelity Taq (Roche Molecular Biochemical) in an express thermal cycler (Hybaid) using the following cycling conditions: one cycle at 95°C for 2 min, followed by 30 cycles at 94°C for 15 sec, 55°C for 20 sec, and 72°C for 3 min, followed by a final extension cycle of 72°C for 10 min. The PCR products from the second amplification were purified using the QIAquick PCR purification kit (QIAGEN) and sequenced on an Applied Biosystems DNA sequencer (ABI 377) at the University of Michigan Core facilities or at the IGH. Some PCR products were cloned in pGEMT-Easy (Promega) prior to sequencing.
Inverse PCR
Inverse PCR was performed essentially as described previously (Morrish et al. 2002). Briefly, 5 μg of genomic DNA derived from G418-resistant colonies was digested to completion with SspI (New England Biolabs) in a total reaction volume of 50 μL. Heating at 65°C for 30 min stopped the reactions. The restricted DNA was ligated using T4 DNA ligase (3200 U; New England Biolabs) in a volume of 600 μL at 16°C for at least 16 h. The ligated DNA was precipitated with ethanol and dissolved in 40 μL of distilled water. Two microliters of DNA were used in the primary PCR reaction in a 50-μL reaction volume containing a 20-nM concentration of each dNTP, 10 pmol of primers U6as2 and 664NEOs, 1× buffer 3, and 2.5 U of enzyme mix in the Expand Long Template PCR system (Roche Molecular Biochemicals). We used a Hybaid Thermocycler programmed as follows: one cycle of 95°C for 2 min, followed by 30 cycles of 94°C for 10 sec, 63°C for 30 sec, and 68°C for 15 min, followed by a final extension step at 68°C for 30 min. The resultant products (1 μL) then were used in a secondary PCR reaction using the same conditions, except that we used primers U6as1 and 1808NEOs. PCR products from the second amplification were purified with QIAquick PCR purification kit (QIAGEN) and sequenced. Sequence flanking the insertion was used as probe in BLAT search (http://genome.cse.ucsc.edu) to identify the preintegration site in the HGWD (Kent 2002).
Acknowledgments
We thank members of the IGH and the University of Michigan Sequencing Core for DNA sequencing assistance. We thank John Goodier for providing the TGf21 “natural” mouse L1, Jef Boeke for providing the synthetic mouse L1 element, and Thierry Heidmann for providing the Alu expression cassette. We thank Matthew Pratt-Hyatt, Laure Boizeau, Aurélie Chanut, and Oussama Meziane for their excellent technical assistance. We thank Lee Lawton and David Burke for assistance with the FluPE experiments. This work was supported in part by grants to N.G. from the Centre National de Recherche Scientifique and the Agence Nationale de la Recherche (JC05 63089). A.J.D. was the recipient of a fellowship from the French government (Ministère de l’Enseignement Supérieur et de la Recherche). J.V.M. was supported in part by the National Institutes of Health (GM60518). J.L.G.P. was supported in part by a MEC/Fulbright postdoctoral grant EX-2003-0881 (MEC, Spain).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5870107
References
- Alisch R.S., Garcia-Perez J.L., Muotri A.R., Gage F.H., Moran J.V., Garcia-Perez J.L., Muotri A.R., Gage F.H., Moran J.V., Muotri A.R., Gage F.H., Moran J.V., Gage F.H., Moran J.V., Moran J.V. Unconventional translation of mammalian LINE-1 retrotransposons. Genes & Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett E.A., Coleman L.E., Tsui C., Pittard W.S., Devine S.E., Coleman L.E., Tsui C., Pittard W.S., Devine S.E., Tsui C., Pittard W.S., Devine S.E., Pittard W.S., Devine S.E., Devine S.E. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Baker P.E., Wilkowski J., Burke D.T., Wilkowski J., Burke D.T., Burke D.T. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–2062. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Wiegand H.L., Hulme A.E., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R., Wiegand H.L., Hulme A.E., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R., Hulme A.E., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R., O'Shea K.S., Moran J.V., Cullen B.R., Moran J.V., Cullen B.R., Cullen B.R. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Retroposons–seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Brosius J. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica. 1999;107:209–238. [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr., Farley A.H., Moran J.V., Kazazian H.H., Jr., Moran J.V., Kazazian H.H., Jr., Kazazian H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdin A., Ustyugova S., Gogvadze E., Vinogradova T., Lebedev Y., Sverdlov E., Ustyugova S., Gogvadze E., Vinogradova T., Lebedev Y., Sverdlov E., Gogvadze E., Vinogradova T., Lebedev Y., Sverdlov E., Vinogradova T., Lebedev Y., Sverdlov E., Lebedev Y., Sverdlov E., Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics. 2002;80:402–406. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- Buzdin A., Gogvadze E., Kovalskaya E., Volchkov P., Ustyugova S., Illarionova A., Fushan A., Vinogradova T., Sverdlov E., Gogvadze E., Kovalskaya E., Volchkov P., Ustyugova S., Illarionova A., Fushan A., Vinogradova T., Sverdlov E., Kovalskaya E., Volchkov P., Ustyugova S., Illarionova A., Fushan A., Vinogradova T., Sverdlov E., Volchkov P., Ustyugova S., Illarionova A., Fushan A., Vinogradova T., Sverdlov E., Ustyugova S., Illarionova A., Fushan A., Vinogradova T., Sverdlov E., Illarionova A., Fushan A., Vinogradova T., Sverdlov E., Fushan A., Vinogradova T., Sverdlov E., Vinogradova T., Sverdlov E., Sverdlov E. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 2003;31:4385–4390. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost G.J., Feng Q., Jacquier A., Boeke J.D., Feng Q., Jacquier A., Boeke J.D., Jacquier A., Boeke J.D., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison R.A., Van Arsdell S.W., Bernstein L.B., Weiner A.M., Van Arsdell S.W., Bernstein L.B., Weiner A.M., Bernstein L.B., Weiner A.M., Weiner A.M. Abundant psuedogenes for small nuclear RNAs are dispersed in the human genome. Proc. Natl. Acad. Sci. 1981;78:810–814. doi: 10.1073/pnas.78.2.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T., Esnault C., Heidmann T., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dombroski B.A., Mathias S.L., Nanthakumar E., Scott A.F., Kazazian H.H., Jr., Mathias S.L., Nanthakumar E., Scott A.F., Kazazian H.H., Jr., Nanthakumar E., Scott A.F., Kazazian H.H., Jr., Scott A.F., Kazazian H.H., Jr., Kazazian H.H., Jr. Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Dombroski B.A., Scott A.F., Kazazian H.H., Jr., Scott A.F., Kazazian H.H., Jr., Kazazian H.H., Jr. Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl. Acad. Sci. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergun S., Buschmann C., Heukeshoven J., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G., Buschmann C., Heukeshoven J., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G., Heukeshoven J., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G., Schnieders F., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G., Chalajour F., Kilic N., Stratling W.H., Schumann G.G., Kilic N., Stratling W.H., Schumann G.G., Stratling W.H., Schumann G.G., Schumann G.G. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- Esnault C., Maestre J., Heidmann T., Maestre J., Heidmann T., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Esnault C., Casella J.F., Heidmann T., Casella J.F., Heidmann T., Heidmann T. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 2002;30:e49. doi: 10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E., Nazarbaghi R., Zomorrodi M., Herrnstadt C., Parker W.D., Davis R.E., Ghosh S.S., Nazarbaghi R., Zomorrodi M., Herrnstadt C., Parker W.D., Davis R.E., Ghosh S.S., Zomorrodi M., Herrnstadt C., Parker W.D., Davis R.E., Ghosh S.S., Herrnstadt C., Parker W.D., Davis R.E., Ghosh S.S., Parker W.D., Davis R.E., Ghosh S.S., Davis R.E., Ghosh S.S., Ghosh S.S. Multiplex fluorescence-based primer extension method for quantitative mutation analysis of mitochondrial DNA and its diagnostic application for Alzheimer's disease. Nucleic Acids Res. 1997;25:3102–3109. doi: 10.1093/nar/25.15.3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Moran J.V., Kazazian H.H., Jr., Boeke J.D., Moran J.V., Kazazian H.H., Jr., Boeke J.D., Kazazian H.H., Jr., Boeke J.D., Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Freeman J.D., Goodchild N.L., Mager D.L., Goodchild N.L., Mager D.L., Mager D.L. A modified indicator gene for selection of retrotransposition events in mammalian cells. Biotechniques. 1994;17:46, 48–49, 52. [PubMed] [Google Scholar]
- Gilbert N., Lutz S., Morrish T.A., Moran J.V., Lutz S., Morrish T.A., Moran J.V., Morrish T.A., Moran J.V., Moran J.V. Multiple fates of l1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogvadze E.V., Buzdin A.A., Sverdlov E.D., Buzdin A.A., Sverdlov E.D., Sverdlov E.D. [Multiple template switches on LINE-directed reverse transcription: The most probable formation mechanism for the double and triple chimeric retroelements in mammals] Bioorg. Khim. 2005;31:82–89. doi: 10.1007/s11171-005-0010-z. [DOI] [PubMed] [Google Scholar]
- Goodier J.L., Ostertag E.M., Du K., Kazazian H.H., Jr., Ostertag E.M., Du K., Kazazian H.H., Jr., Du K., Kazazian H.H., Jr., Kazazian H.H., Jr. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.S., Boeke J.D., Boeke J.D. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- Han K., Xing J., Wang H., Hedges D.J., Garber R.K., Cordaux R., Batzer M.A., Xing J., Wang H., Hedges D.J., Garber R.K., Cordaux R., Batzer M.A., Wang H., Hedges D.J., Garber R.K., Cordaux R., Batzer M.A., Hedges D.J., Garber R.K., Cordaux R., Batzer M.A., Garber R.K., Cordaux R., Batzer M.A., Cordaux R., Batzer M.A., Batzer M.A. Under the genomic radar: The stealth model of Alu amplification. Genome Res. 2005;15:655–664. doi: 10.1101/gr.3492605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohjoh H., Singer M.F., Singer M.F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- Holmes S.E., Singer M.F., Swergold G.D., Singer M.F., Swergold G.D., Swergold G.D. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 1992;267:19765–19768. [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D., LaCava J., Tollervey D., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Hulme A.E., Kulpa D.A., Garcia-Perez J.L., Moran J.V., Kulpa D.A., Garcia-Perez J.L., Moran J.V., Garcia-Perez J.L., Moran J.V., Moran J.V. The impact of LINE-1 retrotransposition on the human genome. In: Lupski J., Stankiewicz P., Stankiewicz P., editors. Genomic disorders: The genomic basis of disease. Humana Press; Totowa, NJ: 2006. [Google Scholar]
- Jensen S., Heidmann T., Heidmann T. An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 1991;10:1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Klonowski P., Kohany O., Walichiewicz J., Kohany O., Walichiewicz J., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H., Jr., Moran J.V., Moran J.V. The impact of L1 retrotransposons on the human genome. Nat. Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT––The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberland M.L., Divoky V., Prchal J., Schwahn U., Berger W., Kazazian H.H., Jr., Divoky V., Prchal J., Schwahn U., Berger W., Kazazian H.H., Jr., Prchal J., Schwahn U., Berger W., Kazazian H.H., Jr., Schwahn U., Berger W., Kazazian H.H., Jr., Berger W., Kazazian H.H., Jr., Kazazian H.H., Jr. Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- Kulpa D.A., Moran J.V., Moran J.V. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- Kulpa D.A., Moran J.V., Moran J.V. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Kunkel G.R., Pederson T., Pederson T. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes & Dev. 1988;2:196–204. doi: 10.1101/gad.2.2.196. [DOI] [PubMed] [Google Scholar]
- Lobo S.M., Hernandez N., Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H., Korman M.H., Jakubczak J.L., Eickbush T.H., Jakubczak J.L., Eickbush T.H., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Martin S.L., Bushman F.D., Bushman F.D. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.L., Branciforte D., Keller D., Bain D.L., Branciforte D., Keller D., Bain D.L., Keller D., Bain D.L., Bain D.L. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.L., Cruceanu M., Branciforte D., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Cruceanu M., Branciforte D., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Branciforte D., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Wai-Lun Li P., Kwok S.C., Hodges R.S., Williams M.C., Kwok S.C., Hodges R.S., Williams M.C., Hodges R.S., Williams M.C., Williams M.C. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Mathias S.L., Scott A.F., Kazazian H.H., Jr., Boeke J.D., Gabriel A., Scott A.F., Kazazian H.H., Jr., Boeke J.D., Gabriel A., Kazazian H.H., Jr., Boeke J.D., Gabriel A., Boeke J.D., Gabriel A., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr., Boeke J.D., Kazazian H.H., Jr., Kazazian H.H., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V., Taccioli G.E., Batzer M.A., Moran J.V., Batzer M.A., Moran J.V., Moran J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Ohshima K., Okada N., Okada N. SINEs and LINEs: Symbionts of eukaryotic genomes with a common tail. Cytogenet. Genome Res. 2005;110:475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- Ostertag E.M., Prak E.T., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr., Prak E.T., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr., Moran J.V., Kazazian H.H., Jr., Kazazian H.H., Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag E.M., Goodier J.L., Zhang Y., Kazazian H.H., Jr., Goodier J.L., Zhang Y., Kazazian H.H., Jr., Zhang Y., Kazazian H.H., Jr., Kazazian H.H., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J., Noel J.F., Briere F., Cousineau B., Lucier J.F., Perreault J.P., Boire G., Noel J.F., Briere F., Cousineau B., Lucier J.F., Perreault J.P., Boire G., Briere F., Cousineau B., Lucier J.F., Perreault J.P., Boire G., Cousineau B., Lucier J.F., Perreault J.P., Boire G., Lucier J.F., Perreault J.P., Boire G., Perreault J.P., Boire G., Boire G. Retropseudogenes derived from the human Ro/SS-A autoantigen-associated hY RNAs. Nucleic Acids Res. 2005;33:2032–2041. doi: 10.1093/nar/gki504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr., Gabriel A., Swergold G.D., Kazazian H.H., Jr., Swergold G.D., Kazazian H.H., Jr., Kazazian H.H., Jr. Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Schmitz J., Churakov G., Zischler H., Brosius J., Churakov G., Zischler H., Brosius J., Zischler H., Brosius J., Brosius J. A novel class of mammalian-specific tail-less retropseudogenes. Genome Res. 2004;14:1911–1915. doi: 10.1101/gr.2720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A.F., Schmeckpeper B.J., Abdelrazik M., Comey C.T., O'Hara B., Rossiter J.P., Cooley T., Heath P., Smith K.D., Margolet L., Schmeckpeper B.J., Abdelrazik M., Comey C.T., O'Hara B., Rossiter J.P., Cooley T., Heath P., Smith K.D., Margolet L., Abdelrazik M., Comey C.T., O'Hara B., Rossiter J.P., Cooley T., Heath P., Smith K.D., Margolet L., Comey C.T., O'Hara B., Rossiter J.P., Cooley T., Heath P., Smith K.D., Margolet L., O'Hara B., Rossiter J.P., Cooley T., Heath P., Smith K.D., Margolet L., Rossiter J.P., Cooley T., Heath P., Smith K.D., Margolet L., Cooley T., Heath P., Smith K.D., Margolet L., Heath P., Smith K.D., Margolet L., Smith K.D., Margolet L., Margolet L. Origin of the human L1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell S.W., Denison R.A., Berstein L.B., Weiner A.M., Manser T., Gesteland R.F., Denison R.A., Berstein L.B., Weiner A.M., Manser T., Gesteland R.F., Berstein L.B., Weiner A.M., Manser T., Gesteland R.F., Weiner A.M., Manser T., Gesteland R.F., Manser T., Gesteland R.F., Gesteland R.F. Direct repeats flank three small nuclear RNA pseudogenes in the human genome. Cell. 1981;26:11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Wallace M.R., Andersen L.B., Saulino A.M., Gregory P.E., Glover T.W., Collins F.S., Andersen L.B., Saulino A.M., Gregory P.E., Glover T.W., Collins F.S., Saulino A.M., Gregory P.E., Glover T.W., Collins F.S., Gregory P.E., Glover T.W., Collins F.S., Glover T.W., Collins F.S., Collins F.S. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- Wang H., Xing J., Grover D., Hedges D.J., Han K., Walker J.A., Batzer M.A., Xing J., Grover D., Hedges D.J., Han K., Walker J.A., Batzer M.A., Grover D., Hedges D.J., Han K., Walker J.A., Batzer M.A., Hedges D.J., Han K., Walker J.A., Batzer M.A., Han K., Walker J.A., Batzer M.A., Walker J.A., Batzer M.A., Batzer M.A. SVA elements: A hominid-specific retroposon family. J. Mol. Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Weber M.J. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Morrish T.A., Alisch R.S., Moran J.V., Morrish T.A., Alisch R.S., Moran J.V., Alisch R.S., Moran J.V., Moran J.V. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Kazazian H.H., Boeke J.D., Moran J.V., Boeke J.D., Moran J.V., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]