Abstract
Theoretical work predicts natural selection to be more efficient in the fixation of beneficial mutations in X-linked genes than in autosomal genes. This “fast-X effect” should be evident by an increased ratio of nonsynonymous to synonymous substitutions (dN/dS) for sex-linked genes; however, recent studies have produced mixed support for this expectation. To make an independent test of the idea of fast-X evolution, we focused on birds, which have female heterogamety (males ZZ, females ZW), where analogous arguments would predict a fast-Z effect. We aligned 2.8 Mb of orthologous protein-coding sequence of zebra finch and chicken from 172 Z-linked and 4848 autosomal genes. Zebra finch data were in the form of EST sequences from brain cDNA libraries, while chicken genes were from the draft genome sequence. The dN/dS ratio was significantly higher for Z-linked (0.110) than for all autosomal genes (0.085; P = 0.002), as well as for genes linked to similarly sized autosomes 1–10 (0.0948; P = 0.04). This pattern of fast-Z was evident even after we accounted for the nonrandom distribution of male-biased genes. We also examined the nature of standing variation in the chicken protein-coding regions. The ratio of nonsynonymous to synonymous polymorphism (pN/pS) did not differ significantly between genes on the Z chromosome (0.104) and on the autosomes (0.0908). In conjunction, these results suggest that evolution proceeds more quickly on the Z chromosome, where hemizygous exposure of beneficial nondominant mutations increases the rate of fixation.
Sex chromosomes can exhibit several unusual properties, including inheritance pattern, reduced recombination, and hemizygosity, which influence the mechanisms of natural selection (Rice 1984; Vicoso and Charlesworth 2006). These differences often make evolutionary comparisons between sex chromosomes and autosomes particularly revealing, as they help answer fundamental questions regarding the signature and pattern of mutation and natural selection, as well as how these forces vary across the genome. For instance, consider new autosomal mutations, which are initially low in frequency and primarily present in heterozygotes. If these mutations are either wholly or partially recessive, they will be obscured by the ancestral allele, and will only rarely be exposed directly to selective forces. In contrast, novel recessive and otherwise nondominant mutations on sex chromosomes are directly exposed to selection in the hemizygous sex. Selection would therefore be expected to act faster on sex chromosomes to fix some types of beneficial mutations (Charlesworth et al. 1987), a situation referred to as fast-X evolution.
The fast-X (or fast-Z in the case of female heterogamety) effect could potentially explain several evolutionary phenomena. For instance, it has been invoked to explain Haldane’s Rule (Haldane 1922), which states that the heterogametic sex suffers higher fitness consequences in interspecific crosses, which could be explained if the X chromosome accumulates epistatic mutations at a faster rate than the autosomes (Charlesworth et al. 1987). Additionally, many studies have found that a disproportionately large percentage of genes relating to speciation, reproductive isolation, and mate choice map to the sex chromosomes (Dobzhansky 1974; Templeton 1977; Coyne 1985; Coyne and Orr 1989; Prowell 1998; Reinhold 1998; Ritchie and Phillips 1998; Presgraves 2002). This may be because sex-linked incompatibility loci evolve at a faster rate than autosomal incompatability sites (Tao and Hartl 2003; Coyne et al. 2004).
However, the theory behind fast-X evolution does not hold for all types of mutations under all forms of selection. If selection acts on standing genetic variation, rather than on novel mutations, the sex-linked genomic regions will have a slower rate of evolution than autosomal regions (Orr and Betancourt 2001). Moreover, if most protein sequence evolution is due to fixation of weakly deleterious alleles through genetic drift, the autosomal genes will show higher rates of evolutionary change than sex-linked genes, where hemizygous exposure would result in faster purging (Charlesworth et al. 1987). Finally, the nature of mutations is also important to consider, as those effects of completely dominant DNA mutations would result in genetic drift fixing mildly deleterious alleles more often on the X.
It is possible to analyze the rate and pattern of nucleotide substitution for the molecular signature of fast-X evolution. Evidence of fast-X would originate from the accelerated accumulation of differences in the coding regions of sex-linked genes, and would manifest in a higher ratio of nonsynonymous (dN) to synonymous (dS) divergence. The ratio of these metrics (dN/dS) can be used as a genomic beacon for fast-X evolution when averaged across large genomic regions. However, several recent tests for fast-X evolution using this or similar methodology have produced mixed results (Betancourt et al. 2002; Thornton and Long 2002, 2005; Torgerson and Singh 2003, 2006; Lu and Wu 2005; Richards et al. 2005), even when overlapping data sets were analyzed (Counterman et al. 2004; Musters et al. 2006; Thornton et al. 2006). This may be because the theoretical requirements for fast-X (beneficial mutations that are novel and nondominant) are too rare to result in a detectable genomic signature, or because other phenomena act to obscure the signal in some of the analyzed taxa.
A novel approach to the question of fast-X evolution is, in contrast to previous molecular evolutionary studies on this topic, to study a system with female heterogamety (males ZZ, females ZW). One obvious advantage with analyzing a ZW system is that it offers an independent test of the theoretical predictions since this type of sex chromosome inheritance allows for the parsing of the effects of maleness from the effects of heterogamety. The theoretical predictions are analogous to those for the X chromosome in male heterogamety; new beneficial and recessive mutations would be more easily fixed on the Z chromosome than on autosomes, manifesting in a higher dN/dS ratio on the Z. Contrasting data from XY and ZW systems have previously proved useful for addressing many aspects of sex chromosome evolution, such as distinguishing between sex- and chromosome-specific effects in the context of male-biased mutation (Ellegren and Fridolfsson 1997), and distinguishing between sexual selection and restricted recombination in explaining the low genetic variability typically seen in the sex-limited chromosome (Berlin and Ellegren 2004).
Female heterogamety is seen in a variety of animals, including birds, butterflies, and some fish, lizards, and snakes. The chicken (Gallus gallus) was the first bird for which a draft genome sequence was presented (International Chicken Genome Sequencing Consortium 2004). Large-scale sequence data for a second bird species, the zebra finch (Taeniopygia guttata), are available in the form of expressed sequence tags (ESTs) from brain cDNA libraries (Clayton 2004). Here we align >5000 zebra finch–chicken 1:1 orthologs, two highly divergent birds that account for roughly 100 million years of avian evolution (van Tuinen et al. 2000), in order to assess the rate of nonsynonymous (dN) and synonymous (dS) substitution over a wide window of avian evolution. We also analyze large-scale polymorphism data from domestic breeds of chicken to be able to examine the pattern of standing genetic diversity in a bird, both the number of nonsynonymous polymorphisms per nonsynonymous site (pN), as well as the number of synonymous polymorphisms per synonymous site (pS). We use these data to examine the nature of coding-region mutations on sex chromosomes and autosomes, and the mechanism by which natural selection acts on those mutations. We then use these metrics to look for evidence of fast-Z evolution in birds and to gain insight into the specific selective processes that produce it.
Results
Divergence data
We found reciprocal best hits and aligned 172 Z-linked and 4848 autosomal zebra finch (unique EST contigs from brain cDNA libraries) and chicken (annotated draft genome sequence) orthologs, which comprised roughly 2.8 Mb of coding sequence (90.6 kb Z-linked and 2.72 Mb autosomal). Data on chromosomal location were obtained from the chicken genome, and there is strong indirect evidence that genes located on the chicken Z chromosome are Z-linked in zebra finch as well (Supplemental Material). It can thus be assumed that sex linkage has been retained in the lineages leading to chicken and zebra finch since they shared a common ancestor.
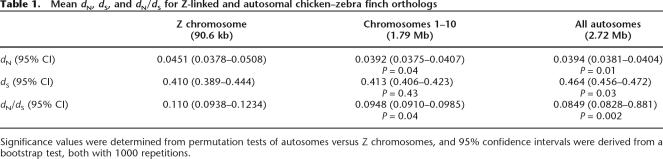
Across all 5020 genes, mean dN = 0.0396 and dS = 0.462. Z-linked genes had a higher rate of nonsynonymous substitutions (0.0451), compared to autosomal (0.0394) regions (permutation test, 1000 repetitions; P = 0.01) (Table 1). The synonymous substitution rate showed the opposite pattern, with the Z-linked average lower (0.410) than the autosomal average (0.464), which was also significantly different (P = 0.03). The dN/dS for Z-linked (0.110) and autosomal (0.0849) genes is significantly different (P = 0.002), with the ratio roughly 30% larger for coding sequences that mapped to the Z chromosome.
Table 1.
Mean dN, dS, and dN/dS for Z-linked and autosomal chicken–zebra finch orthologs
Significance values were determined from permutation tests of autosomes versus Z chromosomes, and 95% confidence intervals were derived from a bootstrap test, both with 1000 repetitions.
The avian karyotype differs from that of many other organisms by showing significant heterogeneity in chromosome size, including a large number of very small (<20 Mb) chromosomes, the “microchromosomes.” The chicken diploid karyotype has 78 chromosomes, with chromosomes 11–38 defined as microchromosomes by the International Chicken Genome Sequencing Consortium (2004), while the zebra finch has 2n = 80 (Pigozzi and Solari 1999). As judged from the analysis of the chicken genome sequence, the microchromosomes differ from the larger chromosomes by showing higher gene density, fewer repeats, shorter introns, higher GC content, and much higher recombination rates (International Chicken Genome Sequencing Consortium 2004). As several of these factors, in particular recombination, could influence the character and efficacy of selection, it is potentially more relevant to compare evolutionary rates in the Z chromosome with those in similarly sized autosomes. This could pose a problem if chromosomal rearrangements have been frequent and differentiated the chicken and zebra finch karyotypes. However, the rates of translocation, fusion, and fission have generally been low during avian genome evolution (Burt et al. 1999; Bourque et al. 2005), and, on the whole (see Supplemental Material), chicken and zebra finch chromosomes 1–10 are syntenic (Griffin et al. 2007). As the Z chromosome is similar in size to autosomes 5 and 6 in chicken, and to obtain a reasonably large set of genes for comparison, we extracted data from genes located on chicken chromosomes 1–10. For 3185 autosomal transcripts (1.79 Mb) from chicken chromosomes 1–10, dN was still smaller (0.0392; P = 0.04) than for the Z chromosome, while dS was no longer significantly different (0.413; P = 0.43) from Z-linked genes. Importantly, the dN/dS ratio was still significantly higher (P = 0.04) in the Z-linked genes (0.110) than in genes that mapped to chicken chromosomes 1–10 (0.0948). The reduction in dS for chromosomes 1–10 compared to all autosomes is consistent with previous observations of a lower mutation rate on macro- than on microchromosomes (International Chicken Genome Sequencing Consortium 2004; Axelsson et al. 2005). This seems largely because of the higher GC content of microchromosomes and the positive correlation between GC and substitution rate in birds (Webster et al. 2006).
There is evidence that the avian Z chromosome has an excess of coding regions that exhibit a male-biased expression pattern (Kaiser and Ellegren 2006; Storchova and Divinia 2006), which has been shown to be positively correlated with the rate of evolution (Meiklejohn et al. 2003; Zhang et al. 2004; Zhang and Parsch 2005). Therefore, we wanted to investigate the relationship between gene expression pattern and dN/dS values in order to determine whether the pattern of fast-Z described above is simply the result of the hyperaccumulation of male-biased genes on the avian Z chromosome. Using data from microarray studies of gene expression in chicken brain (H. Ellengren, L. Hultin-Rosenberg, B. Brunströom, L. Dencker, K. Kultima, and B. Scholz, unpubl.), we identified and then removed 64 coding regions with male-biased expression (32 Z-linked, 32 autosomal) from our analysis; the pattern of fast-Z evolution still remains evident after this (P = 0.02 for Z vs. all autosomal genes; P = 0.005 for Z vs. autosomes 1–10) (data not shown).
Polymorphism data
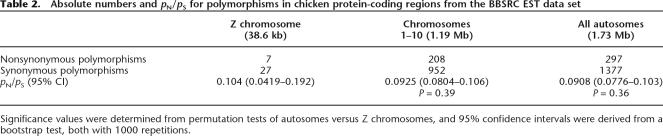
To get a broader picture of the nature of selective forces on autosomes and the Z chromosome, we next turned to the intraspecific diversity data of the domestic chicken. Coding-sequence polymorphisms have been identified through large-scale EST sequencing of multiple cDNA libraries of chicken (>485,000 sequences) (Hubbard et al. 2005) and are available in the BBSRC ChickEST database (http://www.chick.umist.ac.uk/). This database includes high-confidence SNPs predicted from stringent filtering criteria, which included the removal of singleton polymorphisms. To be able to make a direct comparison to chicken–zebra finch divergence data, we focused on the same genes as used for divergence estimates; 1724 nonsynonymous and synonymous substitutions were identified in this gene set.
While the pN/pS ratio for Z-linked genes was higher (0.104) than that for autosomal genes (0.0925), this difference was not statistically significant (randomization test, 1000 permutations; P > 0.3 in all cases) (Table 2). The pN/pS ratios for different genomic regions are similar as well (0.0934 vs. 0.104 for chromosomes 1–10 and the Z chromosome, respectively; P > 0.3 in all cases). The observation of similar pN/pS ratios but a higher dN/dS ratio for sex-linked genes indicates that the rate of fixation of nonsynonymous substitutions is higher on the Z chromosome than on autosomes, as the pattern in the underlying pool of genetic variability does not differ among chromosomal classes.
Table 2.
Absolute numbers and pN/pS for polymorphisms in chicken protein-coding regions from the BBSRC EST data set
Significance values were determined from permutation tests of autosomes versus Z chromosomes, and 95% confidence intervals were derived from a bootstrap test, both with 1000 repetitions.
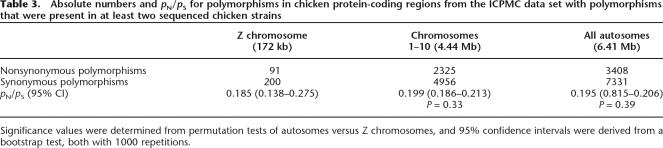
Another source of chicken polymorphism data is from the International Chicken Polymorphism Map Consortium (2004), which identified 2.8 million single nucleotide polymorphisms (SNPs) in light shotgun (0.25× coverage) sequencing of three individuals from different breeds of domestic chicken. This is the most extensive polymorphism data set available for a bird, however, since the majority of SNPs identified in that study have not been validated and typically originate from nonoverlapping reads aligned to the reference genome sequence; a certain fraction likely represent sequencing errors. Such errors would have the largest effect on relative frequency estimates for rare classes of polymorphism, primarily nonsynonymous mutations. Indeed, small-scale validation by resequencing indicates that while 94% of genome-wide SNPs are confirmed, this drops to 80% for nonsynonymous polymorphisms (International Chicken Polymorphism Map Consortium 2004). Additionally, since the Z chromosome harbors less variation in general (Sundström et al. 2004), it would be expected that sequencing errors would disproportionately influence our estimates of Z-linked polymorphisms. We therefore sought to reduce the impact of potential sequencing errors by limiting the International Chicken Polymorphism Map Consortium (ICPMC) data set to only include polymorphisms observed in at least two chicken strains. In this resulting data set, pN/pS ratios of sex-linked and autosomal genes are again very similar (0.185 for the Z chromosome, 0.195 for autosomes, and 0.199 for autosomes 1–10) (Table 3). The overall higher pN/pS ratios in the BBSRC compared to the ICPMC data sets are likely explained by the fact that we could not limit the ICPMC data to only include those genes used for divergence estimates, as this would have resulted in extremely limited sample sizes. This means that the ICPMC polymorphism data were not limited to genes expressed in brain, and include genes predicted from the chicken genome sequence that are not confirmed by zebra finch EST sequencing.
Table 3.
Absolute numbers and pN/pS for polymorphisms in chicken protein-coding regions from the ICPMC data set with polymorphisms that were present in at least two sequenced chicken strains
Significance values were determined from permutation tests of autosomes versus Z chromosomes, and 95% confidence intervals were derived from a bootstrap test, both with 1000 repetitions.
Discussion
Our results show that Z-linked protein-coding genes have evolved at a significantly faster average nonsynonymous-to-synonymous rate than autosomal genes in the lineages leading to zebra finch and chicken. While our analysis is comprised of genes expressed in the brain, there are several reasons to think that these results are more broadly applicable. First, the 5050 coding sequences analyzed here represent roughly a quarter of annotated avian genes. Additionally, while our data set is comprised of ESTs expressed in the brain, most of them are by no means brain-specific, and many are expressed broadly throughout the organism. Both these lines of evidence suggest that these results would apply to the majority of the avian transcriptome.
These birds represent two highly divergent clades in the avian phylogenetic tree. Within Neognathae (all extant birds except the mostly flightless ratites), the first split occurred between Galloanserae (including chicken) and Neoaves (remaining extant orders including song birds to which zebra finch belongs) ∼100 million years ago (van Tuinen et al. 2000). The vast span of evolutionary time separating our study organisms suggests that the results obtained in this analysis may be representative for birds in general.
The increased dN/dS for Z-linked coding regions during avian evolution is compatible with the notion that many novel beneficial mutations are nondominant, and that they are fixed faster through hemizygous exposure to natural selection on the sex chromosomes than they would be in heterozygous exposure on an autosome (Charlesworth et al. 1987). This is the first comparative genomic support for fast-X evolution in a female heterogametic system, an observation that, by analogy, we denote fast-Z evolution.
Two questions arise from this observation. First, are there other explanations to fast-Z than a higher fixation rate of advantageous mutations? Second, are there reasons to expect fast-Z to be more evident than fast-X? The latter question arises from the conflicting evidence for the existence of a fast-X effect in recent studies of male heterogametic systems (Betancourt et al. 2002; Torgerson and Singh 2003; Counterman et al. 2004; Thornton et al. 2006).
It is theoretically possible that the lower effective population size (Ne) of the Z chromosome compared to autosomes could create a fast-Z effect through the fixation of some weakly deleterious nonsynonymous mutations. Under random mating in an ideal population, Ne of the Z is three-quarters as large as that for autosomes. With reproductive skews, common in birds where sexual selection often reduces the number of males contributing to the next generation (Andersson 1994), Z chromosome Ne can be expected to be even less relative to the autosomes. Although Charlesworth et al. (1987) accounted for a difference in Ne due to chromosome counts in their prediction for relative rates of evolution on sex chromosomes and autosomes, these predictions require some very specific assumptions about the nature of mutations, namely, that they are nondominant and in many cases beneficial. When fully dominant, slightly deleterious mutations would more easily drift to fixation on the Z chromosome, leading to an increased dN/dS ratio. While the limiting sample size of our polymorphism data set prevents the drawing of definitive conclusions, it does suggest that the pattern of fast-Z we observe is not due to Ne, as the presence of a significant proportion of weakly deleterious mutations segregating at Z-linked loci would have manifested in a higher pN/pS than for autosomal loci, and we do not observe this pattern in our data set.
The difference in pN and pS levels between the BBSRC and ICPMC polymorphism estimates is likely due to the nature of the underlying data sets. While the genes in our study are by no means brain-specific, the library that forms the basis of the data set is enriched for brain genes, which, on average, seem to be subject to stronger constraint than genes expressed in other tissues (Nielsen et al. 2005). This library is also likely enriched for housekeeping genes, which are more slowly evolving than the transcriptome when taken as a whole (e.g., Duret and Mouchiroud 2000). Because we limited our analysis of the BBSRC data to those coding sequences identified from brain EST libraries, it is likely that this data set is also biased toward slowly evolving housekeeping genes. This would suggest that the reduced level of polymorphism in the BBSRC data set is due to stronger evolutionary constraints acting on housekeeping genes, and the higher level of polymorphism observed in the ICPMC data is derived from the fact that this data set is comprised of all coding regions with sufficient sequencing coverage and does not differentiate according to expression locus. This enrichment in the underlying data set likely results in an underestimate of the true fast-Z effect, but would not be expected to create a spurious correlation. It is therefore doubtful that the nature of the brain EST library affects the overall conclusions of our analysis.
Theoretical predictions on the rate of evolution of sex-linked genes could be violated if genes are nonrandomly distributed across the genome, producing a fast-Z pattern for reasons other than those predicted by Charlesworth et al. (1987). An important conclusion from recent genome-wide gene expression analysis is that a significant proportion of the transcriptome shows sex-biased expression (Reinke et al. 2000; Jin et al. 2001; Ranz et al. 2003; Oliver and Parisi 2004; Yang et al. 2006). Male- and female-biased genes, especially if sex-biased gene expression has arisen from sexual antagonism, are predicted to have nonrandom genomic distributions depending on the heterogametic system (Rice 1984) or incomplete dosage compensation. This could influence studies of fast-Z as male-biased genes evolve more quickly (Zhang et al. 2004; Zhang and Parsch 2005), possibly because of their involvement with sexual selection or male–male competition. If these genes are over- or under-represented on the sex chromosome, this will impact on the mean rates of divergence of sex-linked genes. For male heterogametic organisms such as worms (Reinke et al. 2000; Jiang et al. 2001; Kelly et al. 2002), flies (Meiklejohn et al. 2003; Parisi et al. 2003; Ranz et al. 2003), and mammals (Betran et al. 2004; Khil et al. 2004; Yang et al. 2006), male-biased genes are under-represented on the X chromosome (with the exception of pre-meiotic genes expressed in testis (Wang et al. 2001; Lercher et al. 2003) because of X inactivation during male meiosis. Preliminary studies in birds show the opposite pattern, with male-biased genes over-represented on the chicken Z chromosome (Kaiser and Ellegren 2006; Storchova and Divinia 2006), possibly because of incomplete dosage compensation in birds (Itoh et al. 2007). However, the removal of genes with male-biased expression does not statistically alter our results, suggesting that the nonrandom distribution of male-biased genes is not responsible for the observed pattern of fast-Z, and offering further indirect support for the causative role of positive selection in fast-Z evolution.
There are at least two reasons to suggest that fast-Z is more easily discerned than fast-X. First, the nonrandom genomic distribution of sex-biased genes described above could theoretically mask fast-X in XY organisms because of the observed shortage of male-biased genes on X (Reinke et al. 2000; Meiklejohn et al. 2003; Parisi et al. 2003; Ranz et al. 2003). While we observed no effect of male-biased gene expression in our analysis of fast-Z, it is possible that the dearth of sex-linked male-biased genes has a stronger effect in some XY organisms because of the inactivation of the X chromosome during male meiosis (Lifschytz and Lindsley 1972; Oliver and Parisi 2004). If the under-representation of X-linked male-biased genes is sufficiently large, it could make it very difficult to detect any fast-X signal, as these forces are contradictory.
Additionally, when favorable mutations have additive effects, male-biased mutation will cause Z-linked loci to evolve faster and X-linked loci slower than autosomal genes (Kirkpatrick and Hall 2004a). Moreover, when mutations are partly dominant, male-biased mutation implies that Z-linked genes evolve faster than autosomal loci over a broader range of dominance values than is the case for X-linked genes (Kirkpatrick and Hall 2004a). Admittedly, the excess of paternally derived mutation in birds is relatively weak, with recent estimates of the male-to-female mutation rate ratio (αm) of ∼2.5 (Bartosch-Härlid et al. 2003; Axelsson et al. 2004), suggesting that the role of male-biased mutation is weak in our analysis. However, male-biased mutation is much more pronounced in some mammals, in particular, primates (Makova and Li 2002). Under this premise, the effects of male-biased mutation might also mask evidence of fast-X.
Finally, it is of interest to make the link between fast-Z and large-Z. There are observations that ZW organisms have more conspicuous male secondary sexual traits, and are thus more prone to extreme sexual selection, than XY organisms (Hastings 1994; Prowell 1998; Reinhold 1998). This idea is lent support from theoretical work comparing the likelihood of Fisher’s runaway and the good-genes mechanisms of sexual selection in ZW and XY systems (Reeve and Pfennig 2003; Kirkpatrick and Hall 2004b). Moreover, there is observation of Z-linkage of plumage traits involved in species recognition in birds (Saetre et al. 2003). If fast-Z is more prevalent than fast-X, it could contribute to more rapid evolution of sex-linked genes involved with sexual selection and reproductive isolation in female heterogametic systems, making the large-Z effect stronger than the large-X.
Methods
Alignments of zebra finch–chicken orthologs
Zebra finch (T. guttata) EST contigs (n = 22,638), each thought to represent a unique transcript and generated by the “Songbird Neurogenomics Initiative,” were available at http://titan.biotec.uiuc.edu/songbird/. The transcripts are all derived from multiple cDNA libraries made from telencephalon of embryonic, juvenile, and adult birds. BlastN was used to search for chicken orthologs to these transcripts among all known and ab initio predicted protein-coding chicken genes identified by Ensembl in the WASHUC 1 assembly (Genebuild Ensembl, December 2005; http://www.ensembl.org/Gallus_gallus/index.html). Orthology was established using the principle of best reciprocal hit, given a minimum E-value of 10−30 for a match. A total of 5658 chicken–zebra finch orthologs were thereby identified. Because e-scores are influenced by sequence length, we excluded all alignments <100 bp from further analysis in order to reduce the possibility of false homology between short alignments.
We screened all zebra finch sequences for high complexity contamination using RepeatMasker (A.F.A. Smit, R. Hubley, and P. Green. 1996–2004. RepeatMasker Open-3.0; http://www.repeatmasker.org) using the Gallus-derived repeat library. As the Ensembl gene annotation methodology automatically removes repetitive DNA elements via RepeatMasker (Curwen et al. 2004), it was not necessary to screen the orthologous chicken coding regions for retroviral contamination.
DNA sequences were subsequently translated into amino acid sequences and aligned using DIALIGN2 (Morgenstern 1999), with a minimum fragment weight (thr) of 10. DIALIGN2 was considered suitable for the alignment of EST sequences with full-length transcripts since it has been shown to perform well on local alignments (Morgenstern 1999; Edgar and Batzoglou 2006) and since it tests all possible reading frames. Gaps introduced in the aligned chicken sequence were removed as they are likely to represent erroneous base calls in EST sequencing. Gap-free regions longer than 7 bp interpreted as nonhomologous by DIALIGN2 were also excluded.
Divergence estimates
From the complete set of 5658 chicken–zebra finch orthologs, 92 were excluded as they contained premature stop codons in the zebra finch sequence; these transcripts could represent pseudogenes but could also contain sequencing errors. Furthermore, as too short sequences make parameter estimations unreliable, 63 alignments shorter than 100 bp were excluded from analysis.
Codeml (PAML package version 3.15) (Yang 1997) was used to estimate the pairwise (run mode = −2) nonsynonymous (dN) and synonymous (dS) divergence for each set of orthologs with settings seqtype = 1 and CodonFreq = 2. Since divergence estimates are not reliable for saturated sites, we excluded 73 orthologs with dS > 3. This left 5430 genes with a mean ungapped alignment length of 559 bp for molecular evolutionary analyses.
The inference of chromosomal location was taken from the May 2006 galGal3 assembly of the chicken genome (http://www.ensembl.org/Gallus_gallus/index.html), and 5050 of the aligned ESTs were mapped to specific chromosomes and were used in further analysis. Mean values of dN (defined as the number of nonsynonymous substitutions per nonsynonymous site) and dS (the number of synonymous substitutions per synonymous site) for autosomes and the Z chromosome, respectively, were calculated by dividing the sum of the number of substitutions over genes by the sum of the number of sites over genes. This means that the problem of infinitely high dN/dS values arising from genes with no synonymous substitutions is circumvented and also that data for individual genes are weighted by alignment length.
A permutation test with 1000 repetitions was used to assess significant differences for each metric (dN, dS, and dN/dS) between the Z and autosomal coding regions. The permutation test calculates the probability that the observed difference in dN, dS, and dN/dS is due to chance association alone. Additionally, 95% confidence intervals were calculated based on bootstrap simulations with 1000 repetitions.
We identified male-biased genes from an Affymetrix microarray study (H. Ellegren et al., unpubl.) of gene expression data in chicken brain tissue. Any coding region that exhibited twofold or greater bias in males compared to females was designated as male biased, according to previous studies of the relationship between male-biased gene expression and rates of evolution (Zhang et al. 2004; Pröschel et al. 2005; Zhang and Parsch 2005). We repeated the above described statistical treatments for the data set excluding these identified as coding regions from both the Z and autosomal categories.
Polymorphism analysis
The BBSRC ChickEST Database at http://www.chick.umist.ac.uk/ harbors 11,000 high-quality polymorphisms found in at least two EST sequences. We first placed these polymorphisms in the chicken genome by using the 100-bp sequence surrounding each SNP in BlastN searches against all known and predicted Ensembl chicken protein-coding transcripts, applying an E-value threshold of 1 × 10−30. To make polymorphism data directly comparable to data on divergence, we restricted the data set to only include SNPs from those genes for which a zebra finch–chicken alignment was available. Although this significantly reduced the number of SNPs available for analysis, it was motivated by the fact that while divergence data were limited to genes found expressed in brain, the total BBSRC SNPs data set was derived from several different tissues. This left a set of 1674 autosomal and 34 Z-linked polymorphisms. The number of synonymous and nonsynonymous polymorphisms was obtained using self-written perl code. In case of multiple mutations per codon, the shortest evolutionary pathway was chosen. Codeml (runmode = −2, CodonFreq = 2) was used to calculate genewise N and S values. Mean values of pN and pS for autosomes and the Z chromosome, respectively, were calculated by dividing the sum of the number of polymorphisms over genes by the sum of the number of sites over genes. This was also done for the autosomal subset of genes that mapped to chromosomes 1–10. Significant differences in pN, pS, and pN/pS were assessed with a permutation test of 1000 repetitions, and 95% confidence intervals determined via bootstrap simulations with 1000 repetitions.
Polymorphism data from the International Chicken Polymorphism Map Consortium (2004) (available at http://genome.ucsc.edu/) were analyzed in a similar way, with codeml estimating the N and S values genewise for sequence covered in shotgun reads. Restricting the data set to polymorphisms seen in at least two chicken strains left 11,036 polymorphisms.
All PERL scripts and alignments used in this analysis are available at http://www.egs.uu.se/evbiol/Research/Data/fast-Z/.
Acknowledgments
Financial support was obtained from the Swedish Research Council and the Wenner-Gren Foundation. The useful comments of three anonymous reviewers are acknowledged.
Footnotes
[Supplemental material is available online at www.genome.org and http://www.egs.uu.se/evbiol/Research/Data/fast-Z/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6031907
References
- Andersson M. Sexual selection. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- Axelsson E., Smith N.G.C., Sundstrom H., Berlin S., Ellegren H., Smith N.G.C., Sundstrom H., Berlin S., Ellegren H., Sundstrom H., Berlin S., Ellegren H., Berlin S., Ellegren H., Ellegren H. Male-biased mutation rate and divergence in autosomal, Z-linked and W-linked introns of chicken and turkey. Mol. Biol. Evol. 2004;21:1538–1547. doi: 10.1093/molbev/msh157. [DOI] [PubMed] [Google Scholar]
- Axelsson E., Webster M.T., Smith N.G.C., Burt D.W., Ellegren H., Webster M.T., Smith N.G.C., Burt D.W., Ellegren H., Smith N.G.C., Burt D.W., Ellegren H., Burt D.W., Ellegren H., Ellegren H. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 2005;15:120–125. doi: 10.1101/gr.3021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch-Härlid A., Berlin S., Smith N.G.C., Møller A.P., Ellegren H., Berlin S., Smith N.G.C., Møller A.P., Ellegren H., Smith N.G.C., Møller A.P., Ellegren H., Møller A.P., Ellegren H., Ellegren H. Life history and the male mutation bias. Evolution Int. J. Org. Evolution. 2003;57:2398–2406. doi: 10.1554/03-036. [DOI] [PubMed] [Google Scholar]
- Berlin S., Ellegren H., Ellegren H. Chicken W: A genetically uniform chromosome in a highly variable genome. Proc. Natl. Acad. Sci. 2004;101:15967–15969. doi: 10.1073/pnas.0405126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A.J., Presgraves D.C., Swanson W.J., Presgraves D.C., Swanson W.J., Swanson W.J. A test for faster X evolution in Drosophila. Mol. Biol. Evol. 2002;19:1816–1819. doi: 10.1093/oxfordjournals.molbev.a004006. [DOI] [PubMed] [Google Scholar]
- Betran E., Emerson J.J., Kaessmann H., Long M., Emerson J.J., Kaessmann H., Long M., Kaessmann H., Long M., Long M. Sex chromosomes and male functions—Where do new genes go? Cell Cycle. 2004;3:873–875. [PubMed] [Google Scholar]
- Bourque G., Zdobnov E.M., Bork P., Pevzner P.A., Tesler G., Zdobnov E.M., Bork P., Pevzner P.A., Tesler G., Bork P., Pevzner P.A., Tesler G., Pevzner P.A., Tesler G., Tesler G. Comparative architecture of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Res. 2005;15:98–110. doi: 10.1101/gr.3002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt D.W., Bruley C., Dunn I.C., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Bruley C., Dunn I.C., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Dunn I.C., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Jones C.T., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Ramage A., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Law A.S., Morrice D.R., Paton I.R., Smith J., Windsor D., Morrice D.R., Paton I.R., Smith J., Windsor D., Paton I.R., Smith J., Windsor D., Smith J., Windsor D., Windsor D., et al. The dynamics of chromosome evolution in birds and mammals. Nature. 1999;402:411–413. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Coyne J.A., Barton N.H., Coyne J.A., Barton N.H., Barton N.H. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- Clayton D.F. Songbird genomics—Methods, mechanisms, opportunities, and pitfalls. Ann. N. Y. Acad. Sci. 2004;1016:45–60. doi: 10.1196/annals.1298.028. [DOI] [PubMed] [Google Scholar]
- Counterman B.A., Ortiz-Barrientos D., Noor M.A.F., Ortiz-Barrientos D., Noor M.A.F., Noor M.A.F. Using comparative genomic data to test for fast-X evolution. Evolution Int. J. Org. Evolution. 2004;58:656–660. [PubMed] [Google Scholar]
- Coyne J.A. The genetic basis of Haldane’s rule. Nature. 1985;314:736–738. doi: 10.1038/314736a0. [DOI] [PubMed] [Google Scholar]
- Coyne J.A., Orr H.A., Orr H.A. Two rules of speciation. In: Otte D., Endler J., Endler J., editors. Speciation and its consequences. Sinauer; Sunderland, MA: 1989. pp. 180–207. [Google Scholar]
- Coyne J.A., Elwyn S., Kim S.Y., Llopart A., Elwyn S., Kim S.Y., Llopart A., Kim S.Y., Llopart A., Llopart A. Genetic studies of two species in the Drosophila melanogaster subgroup, D. yakuba and D. santomea. Genet. Res. 2004;84:11–26. doi: 10.1017/s0016672304007013. [DOI] [PubMed] [Google Scholar]
- Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M.J., Clamp M., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M.J., Clamp M., Andrews T.D., Clarke L., Mongin E., Searle S.M.J., Clamp M., Clarke L., Mongin E., Searle S.M.J., Clamp M., Mongin E., Searle S.M.J., Clamp M., Searle S.M.J., Clamp M., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetic analysis of hybrid sterility within the species Drosophila pseudoobscura. Hereditas. 1974;77:81–88. doi: 10.1111/j.1601-5223.1974.tb01356.x. [DOI] [PubMed] [Google Scholar]
- Duret L., Mouchiroud D., Mouchiroud D. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Edgar R.C., Batzoglou S., Batzoglou S. Multiple sequence alignment. Curr. Opin. Struct. Biol. 2006;16:368–373. doi: 10.1016/j.sbi.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Fridolfsson A.K., Fridolfsson A.K. Male-driven evolution of DNA sequences in birds. Nat. Genet. 1997;17:182–184. doi: 10.1038/ng1097-182. [DOI] [PubMed] [Google Scholar]
- Griffin D.K., Robertson L.B.W., Tempest H.G., Skinner B.M., Robertson L.B.W., Tempest H.G., Skinner B.M., Tempest H.G., Skinner B.M., Skinner B.M. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 2007 doi: 10.1159/000103166. (in press) [DOI] [PubMed] [Google Scholar]
- Haldane J.B.S. Sex-ratio and unisexual sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Hastings I.M. Manifestations of sexual selection may depend on the genetic basis of sex determination. Proc. R. Soc. Lond. B Biol. Sci. 1994;258:83–87. doi: 10.1098/rspb.1994.0146. [DOI] [PubMed] [Google Scholar]
- Hubbard S.J., Grafham D.V., Beattie K.J., Overton I.M., McLaren S.R., Croning M.D.R., Boardman P.E., Bonfield J.K., Burnside J., Davies R.M., Grafham D.V., Beattie K.J., Overton I.M., McLaren S.R., Croning M.D.R., Boardman P.E., Bonfield J.K., Burnside J., Davies R.M., Beattie K.J., Overton I.M., McLaren S.R., Croning M.D.R., Boardman P.E., Bonfield J.K., Burnside J., Davies R.M., Overton I.M., McLaren S.R., Croning M.D.R., Boardman P.E., Bonfield J.K., Burnside J., Davies R.M., McLaren S.R., Croning M.D.R., Boardman P.E., Bonfield J.K., Burnside J., Davies R.M., Croning M.D.R., Boardman P.E., Bonfield J.K., Burnside J., Davies R.M., Boardman P.E., Bonfield J.K., Burnside J., Davies R.M., Bonfield J.K., Burnside J., Davies R.M., Burnside J., Davies R.M., Davies R.M., et al. Transcriptome analysis for the chicken based on 19,626 finished cDNA sequences and 485,337 expressed sequence tags. Genome Res. 2005;15:174–183. doi: 10.1101/gr.3011405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Chicken Polymorphism Map Consortium A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature. 2004;432:717–722. doi: 10.1038/nature03156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Melamed E., Yang X., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Melamed E., Yang X., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Yang X., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Kampf K., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Wang S., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Yehya N., Van Nas A., Replogle K., Band M.R., Clayton D.F., Van Nas A., Replogle K., Band M.R., Clayton D.F., Replogle K., Band M.R., Clayton D.F., Band M.R., Clayton D.F., Clayton D.F., et al. Dosage compensation is less effective in birds than in mammals. J. Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Ryu J., Kiraly M., Duke K., Reinke V., Kim S.K., Ryu J., Kiraly M., Duke K., Reinke V., Kim S.K., Kiraly M., Duke K., Reinke V., Kim S.K., Duke K., Reinke V., Kim S.K., Reinke V., Kim S.K., Kim S.K. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Riley R.M., Wolfinger R.D., White K.P., Passador-Gurgel G., Gibson G., Riley R.M., Wolfinger R.D., White K.P., Passador-Gurgel G., Gibson G., Wolfinger R.D., White K.P., Passador-Gurgel G., Gibson G., White K.P., Passador-Gurgel G., Gibson G., Passador-Gurgel G., Gibson G., Gibson G. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- Kaiser V.B., Ellegren H., Ellegren H. Nonrandom distribution of genes with sex-biased expression in the chicken genome. Evolution Int. J. Org. Evolution. 2006;60:1945–1951. [PubMed] [Google Scholar]
- Kelly W.G., Schaner C.E., Dernburg A.F., Lee M.H., Kim S.K., Villeneuve A.M., Reinke V., Schaner C.E., Dernburg A.F., Lee M.H., Kim S.K., Villeneuve A.M., Reinke V., Dernburg A.F., Lee M.H., Kim S.K., Villeneuve A.M., Reinke V., Lee M.H., Kim S.K., Villeneuve A.M., Reinke V., Kim S.K., Villeneuve A.M., Reinke V., Villeneuve A.M., Reinke V., Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil P.P., Smirnova N.A., Romanienko P.J., Camerini-Otero R.D., Smirnova N.A., Romanienko P.J., Camerini-Otero R.D., Romanienko P.J., Camerini-Otero R.D., Camerini-Otero R.D. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat. Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., Hall D.W., Hall D.W. Male-biased mutation, sex linkage, and the rate of adaptive evolution. Evolution Int. J. Org. Evolution. 2004a;58:437–440. [PubMed] [Google Scholar]
- Kirkpatrick M., Hall D.W., Hall D.W. Sexual selection and sex linkage. Evolution Int. J. Org. Evolution. 2004b;58:683–691. doi: 10.1111/j.0014-3820.2004.tb00401.x. [DOI] [PubMed] [Google Scholar]
- Lercher M.J., Urrutia A.O., Hurst L.D., Urrutia A.O., Hurst L.D., Hurst L.D. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol. Biol. Evol. 2003;20:1113–1116. doi: 10.1093/molbev/msg131. [DOI] [PubMed] [Google Scholar]
- Lifschytz E., Lindsley D.L., Lindsley D.L. The role of X-chromosome inactivation during spermatogenesis. Proc. Natl. Acad. Sci. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Wu C.I., Wu C.I. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc. Natl. Acad. Sci. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova K.D., Li W.H., Li W.H. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- Meiklejohn C.D., Parsch J., Ranz J.M., Hartl D.L., Parsch J., Ranz J.M., Hartl D.L., Ranz J.M., Hartl D.L., Hartl D.L. Rapid evolution of male-biased gene expression in Drosophila. Proc. Natl. Acad. Sci. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: Improvements of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Musters H., Huntley M.A., Singh R.S., Huntley M.A., Singh R.S., Singh R.S. A genomic comparison of faster-sex, faster-X, and faster-male evolution between Drosophila melanogaster and Drosophila pseudoobscura. J. Mol. Evol. 2006;62:693–700. doi: 10.1007/s00239-005-0165-5. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Bustamante C., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Clark A.G., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Glanowski S., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Sackton T.B., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Hubisz M.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Fledel-Alon A., Tanenbaum D.M., Civello D., White T.J., Tanenbaum D.M., Civello D., White T.J., Civello D., White T.J., White T.J., et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:976–985. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B., Parisi M., Parisi M. Battle of the Xs. Bioessays. 2004;26:543–548. doi: 10.1002/bies.20034. [DOI] [PubMed] [Google Scholar]
- Orr H.A., Betancourt A.J., Betancourt A.J. Haldane’s sieve and adaptation from the standing genetic variation. Genetics. 2001;157:875–884. doi: 10.1093/genetics/157.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B., Malley J., Andrews J., Eastman S., Oliver B., Andrews J., Eastman S., Oliver B., Eastman S., Oliver B., Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigozzi M.I., Solari A.J., Solari A.J. The ZW pairs of two paleognath birds from two orders show transitional stages of sex chromosome differentiation. Chromosome Res. 1999;7:541–551. doi: 10.1023/a:1009241528994. [DOI] [PubMed] [Google Scholar]
- Presgraves D.C. Patterns of postzygotic isolation in Lepidoptera. Evolution Int. J. Org. Evolution. 2002;56:1168–1183. doi: 10.1111/j.0014-3820.2002.tb01430.x. [DOI] [PubMed] [Google Scholar]
- Pröschel M., Zhang Z., Parsch J., Zhang Z., Parsch J., Parsch J. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2005;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowell D. Sex linkage and speciation in Lepidoptera. In: Berlocher S., Howard D., Howard D., editors. Endless forms: Species and speciation. Oxford University Press; New York: 1998. pp. 309–319. [Google Scholar]
- Ranz J.M., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L., Castillo-Davis C.I., Meiklejohn C.D., Hartl D.L., Meiklejohn C.D., Hartl D.L., Hartl D.L. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Reeve H.K., Pfennig D.W., Pfennig D.W. Genetic biases for showy males: Are some genetic systems especially conducive to sexual selection? Proc. Natl. Acad. Sci. 2003;100:1089–1094. doi: 10.1073/pnas.0337427100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold K. Sex linkage among genes controlling sexually selected traits. Behav. Ecol. Sociobiol. 1998;44:1–7. [Google Scholar]
- Reinke V., Smith H.E., Nance J., Wang J., Van Doren C., Begley R., Jones S.J.M., Davis E.B., Scherer S., Ward S., Smith H.E., Nance J., Wang J., Van Doren C., Begley R., Jones S.J.M., Davis E.B., Scherer S., Ward S., Nance J., Wang J., Van Doren C., Begley R., Jones S.J.M., Davis E.B., Scherer S., Ward S., Wang J., Van Doren C., Begley R., Jones S.J.M., Davis E.B., Scherer S., Ward S., Van Doren C., Begley R., Jones S.J.M., Davis E.B., Scherer S., Ward S., Begley R., Jones S.J.M., Davis E.B., Scherer S., Ward S., Jones S.J.M., Davis E.B., Scherer S., Ward S., Davis E.B., Scherer S., Ward S., Scherer S., Ward S., Ward S., et al. A global profile of germline gene expression in C. elegans. Mol. Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rice W.R. Sex chromosomes and the evolution of sexual dimorphism. Evolution Int. J. Org. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Liu Y., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Bettencourt B.R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Letovsky S., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Nielsen R., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Thornton K., Hubisz M.J., Chen R., Meisel R.P., Hubisz M.J., Chen R., Meisel R.P., Chen R., Meisel R.P., Meisel R.P., et al. Comparative genome sequencing of Drosophila pseudoobscura: Chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.G., Phillips S.D.F., Phillips S.D.F. The genetics of sexual isolation. In: Berlocher S., Howard D., Howard D., editors. Endless forms: Species and speciation. Oxford University Press; New York: 1998. pp. 291–308. [Google Scholar]
- Saetre G.P., Borge T., Lindroos K., Haavie J., Sheldon B.C., Primmer C., Syvanen A.C., Borge T., Lindroos K., Haavie J., Sheldon B.C., Primmer C., Syvanen A.C., Lindroos K., Haavie J., Sheldon B.C., Primmer C., Syvanen A.C., Haavie J., Sheldon B.C., Primmer C., Syvanen A.C., Sheldon B.C., Primmer C., Syvanen A.C., Primmer C., Syvanen A.C., Syvanen A.C. Sex chromosome evolution in Ficedula flycatchers. Proc. Roy. Soc. Lond. B Biol. Sci. 2003;270:53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova R., Divinia P., Divinia P. Nonrandom representation of sex-biased genes on the chicken Z chromosome. J. Mol. Evol. 2006;63:676–681. doi: 10.1007/s00239-006-0022-1. [DOI] [PubMed] [Google Scholar]
- Sundström H., Webster M.T., Ellegren H., Webster M.T., Ellegren H., Ellegren H. Reduced variation on the chicken Z chromosome. Genetics. 2004;167:377–385. doi: 10.1534/genetics.167.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Hartl D.L., Hartl D.L. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane’s rule. Evolution Int. J. Org. Evolution. 2003;57:2580–2598. doi: 10.1111/j.0014-3820.2003.tb01501.x. [DOI] [PubMed] [Google Scholar]
- Templeton A.R. Analysis of head shape differences between two infertile species of Hawaiian Drosophila. Evolution Int. J. Org. Evolution. 1977;31:630–641. doi: 10.1111/j.1558-5646.1977.tb01052.x. [DOI] [PubMed] [Google Scholar]
- Thornton K., Long M., Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol. Biol. Evol. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- Thornton K., Long M., Long M. Excess of amino acid substitutions relative to polymorphism between X-linked duplications in Drosophila melanogaster. Mol. Biol. Evol. 2005;22:273–284. doi: 10.1093/molbev/msi015. [DOI] [PubMed] [Google Scholar]
- Thornton K., Bachtrog D., Andolfatto P., Bachtrog D., Andolfatto P., Andolfatto P. X chromosomes and autosomes evolve at similar rates in Drosophila: No evidence for faster-X protein evolution. Genome Res. 2006;16:498–504. doi: 10.1101/gr.4447906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson D.G., Singh R.S., Singh R.S. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol. Biol. Evol. 2003;20:1705–1709. doi: 10.1093/molbev/msg193. [DOI] [PubMed] [Google Scholar]
- Torgerson D.G., Singh R.S., Singh R.S. Enhanced adaptive evolution of sperm-expressed genes on the mammalian X chromosome. Heredity. 2006;96:39–44. doi: 10.1038/sj.hdy.6800749. [DOI] [PubMed] [Google Scholar]
- van Tuinen M., Sibley C.G., Hedges S.B., Sibley C.G., Hedges S.B., Hedges S.B. The early history of modern birds inferred from DNA sequences of nuclear and mitochondrial ribosomal genes. Mol. Biol. Evol. 2000;17:451–457. doi: 10.1093/oxfordjournals.molbev.a026324. [DOI] [PubMed] [Google Scholar]
- Vicoso B., Charlesworth B., Charlesworth B. Evolution on the X chromosome: Unusual patterns and processes. Nat. Rev. Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Wang P.J., McCarrey J.R., Yang F., Page D.C., McCarrey J.R., Yang F., Page D.C., Yang F., Page D.C., Page D.C. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Webster M.T., Axelsson E., Ellegren H., Axelsson E., Ellegren H., Ellegren H. Strong regional biases in nucleotide substitution in the chicken genome. Mol. Biol. Evol. 2006;23:1203–1216. doi: 10.1093/molbev/msk008. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J., Ingram-Drake L., Drake T.A., Lusis A.J., Drake T.A., Lusis A.J., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Parsch J., Parsch J. Positive correlation between evolutionary rate and recombination rate in Drosophila genes with male-biased expression. Mol. Biol. Evol. 2005;22:1945–1947. doi: 10.1093/molbev/msi189. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hambuch T.M., Parsch J., Hambuch T.M., Parsch J., Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]