Abstract
Large-scale cDNA sequencing projects and tiling array studies have revealed the presence of many unannotated genes. For protein coding genes, small coding sequences may not be identified by gene finders because of the conservative nature of prediction algorithms. In this study, we identified small open reading frames (sORFs) with high coding potential by a simple gene finding method (Coding Index, CI) based on the nucleotide composition bias found in most coding sequences. Applying this method to 18 Arabidopsis thaliana and 84 yeast sORF genes with evidence of expression at the protein level gives 100% accurate prediction. In the A. thaliana genome, we identified 7159 sORFs that are likely coding sequences (coding sORFs) with the CI measure at the 1% false-positive rate. To determine if these coding sORFs are parts of functional genes, we evaluated each coding sORF for evidence of transcription or evolutionary conservation. At the 5% false-positive rate, we found that 2996 coding sORFs are likely expressed in at least one experimental condition of the A. thaliana tiling array data. In addition, the evolutionary conservation of each A. thaliana sORF was examined within A. thaliana or between A. thaliana and five plants with complete or partial genome sequences. In 3997 coding sORFs with readily identifiable homologous sequences, 2376 are subject to purifying selection at the 1% false-positive rate. After eliminating coding sORFs with similarity to known transposable elements and those that are likely missing exons of known genes, the remaining 3241 coding sORFs with either evidence of transcription or purifying selection likely belong to novel coding genes in the A. thaliana genome.
Transcriptome sequencing and whole genome tiling array studies have revealed significant levels of expression in numerous intergenic regions in human (Kapranov et al. 2002; Rinn et al. 2003), fly (Stolc et al. 2004; Manak et al. 2006), sea urchin (Samanta et al. 2006), Arabidopsis thaliana (Yamada et al. 2003; Stolc et al. 2005b), and rice (Stolc et al. 2005a), suggesting the presence of genic sequences in unannotated “intergenic” regions. However, in most cases it remains an open question if these transcripts represent unannotated protein coding or RNA genes. Some studies assumed that these sequences represent noncoding RNA genes because the sequences had not been annotated as coding regions (Stolc et al. 2005b; Yamada et al. 2003). In several other cases, the designation of coding regions tends to be arbitrary with an ad hoc length threshold without distinguishing coding from noncoding sequences experimentally or computationally with gene finders (Ota et al. 2004).
Most ab initio gene prediction programs distinguish coding (CDS) and noncoding sequences (NCDS) with their differences in nucleotide composition, intron splice sites, promoters, translational start/stop sites, and polyadenylation signals. These signals are generally integrated for evaluating the coding likelihood of a sequence (Brent and Guigo 2004). The integration of multiple criteria decreases the chance that false exons are predicted as true (low false-positive rate) but likely increases the chance that true exons are not predicted (high false-negative rate) (Claverie 1997). The issue of false-negative prediction is particularly serious for smaller CDSs (≤300 nucleotides) due to the difficulty in distinguishing the relatively few biologically meaningful sequences from the very large pool of small ORFs (sORFs) (Basrai et al. 1997; Wang et al. 2003). Despite the difficulties in their prediction, proteins translated from sORFs include several classes of important genes. In yeast, these small proteins include mating pheromones, proteins involved in energy metabolism, proteolipids, chaperonins, stress proteins, transporters, transcriptional regulators, nucleases, ribosomal proteins, thioredoxins, and metal ion chelators (Basrai et al. 1997). In addition, many yeast sORF genes missed by ab initio prediction methods but supported by evidence of expression have been shown to be translated and functional in many cases (Ghaemmaghami et al. 2003; Huh et al. 2003; Kastenmayer et al. 2006). In human, 997 known genes from Ensembl (Hubbard et al. 2002) are coding sORFs, and 593 of them are annotated in Refseq (Pruitt et al. 2005) or Swissprot (Boeckmann et al. 2005). In A. thaliana, relatively little is known about sORF genes, but a number of small, secreted proteins that likely act as receptor ligands are identified not by gene finding programs but by similarity searches and/or functional studies (Cock and McCormick 2001; Butenko et al. 2003).
Noting the relatively high false-negative rate of current gene finding algorithms and the difficulty to identify small protein genes, we developed the Coding Index (CI) measure for coding sORF prediction based only on the hexamer composition bias, which has been established as a general measure for distinguishing CDS from NCDS (Farber et al. 1992; Fickett and Tung 1992). After validating the CI measure using known small coding genes from yeast and A. thaliana, it is applied to predict coding sORFs ranging from 90 to 300 bp in the intergenic regions of the A. thaliana genome. We then looked for evidence of expression with the A. thaliana genome tiling array data. Finally, since most genes are subject to purifying selection, a functional coding sORF is expected to undergo stronger selective constraints on nonsynonymous sites than for synonymous ones (Li 1997; Makalowski and Boguski 1998). Therefore, we examined the signature of purifying selection among predicted coding sORFs. The analysis procedures and findings are summarized in Figure 1.
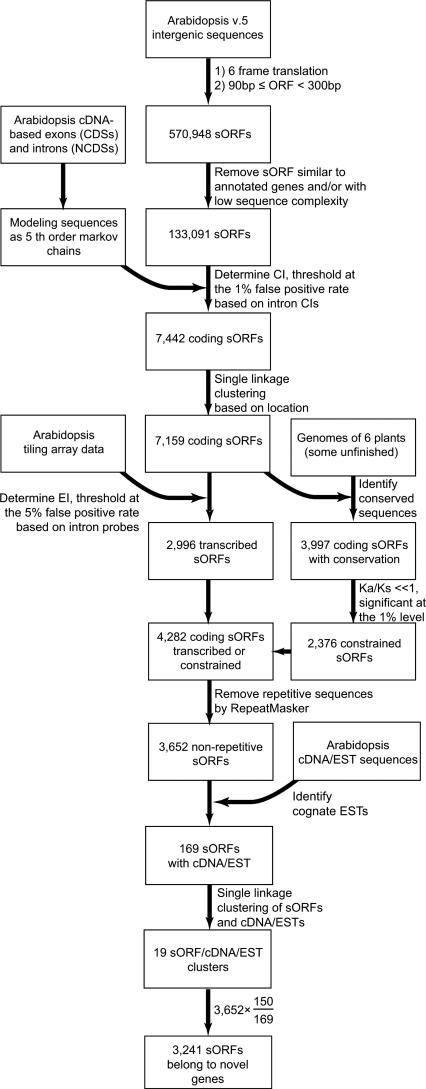
Figure 1.
Analysis procedures and summary of results The overall procedures for identifying sORFs (between 90 and 300 bp) that have qualifying Coding Index (CI) values, above background tiling array hybridization intensities, evidence of purifying selection, and cognate cDNA/ESTs.
Results
The CI measure
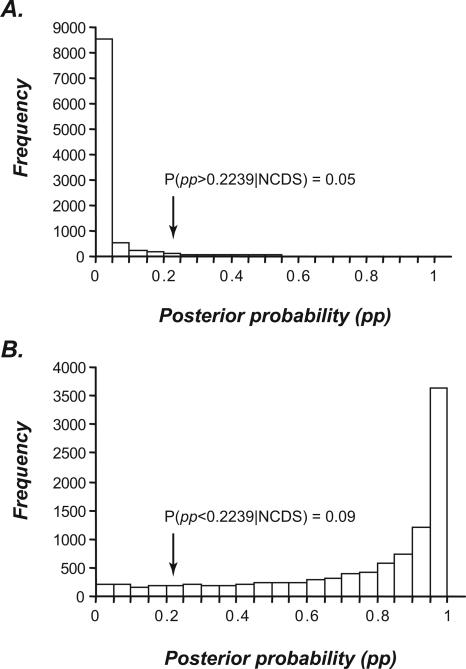
We first derived the posterior probability (pp) that a particular reading frame is coding according to the hexamer composition of CDS and NCDS, using Bayes’ theorem applied along a Markov Chain (see Methods). We conducted simulation studies with various sequence lengths and found 75 bp to be the shortest length providing acceptable statistical power for distinguishing between CDS and NCDS (the Kolmogorov-Smirnov [K-S] test, P < 2 × 10−16; Fig. 2). Since 95% of the NCDS-like random sequences have pp < 0.2239, any 75-bp sequence window with a pp ≥ 0.2239 is regarded as coding. We examined the pp values of 75-bp windows within the exons and introns of A. thaliana with 3-bp steps. In average, 84.95% of the windows in exons and only 2.30% of windows in introns are above the threshold.
Figure 2.
Frequency distributions of posterior probabilities for simulated coding and noncoding sequences. (A) Distribution of posterior probability (pp) of sequences resembling noncoding sequence (NCDS). Ten-thousand random sequences were generated based on the hexamer and pentamer frequencies of intronic ORFs. The great majority of simulated sequences have very small pp, and only 5% of the pp values are >0.2239. (B) The pp distribution of sequences resembling coding sequences (CDSs). Random sequences were generated according to cDNA CDSs. Approximately 10% of the CDS-like random sequences have pp values <0.2239.
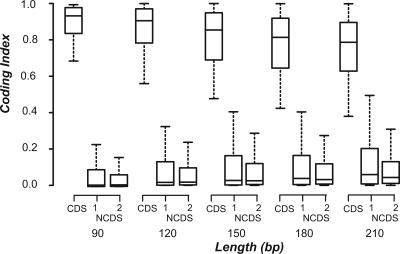
In addition to annotated genes, many intergenic regions in the genome contain windows with high pp values. An example (discussed in a later section) is shown in Figure 3. These tracks with high pp values differ in widths and shorter tracks are more likely to be spurious than longer ones. Two measures are necessary for evaluating the coding likelihood of these high pp regions: (1) a summary statistic describing the coding potential of the region and (2) a threshold definition that takes region lengths into account. We devised the CI measure that is the averaged pp values over all windows (75-bp window with 3-bp increments) in a given sequence. The regions we examined contain at least six windows since we focused on sORFs from 90 to 300 bp. Simulation studies were conducted to determine the CI thresholds at different NCDS (random sequences generated according to intron hexamer compositions) lengths ranging from 90 to 300 bp. Although the median CI values of different sequence lengths are similar, the distributions of shorter sequences are significantly skewed toward higher CI values compared with longer sequences (Supplement A). We have also calculated CI values of exon CDS, intronic ORFs, and intergenic ORFs at different sequence lengths. In contrast to simulated sequences, the distribution of longer NCDS sequences is skewed toward higher CI values (Fig. 4). Since longer ORFs are less likely to be expected randomly compared to shorter ORFs, longer ORFs from intergenic regions and introns are more likely to be true coding sequences. In addition, the thresholds defined with simulated sequences are more conservative (with higher CI threshold values) than thresholds defined based on intronic or intergenic ORFs regardless of sequence length. Therefore, a threshold curve was generated by fitting the 99 percentile CI values of simulated sequence at various lengths with power law (r2 = 0.989) and the fitted line was used to infer the CI thresholds at the 1% false-positive rate.
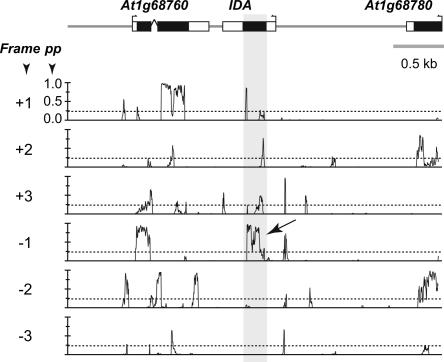
Figure 3.
Sliding window calculation of pp in genomic sequences surrounding IDA. The pp values were determined in 75-bp windows with 3-bp steps for A. thaliana chromosome sequences. The pp values in a region containing the small protein gene IDA and flanking sequences are shown. The diagram on top indicates the locations of exons (white box, untranslated regions; black box, CDS), introns (bent lines), transcriptional starts (small arrows), and intergenic sequences (thick gray lines). The six plots below the annotation diagram are the results of pp calculations in six reading frames (forward, +; reverse, −). The dotted line indicates pp = 0.2239, the threshold value for calling whether a 75-bp window is likely a CDS or not. The shaded areas highlight the overlap between IDA CDS and regions with a high pp. The arrow indicates the correct frame for the IDA CDS.
Figure 4.
Distributions of CI values of CDS and NCDS The CI value distributions are shown as box plots with the solid horizontal line indicating the median CI value, the box representing the inter quartile range (25%–75%), and the dotted line indicating the first to the 99th percentile. CDS refers to the exon coding sequences derived from full-length cDNAs. sORFs of NCDSs are obtained from two types of sequences: (1) annotated intergenic regions and (2) intron sequences derived from full-length cDNAs.
Performance of the CI measure
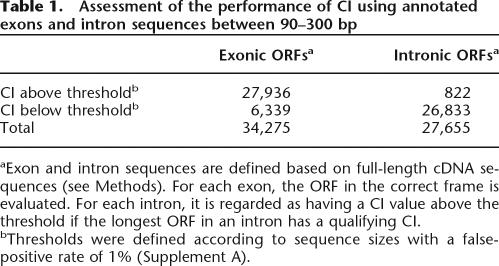
To evaluate the performance of CI and the thresholds defined, first we calculated and compared the CI values of exons and introns from genes with full-length cDNAs from A. thaliana. Of the 34,275 exons in the length range 90–300 bp, 27,936 (81.50%) have above-threshold CI values (Table 1). For the same size range, only 3% of the sORFs in introns were called as CDSs. Although the CI measure is strongly influenced by ORF sizes, the CI distributions of exon CDSs are always clearly separated from intergenic and intronic sequences (Fig. 4). To further evaluate the feasibility of using the CI measure to identify novel small protein genes, we applied our method to two small protein gene data sets. The first is a collection of 18 A. thaliana genes that code for small, secreted proteins (90–300 bp). These genes were not annotated in the original release of the A. thaliana genome but were identified by similarity searches and/or functional studies (Cock and McCormick 2001; Butenko et al. 2003). The CDSs of these small protein-coding genes are located in regions with high pp values (e.g., IDA shown in Fig. 3). Most importantly, the CI values of all 18 A. thaliana small protein-coding genes are above the CI thresholds (Supplement B). It should be noted that the training data are based on sequences without substantial low complexity regions. Most of the 18 A. thaliana small proteins, however, would have been excluded after filtering for low complexity regions. This finding indicates that there is likely coding sORFs with substantial low complexity sequences that we failed to identify.
Table 1.
Assessment of the performance of CI using annotated exons and intron sequences between 90–300 bp
aExon and intron sequences are defined based on full-length cDNA sequences (see Methods). For each exon, the ORF in the correct frame is evaluated. For each intron, it is regarded as having a CI value above the threshold if the longest ORF in an intron has a qualifying CI.
bThresholds were defined according to sequence sizes with a false-positive rate of 1% (Supplement A).
The second small protein gene data set is a collection of yeast small coding genes with evidence of expression at the protein level based on genome-wide TAP- and GFP-tagging experiments (Ghaemmaghami et al. 2003; Huh et al. 2003). Again, many of these small coding genes are discovered through expression-based analysis instead of ab initio gene finding (Kastenmayer et al. 2006). Using the same procedure applied to the A. thaliana genome, a CI threshold curve was generated based on yeast intron sequences (Supplement A). After eliminating those with significant low complexity regions, the CI values for 84 small coding genes were determined, and all 84 genes have above-threshold CIs (Supplement B). Taken together, our method can predict correctly all the small protein benchmark data sets from A. thaliana and yeast. Therefore, the CI measure is a good predictor of coding regions and is suitable for predicting small protein coding genes in the A. thaliana and yeast genome.
Identification of novel coding sORFs in intergenic regions of A. thaliana genome
To uncover novel coding sORFs in the A. thaliana genome, we first retrieved ORF sequences 90–300 bp started with ATG in the intergenic regions. After removing ORFs that overlap with or are similar to annotated A. thaliana genes, pseudogenes, transposons, simple sequence repeats, and sequences with low complexity (see Methods), the CI values for the remaining 133,091 sORFs were determined and the 7442 sORFs were predicted to be coding sORFs at the 1% false-positive rate (Supplement A). These 7442 coding sORFs form 7159 non-overlapping clusters, representing 7159 regions that belong to potential novel coding genes. From this point on, coding sORFs are referred to as the sORFs with the highest qualifying CI value in each cluster.
Since there are 133,091 sORFs meeting the filtering criteria, the false-positive rate of 3% based on introns derived from full-length cDNAs (Table 1) indicates there could be ∼4000 false positives. However, many introns we looked into may contain coding sequences that elevated the CI values of introns. Using introns defined based on cDNA evidence to search the A. thaliana protein annotation, we defined any intron after translation as alternatively spliced if it has ≥80% amino acid identity to any protein sequence and the alignment spans ≥80% intron length. We found 480 alternatively spliced introns that meet these criteria, and 318 (66%) of these introns are above the threshold CI. This finding indicates that some of the introns with the above-threshold CIs may in fact contain CDSs. Nonetheless, we cannot rule out the possibility that some of these coding sORFs are false positives. Therefore, we further evaluated their functional significance by searching for evidence of their transcription and/or purifying selection.
Transcription of novel coding sORFs
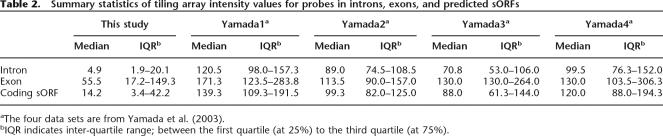
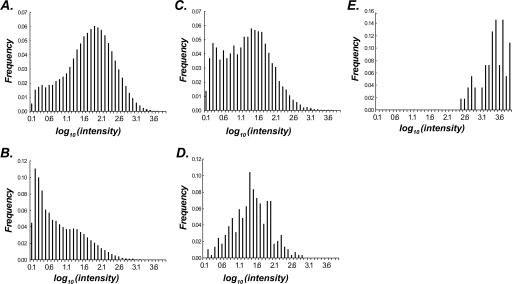
Since the CI measure is based solely on hexamer composition bias, we expect the false-positive rate will be higher than that of more complicated gene finders. To address this concern, we applied two independent criteria to uncover coding sORFs that are likely functional. Assuming expressed sequences are more likely to be functional, the first criterion is whether there is evidence that the coding sORF is expressed. To assess which intergenic regions have transcriptional activities, we isolated four independent RNA samples from 3-d-old seedlings and hybridized cDNA generated from these RNA samples to the A. thaliana genome tiling array. Since exonic sequences are much more likely to be represented in an mRNA pool than intronic sequences, we compared the hybridization signal intensity distribution of features in known exons, introns, and coding sORFs (Table 2; Fig. 5). We found that the intensity distribution of coding sORF probes (Fig. 5C) was more similar to the exon probe intensity distribution (Fig. 5A) than to the intron one (Fig. 5B), indicating that some of coding sORFs are expressed. In addition to the replicated tiling array experiments we have conducted, we analyzed publicly available tiling array data in another four conditions (Yamada et al. 2003) and reached the same conclusion (Table 2). We also compared intensity distribution of coding sORFs and annotated genes to those of tRNA and rRNA (Fig. 5D,E). It is known that rRNAs are expressed constitutively at very high levels as reflected by the skewed intensity distribution of rDNA probes toward higher hybridization signals (Fig. 5E). On the other hand, tRNAs have lower median intensity that is similar to that of coding sORFs (Fig. 5C,D). Taken together, these findings indicate that a substantial number of coding sORFs are expressed, although at lower levels compared to annotated genes in general.
Table 2.
Summary statistics of tiling array intensity values for probes in introns, exons, and predicted sORFs
aThe four data sets are from Yamada et al. (2003).
bIQR indicates inter-quartile range; between the first quartile (at 25%) to the third quartile (at 75%).
Figure 5.
Distributions of hybridization intensities values for probes in intron, coding sORFs and exons. The distribution of intensities values from the 7-d-old seeding tiling array expression data for probes in exons (A), introns (B), coding sORFs (C), tRNA genes (D), and rRNA genes (E). X-axis and Y-axis indicate logarithmically transformed intensity values (base 10) of expression and frequency of probes in different intensity bins, respectively.
To determine whether a coding sORF is expressed or not, we devised the Expression Index (EI) (see Supplement A-3) that is the average intensity over all probes in a given sequence. Since coding sORFs have variable numbers of probes, simulation studies were conducted to determine the EI threshold at different probe numbers according to the intensities and numbers of probes located in introns (Supplement A-4). At the 5% false-positive rate, the EI values of 2996 coding sORFs are significant higher than EIs of introns in at least one experimental condition (Supplement C), consistent with the notion that a substantial number of A. thaliana sORFs are expressed. We refer to these sORFs as transcribed coding sORFs.
Conservation across species and signature of purifying selection
The second independent criterion for assessing the functionality of coding sORF is signature of purifying selection. For a CDS, a significantly lower nonsynonymous substitution rate than the synonymous substitution rate indicates the sequences have experienced purifying selection or functional constraint. To assess the degree of functional constraints on coding sORFs, we first identified sequences that are likely homologous to coding sORFs within A. thaliana or between A. thaliana and five other plants including Brassica oleracea (Ayele et al. 2005), Oryza sativa var. japanica (IRGSP 2005), Medicago truncatula (Bell et al. 2001), Lotus japonicus (Sato et al. 2001; Nakamura et al. 2002; Kaneko et al. 2003; Asamizu et al. 2003), and Populus trichocarpa (Tuskan et al. 2006). Among 7159 coding sORFs, 3997 have ≥1 matches with ≥30% identity to the plant genomes. Applying a likelihood ratio test (Nekrutenko et al. 2002) to these 3997 coding sORFs, we found that 2376 of them show signature of purifying selection (referred to as constrained coding sORFs, Supplement C).
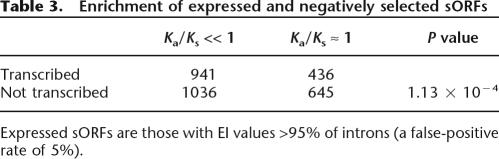
Taken together, we found 4282 coding sORFs with the evidence of either transcription or purifying selection. After eliminating repetitive sequences including transposons with RepeatMasker, 3652 coding sORFs are defined to be small coding regions. Among these 3652 coding sORFs, 941 coding sORFs have evidence both of purifying selection and transcription. The number of transcribed sORFs with purifying selection is significantly larger than that of nontranscribed sORFs with purifying selection (Table 3, χ2 test, P < 0.01), indicating that coding sORFs that are transcribed tend to be subject to purifying selection as well. Many transcribed sORFs, however, do not have evidence of purifying selection.
Table 3.
Enrichment of expressed and negatively selected sORFs
Expressed sORFs are those with EI values >95% of introns (a false-positive rate of 5%).
To determine how many of these transcribed and/or constrained sORFs may belong to gene families, we generated similarity clusters of these sORFs and found 1903 clusters and 936 singletons (Supplement C). Therefore, approximately two thirds of small coding genes have potential paralogs, indicating that these sORFs may belong to novel gene families.
Number of novel coding genes represented by transcribed and/or constrained coding sORFs
The transcribed and/or constrained sORFs may be missing exons of known genes or exons of novel genes. To estimate the number of sORFs belong to truly novel transcription units, we determined the number of sORF-matching ESTs that are part of annotated genes. The assumption is that if an sORF S has an EST match X, which is matched to a known gene G, then S and G likely belong to the same gene. Among 169 transcribed and/or constrained sORFs with ≥97% identity to full-length cDNA/ESTs, 19 were associated with neighboring annotated genes via cDNA/ESTs (>80% identity between the annotated genes and cognate ESTs of sORFs; Supplement C). These 19 sORFs are regarded as potentially missing exons of annotated genes. A less stringent identity threshold (80%) between cDNA/EST and known genes is chosen since we intend to reduce the false-positive prediction of novel small protein genes that are in fact part of annotated genes. The ESTs of the rest 150 predicted sORFs (88.7%, 150/169) seems to belong to truly novel transcription units. Under the assumption that predicted sORFs with cDNA/ESTs that do not overlap with annotated genes are parts of novel genes, the 3652 predicted sORFs with evidence of expression or purifying selection likely belong to 3241[3652 × (150/169)] novel protein coding genes in the A. thaliana genome.
Discussion
Using the CI measure based on CDS nucleotide composition bias, we found a large number of sORFs with significantly higher than expected coding potential in the intergenic regions of the A. thaliana genome. Based on cognate cDNA/EST of coding sORFs, we estimated that >3000 coding sORFs with either evidence of transcription or purifying selection likely belong to novel coding genes in the A. thaliana genome. In addition to the cDNA/EST-based estimate, 2341 sORFs >850 bp away from their neighboring genes likely belong to novel protein coding genes since 95% of introns are ≤850 bp in A. thaliana. The CI measure performed well in distinguishing most CDSs in exons from NCDSs derived from introns. In addition, two benchmark small protein data sets from A. thaliana and yeast were correctly identified as coding sORF by the CI measure. These findings indicate that the CI measure can be applied to find coding sORFs in eukaryotes. Application of the CI measure led to the identification of 7159 coding sORFs in the intergenic regions of the A. thaliana genome, 4282 of which had evidence of either expression or purifying selection. Based on a detailed comparison between the A. thaliana and B. oleracea genomes for nucleotide sequence conservation (Ayele et al. 2005), it is predicted that thousands of protein coding or RNA genes remain unannotated in A. thaliana although our prediction does not overlap significantly with the Brassica–Arabidopsis conservation data (Supplement C). Nonetheless, with completely different approaches and goals, our findings similarly indicate the presence of many unannotated small protein genes in the A. thaliana genome.
It should be noted that the CI measure assumes coding regions should be similar to the training CDS but different from the training NCDS in sequence composition. Since we use exons and introns of genes with full-length cDNAs as the training sets for CDS and NCDS, it is likely that we miss true CDS with very different nucleotide composition from CDS in full-length cDNAs. This compositional difference can be affected by both expression level and amino acid composition. It is well established that gene expression level correlates with codon usage bias in prokaryotes and in a number of eukaryotes (Grantham et al. 1981; Bennetzen and Hall 1982; Gouy and Gautier 1982; Grosjean and Fiers 1982; Duret and Mouchiroud 1999). Since genes with higher expression levels have a better chance to be present in the cDNA pool, the CI measure is likely biased toward a codon usage pattern of genes with relatively higher expression levels. However it has been shown that such correlation is not pronounced in A. thaliana (Mathe et al. 1999). Therefore, it is unclear how much expression level will affect our predictions. Nucleotide composition is also affected by codon compositions. In particular, regions with low amino acid complexity, such as signal sequences and trans-membrane regions, seem to have different codon composition and reduced CI (in Fig. 1, for example, the region corresponding to IDA signal peptide has low pp). Nearly all of the 18 A. thaliana small secreted protein genes used for verifying the effectiveness of CI would have been excluded because signal peptides occupy a major fraction of their sequence lengths. In our analysis of A. thaliana genome, we excluded sequences with low amino acid complexity and likely leave a number of truly novel sORF genes out from our predictions.
Many coding sORFs are likely transcribed, and some of them in fact have cDNAs and/or ESTs. However, relatively much fewer of our predictions have cognate ESTs compared to annotated genes. This is likely due to the fact that coding sORFs tend to have significantly lower expression levels than genes with cDNA/EST evidence (Fig. 5; Table 2). There are >4000 coding sORFs without expression evidence. One explanation is that some may be expressed under conditions not covered. Another potential reason is because the EI thresholds are too conservative. Quite a few introns are alternatively sliced and expressed (supported by the long tail of intron probe intensity distribution in Fig. 5B). Since we estimated EI thresholds using intron probe intensities that include those from expressed sequences, the false-negative rate will be higher than the expected 5%, leading to the underestimation of the number of expressed coding sORFs. Another question regarding sORF transcription is whether transcription of a sequence necessarily lends support to their functionality. Since a true sORF gene has to be transcribed, coding sORFs with evidence of transcription is more likely to be functional sORF genes than those without transcriptional evidence. Nonetheless, evidence of expression is not itself evidence of protein coding potential. We cannot rule out the possibility that some of the transcribed coding sORFs are RNA genes. However, the training models for CI determination explicitly incorporate both CDS and NCDS information. The coding sORFs therefore have to be dissimilar from intron NCDS but similar to CDS, reducing their likelihood to be RNA genes.
Approximately 55% of the coding sORFs have related sequences in five non–A. thaliana plant genomes and within the A. thaliana genome. For those coding sORFs without detectable conservation, the possible explanations are (1) false-positive coding sORFs, (2) incomplete plant genome sequences used, or (3) high evolutionary rates. Although we have used stringent criteria for identifying coding sORFs, we cannot rule out the possibility that some of our predictions are false positives. We find ∼3% of annotated intronic sequence are above the threshold CI. Taking this as a false-positive rate estimate for the CI measure, ∼4000 sORFs will have higher-than-threshold CI values by chance. However, in A. thaliana, 3161 (∼12%) annotated genes have more than one splice variants based on available cDNA/EST data. The true number is likely higher since only ∼60% A. thaliana genes have more than one full-length cDNAs. Therefore, some introns will contain coding sequences that contribute to high CI values. Importantly, our coding sORF prediction is significantly enriched in expressed sequences compared to all sORFs indicating the nonrandom nature of our prediction. Three of the dicotyledon genomes used, L. japonicus, M. truncatula, and B. oleracea, are incomplete. Based on the same criteria in establishing sORF-genome translation pairs, we found that 38.8% (10,179/26207) A. thaliana and 71.0% (41,132/57915) rice annotated genes do not have cross-genome match (S.H. Shiu, unpubl.), although the rice genome is 95% complete (IRGSP 2005). Therefore, some coding sORFs without cross-species matches may be the consequences of incomplete coverage or fast evolving or novel sequences. There are also quite a few coding sORFs that have conservation to sequences in other plant genomes but without evidence of purifying selection. Note that the false-positive rate of the likelihood ratio test procedure for determining the departure of Ka/Ks from one is estimated to be ∼2.5% depending on the sequence lengths and divergence (Nekrutenko et al. 2002). In fact, the test tends to be more stringent for shorter sequences such as sORFs. Therefore, we likely underestimated the number of sORFs with signature of purifying selection. This lack of sensitivity may explain why many transcribed sORFs do not have evidence of purifying selection. In addition, some of these conserved sequences may represent pseudogenes that still possess the nucleotide composition of true CDS. We have filtered out intergenic regions with similarity to annotated A. thaliana genes with a very liberal criterion. If some of these sORFs were pseudogenes, they would mostly belong to single copy genes that become pseudogenes recently.
More and more comparative and functional genomics studies reveal the importance of intergenic “dark matter” (Yamada et al. 2003). Although various studies show that substantial intergenic regions are expressed in various genomes, it is not known what the relative abundance of coding and noncoding RNA is. Our method is the first step not only for identifying sequences with a high coding potential in intergenic regions but also for estimating how much intergenic transcription can be attributed to protein coding genes. Further studies aimed at verifying the translation of these predictions experimentally will be an important next step.
Methods
Bayes’ estimation of coding likelihood
For a given sequence segment F, the pp that F appears in the coding regions of a genome can be given by Bayes’ theorem as follows:
 |
where i = 1∼6 is the six possible coding frames, P(F|codingi) and P(F|noncoding) are the probabilities that F is derived from the coding and noncoding regions in a genome, respectively. P(F|codingi) and P(F|noncoding) are estimated using training models of A. thaliana coding and noncoding (intron) sequences, respectively, according to experimentally determined full-length cDNA sequences (Yamada et al. 2003). Coding regions in cDNAs were identified by similarity searches against the annotated CDS of the A. thaliana TIGR v.5 annotation. cDNA sequences with <100% identity to annotated CDS were excluded from further analyses. The remaining CDS in cDNAs were then used to search against the genome assembly to identify introns. The introns are identified by searching the CDSs of full-length cDNAs against the A. thaliana genome. The genomic sequences corresponding to the alignment gaps in CDS were regarded as introns. The intron data set was then used to search the A. thaliana CDSs. For model training purpose, any intron sequence with 100% identity to CDS of the same gene is regarded as an alternatively spliced intron containing CDS. Training data of coding and noncoding sequences for Saccharomyces cerevisiae were generated based on annotation in NCBI (version 22-05-06).
CDS were translated into amino acid sequences, and low complexity regions of the amino acid sequences were defined with PSEG (Wootton and Federhan 1996). After excluding sequences with substantial low complexity regions, pentamer, and hexamer composition frequency tables (CDS tables) were generated for six reading frames with the sense strand first frame as the true reading frame. For introns, we first identified contiguous translatable regions (intronic “ORFs”) in six frames. Intronic ORFs were also subject to low-complexity filtering, and those that passed were used to generate pentamer and hexamer frequency tables (NCDS tables). With these frequency tables, we modeled the sequences with fifth-order Markov chains. P(F|coding) was calculated by multiplying the frequencies of the first pentamer (taken as the initiation probability) and all subsequent hexamers (taken as the transition probabilities) in six different frames using the CDS tables. P(F|noncoding) was similarly determined using the NCDS tables, but only the first reading frame was calculated. P(coding) and P(noncoding) of prior probabilities were set to be 0.3 and 0.7 in A. thaliana and as 0.5 and 0.5 in S. cerevisiae. These prior probabilities were defined based on the proportion of total base pairs that are annotated coding sequences and noncoding portions of the respective genomes. The prior probability of coding in each frame was P(coding)/6.
CI calculation and threshold determination
For each sequence, pp was calculated on consecutive windows of 75 bp with a step size of 3 nt. The CI for a given sequence is the averaged posterior probabilities of all windows within a sequence. If the CI of a sequence is higher than the threshold value determined by simulation, it is regarded as a coding sequence. Since the predictive power drops with reduced sequence length, a single CI threshold is not feasible. We generated 100,000 random sequences based on the NCDS training data (intron sequences) for each sequence length class with 3-nt increments from 90 to 300 nt. CI values were calculated for each sequence, and the frequency distribution of CI values for each length class was generated. For each size class (e.g., 90 nt), the CI value of randomly generated NCDS of the same length at 99 percentile was used as the threshold. After plotting the CI thresholds according to their size classes, the data points were best fitted by the power law (Supplement A). The threshold value for each size class was determined based on this power law fit and used to determine if a sequence is likely coding or not. Because the false-positive rate increases dramatically as sequence length decreases, the CI measure was applied only to sequences ≥90 bp (six windows). The analysis procedures are illustrated in Supplement A.
ORF processing
The A. thaliana chromosome pseudo-molecule version 5 was obtained from TIGR (http://www.tigr.org/tdb/e2k1/ath1/). The chromosome sequences were translated in all six frames. An ORF was analyzed further if (1) it started with methionine codon and was between 30 and 100 amino acids (aa) long or (2) it was >100 aa with multiple ORF and the longest ORF was 30–100 aa long. In case 2, the longest ones were used for further analysis. There were 570,948 such ORF sequences. Because we are interested in novel coding sequences, an ORFs was excluded if it (1) overlapped at least 1 bp with a known gene or pseudogene sequence according to A. thaliana genome annotation version 5 from TIGR, (2) overlapped at least 1 bp with an unannotated region similar to a known gene, (3) matched a known gene with >40% protein sequence identity over 80% length of ORFs, or (4) contained substantial low complexity regions. Regions in case 2 were defined by using the A. thaliana predicted protein sequences as query to search against A. thaliana chromosome sequences with BLAST (Altschul et al. 1997) using default setting with E value threshold of one. After linking contiguous regions of matches in the genome that are collinear with a query sequence, this genome sequence chain was regarded as a pseudogene if they do not overlap with known genes. Low complexity regions in case 4 were defined with PSEG to filter the sequences and A. thaliana predicted protein sequences. Since 95% of predicted protein sequences have <26.11% of their lengths identified as low-complexity region, ORFs with >26.11% low complexity region in their sequences were excluded from further analysis.
Tiling array sample, hybridization, data acquisition, and analysis
Four plants (lines) of A. thaliana accession Col-0 were grown, and seeds from each line were collected independently. The seeds from each line were stratified for 5 d and grown horizontally in a growth chamber (Percival Scientific Inc., model E361) for 3 d. About 20 μg of total RNA was isolated from 120 seedlings from each line using RNeasy plant mini kit (Qiagen). Poly(A) RNA was further enriched from total RNA using Oligotex mRNA mini kit (Qiagen). For first-strand cDNA synthesis, poly(A) RNA was mixed with 166ng random hexamers, 8 μL of 5× first-strand buffer, 4 μL of 0.1 M DTT, 2 μL of 10 mM dNTP mix, and 400 units of superscript II reverse transcriptase (Invitrogen) in a total volume of 40 μL for 1 h at 42°C. For second-strand cDNA synthesis, the 40 μL first-strand reaction was mixed with 60 μL of 5× second-strand reaction buffer, 6 μL of 10 mM dNTP mix, 20 units of Escherichia coli DNA ligase, 80 units of E. coli DNA polymerase I (Invitrogen), and 4 units of E. coli RNase H (Epicentre) in a total volume of 300 μL for 2 h at 16°C. Double-stranded cDNA was further purified using Qiaquick PCR purification kit (Qiagen), and then labeled using BioPrime DNA labeling system (Invitrogen) with conditions modified as previously described (Borevitz et al. 2003); 95 μL labeling reaction from each of four cDNA samples was subjected to hybridization to AtTILE1 forward chips (Affymetrix), using hybridization protocol for eukaryotic cRNA target (Affymetrix). Chips were scanned by an Affymetrix Scanner 7G with GeneChip Operational System (Affymetrix).
The array intensities were processed using the Bioconductor (http://www.bioconductor.org) affy package in the R software environment (http://www.r-project.org). Specifically, the intensities were adjusted to reduce background with the bg.adjust function, and the normalize.quantiles function was used for between array normalization. The background corrected and normalized intensities were used for further analysis. To reduce the impacts of cross hybridization, the probe sequences were queried against the TIGR A. thaliana genome v.5 assembly with BLAST. Probes with no perfect match or with a second match that has an identity ≥80% (potentially cross-hybridizing) were excluded from further analysis. In addition to our tiling array data, additional array data in four experimental conditions (Yamada et al. 2003) was retrieved from Gene Expression Omnibus (GEO) (Barrett et al. 2005). Since our tiling array data has four replicates in each probe, the median of expression intensity in four replications was used as the representative intensity of each probe. Our tiling array data were generated using cDNA and forward chip only, so direction of transcription is unknown. On the other hand, the publicly available tiling array data have only one data point for each probe for each condition. But cRNA was used to assess the directionality of transcription. Therefore, these two data sets are complementary and were both used in our studies.
EI calculation and threshold determination
The EI for a given sORF is the averaged expression intensity of all probes within the sORF. If the EI of the sORF is higher than the threshold value determined by random sampling from probes of introns, it is regarded as evidence of transcription of the sORF. Since the predictive power drops with reduced number of probes, we conducted simulation studies to determine the proper thresholds for sequences with varying numbers of probes. We randomly sampled intron sequences with one to 10 probes each for 10,000 times to generate intensity distributions of sequences that are expected to be expressed. For each number of probes, the EI >95% of the randomly sampled EIs was used as the threshold. The analysis procedures are illustrated in Supplement A.
Estimation of purifying selection
In addition to the A. thaliana genome, the following genomes were used to assess conservation of the putative novel genes: (1) B. oleracea shotgun sequences (Ayele et al. 2005), (2) O. sativa subsp. japonica chromosome pseudo-molecule version 3 from TIGR (http://www.tigr.org/tdb/e2k1/osa1/), (3) P. trichocarpa shotgun assembly from JGI (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html), (4) M. truncatula genomic sequences including BACs from NCBI (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=3880), and (5) Lotus corniculatus var. japonicus genomic sequences including BACs from NCBI (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=34305). Sequence pairs are regarded as conserved if they have (1) ≥30% identity at the protein level, (2) alignments ≥20 codons, (3) alignment ≥70% of the query putative sORFs, and (4) no stop codon in the translated genomic sequence matches. These conserved pairs were aligned, and the synonymous and nonsynonymous substitution rates (Ka and Ks) were calculated using PAML (Yang 1997). To determine if the Ka/Ks values were significantly smaller than one, a likelihood ratio-based procedure was applied to sequence pairs with a proper Ks value range as described previously (Nekrutenko et al. 2002). For each pair, two maximum likelihood values were calculated with the Ka/Ks ratio fixed at 1 and with the Ka/Ks ratio as a free parameter. The ratio of the maximum likelihood values was then compared to the χ2 distribution.
Comparing sORFs and ESTs
ESTs of A. thaliana were obtained from GenBank as of April 1, 2006. The sORF-At nucleotide sequences were used to search against the Brassicaceae ESTs. An EST is assigned to an sORF if (1) its sequence identity is >97%, (2) the alignment covers >90% of the sORF, and (3) the alignment covers the start position of the sORF. For sORF that match to several regions of the same EST, the different regions will be concatenated together if the gap size is no more than 1 nt. The concatenated sequence will then be examined based on the above criteria. To determine if an sORF is part of a transcriptional unit that includes a known gene, the sORF-matching ESTs was used to search annotated CDS. The matches between an annotated CDS and ESTs were concatenated together with similar identity criteria used for establishing sORF-EST matches but with a maximal gap of 10 kb.
Acknowledgment
We thank Melissa D. Lehti-Shiu and Arnar Palsson for reading the manuscript and discussion, Yoji Nakamura for useful comments for Bayesian estimation, and Hank Wu and Chris Town for discussion and providing the intergenic gene predictions based on Brassica–A. thaliana comparison. We also thank Phil Green and two anonymous reviewers for their extremely valuable and constructive comments. This work is supported by the startup funds from Michigan State University and a National Science Foundation grant (DBI-0638591) to S.-H.S. and National Institute of Health grants to W.-H.L.
Footnotes
[Supplemental material is available online at www.genome.org. The replicated tiling array experiment data has been deposited in Gene Expression Omnibus (GEO) with accession no. GSE6562.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5836207
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generate of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E., Kato T., Sato S., Nakamura Y., Kaneko T., Tabata S., Kato T., Sato S., Nakamura Y., Kaneko T., Tabata S., Sato S., Nakamura Y., Kaneko T., Tabata S., Nakamura Y., Kaneko T., Tabata S., Kaneko T., Tabata S., Tabata S. Structural analysis of a Lotus japonicus genome. IV. Sequence features and mapping of seventy-three TAC clones which cover the 7.5 Mb regions of the genome. DNA Res. 2003;10:115–122. doi: 10.1093/dnares/10.3.115. [DOI] [PubMed] [Google Scholar]
- Ayele M., Haas B.J., Kumar N., Wu H., Xiao Y., Van Aken S., Utterback T.R., Wortman J.R., White O.R., Town C.D., Haas B.J., Kumar N., Wu H., Xiao Y., Van Aken S., Utterback T.R., Wortman J.R., White O.R., Town C.D., Kumar N., Wu H., Xiao Y., Van Aken S., Utterback T.R., Wortman J.R., White O.R., Town C.D., Wu H., Xiao Y., Van Aken S., Utterback T.R., Wortman J.R., White O.R., Town C.D., Xiao Y., Van Aken S., Utterback T.R., Wortman J.R., White O.R., Town C.D., Van Aken S., Utterback T.R., Wortman J.R., White O.R., Town C.D., Utterback T.R., Wortman J.R., White O.R., Town C.D., Wortman J.R., White O.R., Town C.D., White O.R., Town C.D., Town C.D. Whole genome shotgun sequencing of Brassica oleracea and its application to gene discovery and annotation in Arabidopsis. Genome Res. 2005;15:487–495. doi: 10.1101/gr.3176505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Rudnev D., Lash A.E., Fujibuchi W., Edgar R., Lash A.E., Fujibuchi W., Edgar R., Fujibuchi W., Edgar R., Edgar R. NCBI GEO: Mining millions of expression profiles—Database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai M.A., Hieter P., Boeke J.D., Hieter P., Boeke J.D., Boeke J.D. Small open reading frames: Beautiful needles in the haystack. Genome Res. 1997;7:768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- Bell C.J., Dixon R.A., Farmer A.D., Flores R., Inman J., Gonzales R.A., Harrison M.J., Paiva N.L., Scott A.D., Weller J.W., Dixon R.A., Farmer A.D., Flores R., Inman J., Gonzales R.A., Harrison M.J., Paiva N.L., Scott A.D., Weller J.W., Farmer A.D., Flores R., Inman J., Gonzales R.A., Harrison M.J., Paiva N.L., Scott A.D., Weller J.W., Flores R., Inman J., Gonzales R.A., Harrison M.J., Paiva N.L., Scott A.D., Weller J.W., Inman J., Gonzales R.A., Harrison M.J., Paiva N.L., Scott A.D., Weller J.W., Gonzales R.A., Harrison M.J., Paiva N.L., Scott A.D., Weller J.W., Harrison M.J., Paiva N.L., Scott A.D., Weller J.W., Paiva N.L., Scott A.D., Weller J.W., Scott A.D., Weller J.W., Weller J.W., et al. The Medicago Genome Initiative: A model legume database. Nucleic Acids Res. 2001;29:114–117. doi: 10.1093/nar/29.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L., Hall B.D., Hall B.D. Codon selection in yeast. J. Biol. Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- Boeckmann B., Blatter M., Famiglietti L., Hinz U., Lane L., Roechert B., Bairoch A., Blatter M., Famiglietti L., Hinz U., Lane L., Roechert B., Bairoch A., Famiglietti L., Hinz U., Lane L., Roechert B., Bairoch A., Hinz U., Lane L., Roechert B., Bairoch A., Lane L., Roechert B., Bairoch A., Roechert B., Bairoch A., Bairoch A. Protein variety and functional diversity: Swiss-Prot annotation in its biological context. C. R. Biol. 2005;328:882–899. doi: 10.1016/j.crvi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Liang D., Plouffe D., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Liang D., Plouffe D., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Plouffe D., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Chang H.S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Weigel D., Berry C.C., Winzeler E., Chory J., Berry C.C., Winzeler E., Chory J., Winzeler E., Chory J., Chory J. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 2003;13:513–523. doi: 10.1101/gr.541303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M., McIninch J.D., Koonin E.V., Rudd K.E., Medigue C., Danchin A., McIninch J.D., Koonin E.V., Rudd K.E., Medigue C., Danchin A., Koonin E.V., Rudd K.E., Medigue C., Danchin A., Rudd K.E., Medigue C., Danchin A., Medigue C., Danchin A., Danchin A. Detection of new genes in a bacterial genome using Markov models for three gene classes. Nucleic Acids Res. 1995;23:3554–3562. doi: 10.1093/nar/23.17.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent M.R., Guigo R., Guigo R. Recent advances in gene structure prediction. Curr. Opin. Struct. Biol. 2004;14:264–272. doi: 10.1016/j.sbi.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Butenko M.A., Patterson S.E., Grini P.E., Stenvik G.E., Amundsen S.S., Mandal A., Aalen R.B., Patterson S.E., Grini P.E., Stenvik G.E., Amundsen S.S., Mandal A., Aalen R.B., Grini P.E., Stenvik G.E., Amundsen S.S., Mandal A., Aalen R.B., Stenvik G.E., Amundsen S.S., Mandal A., Aalen R.B., Amundsen S.S., Mandal A., Aalen R.B., Mandal A., Aalen R.B., Aalen R.B. Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell. 2003;15:2296–2307. doi: 10.1105/tpc.014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J.M. Computational methods for the identification of genes in vertebrate genomic sequences. Hum. Mol. Genet. 1997;6:1735–1744. doi: 10.1093/hmg/6.10.1735. [DOI] [PubMed] [Google Scholar]
- Cock J.M., McCormick S., McCormick S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001;126:939–942. doi: 10.1104/pp.126.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber R., Lapedes A., Sirotkin K., Lapedes A., Sirotkin K., Sirotkin K. Determination of eukaryotic protein coding regions using neural networks and information theory. J. Mol. Biol. 1992;226:471–479. doi: 10.1016/0022-2836(92)90961-i. [DOI] [PubMed] [Google Scholar]
- Fickett J.W., Tung C.S., Tung C.S. Assessment of protein coding measures. Nucleic Acids Res. 1992;20:6441–6450. doi: 10.1093/nar/20.24.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S., Belle A., Dephoure N., O'Shea E.K., Weissman J.S., Dephoure N., O'Shea E.K., Weissman J.S., O'Shea E.K., Weissman J.S., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C., Gautier C. Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R., Gautier C., Gouy M., Jacobzone M., Mercier R., Gouy M., Jacobzone M., Mercier R., Jacobzone M., Mercier R., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W., Fiers W. Preferential codon usage in prokaryotic genes: The optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982;18:199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., Clark L., Cox T., Cuff J., Curwen V., Down T., Cox T., Cuff J., Curwen V., Down T., Cuff J., Curwen V., Down T., Curwen V., Down T., Down T., et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K., Howson R.W., Weissman J.S., O'Shea E.K., Weissman J.S., O'Shea E.K., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- IRGSP The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Asamizu E., Kato T., Sato S., Nakamura Y., Tabata S., Asamizu E., Kato T., Sato S., Nakamura Y., Tabata S., Kato T., Sato S., Nakamura Y., Tabata S., Sato S., Nakamura Y., Tabata S., Nakamura Y., Tabata S., Tabata S. Structural analysis of a Lotus japonicus genome. III. Sequence features and mapping of sixty-two TAC clones which cover the 6.7 Mb regions of the genome. DNA Res. 2003;10:27–33. doi: 10.1093/dnares/10.1.27. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kastenmayer J.P., Ni L., Chu A., Kitchen L.E., Au W.C., Yang H., Carter C.D., Wheeler D., Davis R.W., Boeke J.D., Ni L., Chu A., Kitchen L.E., Au W.C., Yang H., Carter C.D., Wheeler D., Davis R.W., Boeke J.D., Chu A., Kitchen L.E., Au W.C., Yang H., Carter C.D., Wheeler D., Davis R.W., Boeke J.D., Kitchen L.E., Au W.C., Yang H., Carter C.D., Wheeler D., Davis R.W., Boeke J.D., Au W.C., Yang H., Carter C.D., Wheeler D., Davis R.W., Boeke J.D., Yang H., Carter C.D., Wheeler D., Davis R.W., Boeke J.D., Carter C.D., Wheeler D., Davis R.W., Boeke J.D., Wheeler D., Davis R.W., Boeke J.D., Davis R.W., Boeke J.D., Boeke J.D., et al. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16:365–373. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Sato S., Nakamura Y., Kaneko T., Asamizu E., Tabata S., Sato S., Nakamura Y., Kaneko T., Asamizu E., Tabata S., Nakamura Y., Kaneko T., Asamizu E., Tabata S., Kaneko T., Asamizu E., Tabata S., Asamizu E., Tabata S., Tabata S. Structural analysis of a Lotus japonicus genome. V. Sequence features and mapping of sixty-four TAC clones which cover the 6.4 Mb regions of the genome. DNA Res. 2003;10:277–285. doi: 10.1093/dnares/10.6.277. [DOI] [PubMed] [Google Scholar]
- Li W.-H. Molecular evolution. Sinauer Associates; Sunderland: 1997. [Google Scholar]
- Makalowski W., Boguski M.S., Boguski M.S. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Cheng J., Bell I., Ghosh S., Piccolboni A., Bell I., Ghosh S., Piccolboni A., Ghosh S., Piccolboni A., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Mathe C., Peresetsky A., Dehais P., Van Montagu M., Rouze P., Peresetsky A., Dehais P., Van Montagu M., Rouze P., Dehais P., Van Montagu M., Rouze P., Van Montagu M., Rouze P., Rouze P. Classification of Arabidopsis thaliana gene sequences: Clustering of coding sequences into two groups according to codon usage improves gene prediction. J. Mol. Biol. 1999;285:1977–1991. doi: 10.1006/jmbi.1998.2451. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Kaneko T., Asamizu E., Kato T., Sato S., Tabata S., Kaneko T., Asamizu E., Kato T., Sato S., Tabata S., Asamizu E., Kato T., Sato S., Tabata S., Kato T., Sato S., Tabata S., Sato S., Tabata S., Tabata S. Structural analysis of a Lotus japonicus genome. II. Sequence features and mapping of sixty-five TAC clones which cover the 6.5-Mb regions of the genome. DNA Res. 2002;9:63–70. doi: 10.1093/dnares/9.2.63. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A., Makova K.D., Li W.H., Makova K.D., Li W.H., Li W.H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002;12:198–202. doi: 10.1101/gr.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Wakamatsu A., Hayashi K., Sato H., Nagai K., Hayashi K., Sato H., Nagai K., Sato H., Nagai K., Nagai K., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R., Tatusova T., Maglott D.R., Maglott D.R. NCBI Reference Sequence (RefSeq): Acurated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Nelson F.K., Miller P., Gerstein M., Miller P., Gerstein M., Gerstein M., et al. The transcriptional activity of human chromosome 22. Genes & Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta M.P., Tongprasit W., Istrail S., Cameron R.A., Tu Q., Davidson E.H., Stolc V., Tongprasit W., Istrail S., Cameron R.A., Tu Q., Davidson E.H., Stolc V., Istrail S., Cameron R.A., Tu Q., Davidson E.H., Stolc V., Cameron R.A., Tu Q., Davidson E.H., Stolc V., Tu Q., Davidson E.H., Stolc V., Davidson E.H., Stolc V., Stolc V. The transcriptome of the sea urchin embryo. Science. 2006;314:960–962. doi: 10.1126/science.1131898. [DOI] [PubMed] [Google Scholar]
- Sato S., Kaneko T., Nakamura Y., Asamizu E., Kato T., Tabata S., Kaneko T., Nakamura Y., Asamizu E., Kato T., Tabata S., Nakamura Y., Asamizu E., Kato T., Tabata S., Asamizu E., Kato T., Tabata S., Kato T., Tabata S., Tabata S. Structural analysis of a Lotus japonicus genome. I. Sequence features and mapping of fifty-six TAC clones which cover the 5.4 Mb regions of the genome. DNA Res. 2001;8:311–318. doi: 10.1093/dnares/8.6.311. [DOI] [PubMed] [Google Scholar]
- Stolc V., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Hua S., Herreman T., Tongprasit W., Barbano P.E., Herreman T., Tongprasit W., Barbano P.E., Tongprasit W., Barbano P.E., Barbano P.E., et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Stolc V., Li L., Wang X., Li X., Su N., Tongprasit W., Han B., Xue Y., Li J., Snyder M., Li L., Wang X., Li X., Su N., Tongprasit W., Han B., Xue Y., Li J., Snyder M., Wang X., Li X., Su N., Tongprasit W., Han B., Xue Y., Li J., Snyder M., Li X., Su N., Tongprasit W., Han B., Xue Y., Li J., Snyder M., Su N., Tongprasit W., Han B., Xue Y., Li J., Snyder M., Tongprasit W., Han B., Xue Y., Li J., Snyder M., Han B., Xue Y., Li J., Snyder M., Xue Y., Li J., Snyder M., Li J., Snyder M., Snyder M., et al. A pilot study of transcription unit analysis in rice using oligonucleotide tiling-path microarray. Plant Mol. Biol. 2005a;59:137–149. doi: 10.1007/s11103-005-6164-5. [DOI] [PubMed] [Google Scholar]
- Stolc V., Samanta M.P., Tongprasit W., Sethi H., Liang S., Nelson D.C., Hegeman A., Nelson C., Rancour D., Bednarek S., Samanta M.P., Tongprasit W., Sethi H., Liang S., Nelson D.C., Hegeman A., Nelson C., Rancour D., Bednarek S., Tongprasit W., Sethi H., Liang S., Nelson D.C., Hegeman A., Nelson C., Rancour D., Bednarek S., Sethi H., Liang S., Nelson D.C., Hegeman A., Nelson C., Rancour D., Bednarek S., Liang S., Nelson D.C., Hegeman A., Nelson C., Rancour D., Bednarek S., Nelson D.C., Hegeman A., Nelson C., Rancour D., Bednarek S., Hegeman A., Nelson C., Rancour D., Bednarek S., Nelson C., Rancour D., Bednarek S., Rancour D., Bednarek S., Bednarek S., et al. Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays. Proc. Natl. Acad. Sci. 2005b;102:4453–4458. doi: 10.1073/pnas.0408203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A., Putnam N., Ralph S., Rombauts S., Salamov A., Ralph S., Rombauts S., Salamov A., Rombauts S., Salamov A., Salamov A., et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Wang J., Li S., Zhang Y., Zheng H., Xu Z., Ye J., Yu J., Wong G.K., Li S., Zhang Y., Zheng H., Xu Z., Ye J., Yu J., Wong G.K., Zhang Y., Zheng H., Xu Z., Ye J., Yu J., Wong G.K., Zheng H., Xu Z., Ye J., Yu J., Wong G.K., Xu Z., Ye J., Yu J., Wong G.K., Ye J., Yu J., Wong G.K., Yu J., Wong G.K., Wong G.K. Vertebrate gene predictions and the problem of large genes. Nat. Rev. Genet. 2003;4:741–749. doi: 10.1038/nrg1160. [DOI] [PubMed] [Google Scholar]
- Wootton J.C., Federhen S., Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Yamada K., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., Southwick A.M., Wu H.C., Kim C., Nguyen M., Wu H.C., Kim C., Nguyen M., Kim C., Nguyen M., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]