Abstract
A maternally established gradient of nuclear Dorsal protein is the first step in subdivision of the Drosophila neurectoderm into stripes of homeodomain gene expression. Dorsal in combination with the EGF and TGFβ signaling pathways are key regulators of the expression of the genes ventral nervous system defective (vnd), intermediate neuroblasts defective (ind), and muscle segment homeobox (msh) in the developing neurectoderm. These three genes encode homeodomain transcription factors that can repress each other, which ensures adjacent, non-overlapping expression domains. Expression of vnd, ind, and msh is maintained after decline in EGF and TGFβ signaling, but the relevant positive transcriptional regulators have not yet been defined. Here we show that Ind can bind DNA with the same sequence specificity as its murine ortholog Gsh1. We have identified a novel upstream regulatory element at the ind locus containing predicted Ind binding sites, and we show that Ind activity is both necessary and sufficient for reporter gene expression from this element. We conclude that Ind can act as a transcriptional activator, and that positive autoregulation of Ind is a mechanism for persistent ind expression within the developing embryonic nervous system.
Keywords: Drosophila, Intermedate neuroblast defective, CNS, Enhancers
1. Introduction
Development of the embryonic central nervous system (CNS) in Drosophila initiates with the formation of the neural stem cells, called neuroblasts. Formation of the neuroblasts is a precise process that is repeated in each segment of the embryo (Broadus et al., 1995; Campos-Ortega and Hartenstein, 1985; Doe, 1992). Initially, these neuroblasts form in three columns on either site of the ventral midline. These columns are defined by the expression of three homeodomain transcription factors encoded by the ventral nervous system defective (vnd), intermediate neuroblasts defective (ind) and muscle segment homeobox (msh) genes.
Subdivision of the CNS across the Dorsoventral axis appears to be a two step process. Initially the ventral neuroectoderm in Drosophila is subdivided into three domains, established by signaling pathways that pattern the embryo on a global basis (Von Ohlen and Doe, 2000). Specifically, the Dorsal morphogen gradient is required for Vnd expression (Mellerick and Nirenberg, 1995; Von Ohlen and Doe, 2000). Both the Dorsal and Epidermal Growth Factor Receptor (Egfr) signaling pathways are required for initiation of Ind (Von Ohlen and Doe, 2000). In the Msh domain, Dorsal is required to keep Decapentaplegic (Dpp) signaling low (Von Ohlen and Doe, 2000). The second step involves an interaction between these three proteins and a transcriptional repression hierarchy (Cowden and Levine, 2003; Von Ohlen and Doe, 2000). In this hierarchy, Vnd represses Ind in the ventral column and Ind represses Msh in the intermediate column (Mc Donald et al., 1998; Von Ohlen and Doe, 2000; Weiss et al., 1998).
The product of the Ind gene is exclusively expressed in the intermediate column during the early stages of CNS development. This homeodomain protein is required for proper formation and specification of the neuroblasts that derive from this domain (Weiss et al., 1998). As described above initiation of the stripes of Vnd, Ind and Msh expression is driven by the maternal Dorsal gradient. However, how these stripes are maintained is not as well understood. Vnd has been shown to respond to the Dorsal gradient via a neurectodermal specific enhancer, located in the first intron of the gene (Markstein et al., 2004; Stathopolous et al., 2002). In addition, it has been suggested that Vnd is capable of activating its own expression (Yu et al., 2005). Although there is no in vivo evidence of Vnd auto-regulation, in cultured cells Vnd is capable of binding to and activating transcription from an enhancer located upstream of its coding sequences (Wang et al., 2005; Yu et al., 2005). This suggests that regulation of Vnd expression may involve modular activity of multiple enhancer elements. Specifically, both an initiator element, that is capable of responding to the Dorsal gradient and other global patterning signals, and a maintenance element that is regulated by Vnd itself are required for complete Vnd expression.
Initial expression of Ind is controlled by an enhancer element located downstream of the ind coding sequence. This element contains known Vnd, Ets and Dorsal binding sites (TVO unpublished; (Cowden and Levine, 2003; Stathopolous and Levine, 2005; Weiss et al., 1998). While direct mutagenesis of these sites has not been performed, expression from this construct does respond to mutations in components of these pathways in a manner consistent with their regulation of Ind (TVO unpublished observation). Here we have identified a novel regulatory element located upstream of the ind coding sequence that is capable of directing ind dependent reporter gene expression. We also present data suggesting that in addition to its role as a transcriptional repressor, Ind also plays a key role in maintaining its own expression. Specifically, we show that Ind is not only required for but is also sufficient to activate expression from this upstream element.
2. Results
2.1 The Ind homeodomain binds DNA in a sequence specific manner
Ind is a homeodomain-containing protein and we predicted that it is a sequence specific DNA binding protein. Therefore, we wanted to determine the DNA binding specificity for Ind. Previously, (Valerius et al., 1995) demonstrated that the mouse Ind ortholog, Gsh1, was a sequence specific DNA binding protein, with the sequence specificity (GC T/C A/C ATTA G/A). An alignment of Ind and Gsh1 revealed a high degree of sequence identity within the homeodomains, particularly within the domains known to make contact with the DNA (Fig 1A). Thus, we predicted that Ind would bind DNA with the same sequence specificity as Gsh1. To test this we designed dsDNA oligos with either the consensus Gsh1 binding sites or a mutagenized oligo with a four base pair substitution of GGGG in place of the ATTA core sequence. We tested whether Ind would bind to these oligonucleotides using electrophoretic mobility shift assays with a bacterially expressed his6 tagged Ind homeodomain (His6IndHD) protein. We found that the His6Ind HD protein was able to shift the fragment of DNA containing the Gsh1 like binding site. We also find that unlabeled wild type Gsh1 fragments were efficiently able to compete for binding in a concentration dependent manner whereas the mutant Gsh1 fragment did not compete (Figure 1B). Therefore, we concluded that Ind can bind DNA with similar sequence specificity as its mouse counterpart Gsh1.
Figure 1.
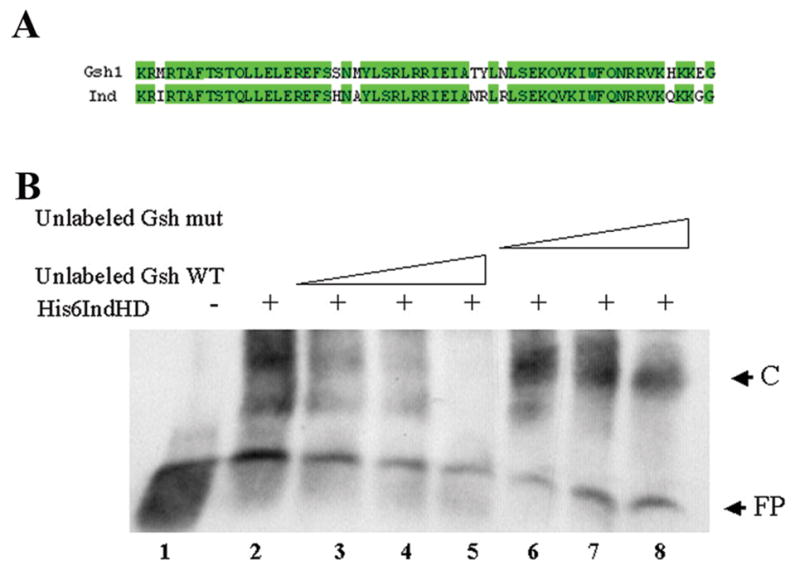
A) Aligment of homeodomain sequences for Ind and Gsh1. Areas shaded green indicate regions of amino acid identity. Overall sequence identity for the homeodomains is 85%. B) Electrophoretic mobility shift assay demonstrating Ind binding to Gsh1 binding site. FP indicates the position of the free probe. C indicates position of the complex formed in the presence of His6-Ind homeodomain. Lane1) Free Gsh1 oligo. Lanes 2–8) + 0.5 mg His6IndHD, Lane 2) no competitor. Lane 3) 5ng unlabeled Gsh1. Lane 4) 10 ng unlabeled Gsh1. Lane5) 20 ng unlabeled Gsh1. Lane 6) 5ng unlabeled mutant Gsh1. Lane 7) 10 ng unlabeled mutant Gsh1. Lane 8) 20 ng unlabeled mutant Gsh1.
2.2 Ind activity is required to maintain ind expression
While initiation of ind transcription is known to be controlled by the early patterning signals, it is less clear what proteins are required for maintenance of expression. One candidate for a gene product controlling maintenance is Ind itself. In order to determine if maintenance of ind expression is dependent on Ind activity, we examined the expression of ind mRNA in ind mutant embryos. Of the collection of ind alleles available, most were generated by P-element excision and delete most, if not all, the coding sequence of the gene (Weiss et al., 1998). However, one allele, indRR108, was generated by EMS mutagenesis and thus had the potential to produce transcript. When we examined indRR108 homozygous mutant embryos for presence of ind transcripts, we found that they did indeed express transcript (Figure 2). ind mRNA expression is initiated normally in these embryos (Figure 2A–B). However, expression of ind mRNA is not maintained properly in later stage indRR108 mutant embryos. Specifically, as early as stage 7 we saw reduced expression of ind mRNA in the neurectoderm (Figure 2D). By stage 11, expression of ind transcripts is completely gone from the presumptive central nervous system (Figure 2G&H). However, expression is maintained in the head, demonstrating that the loss of expression is not due to failed staining. We concluded from this that ind expression in the head region is regulated independently of expression in the trunk regions. These data are consistent with the hypothesis that maintenance of ind transcription during CNS formation is dependent on the ability of Ind to positively regulate its own expression.
Figure 2.
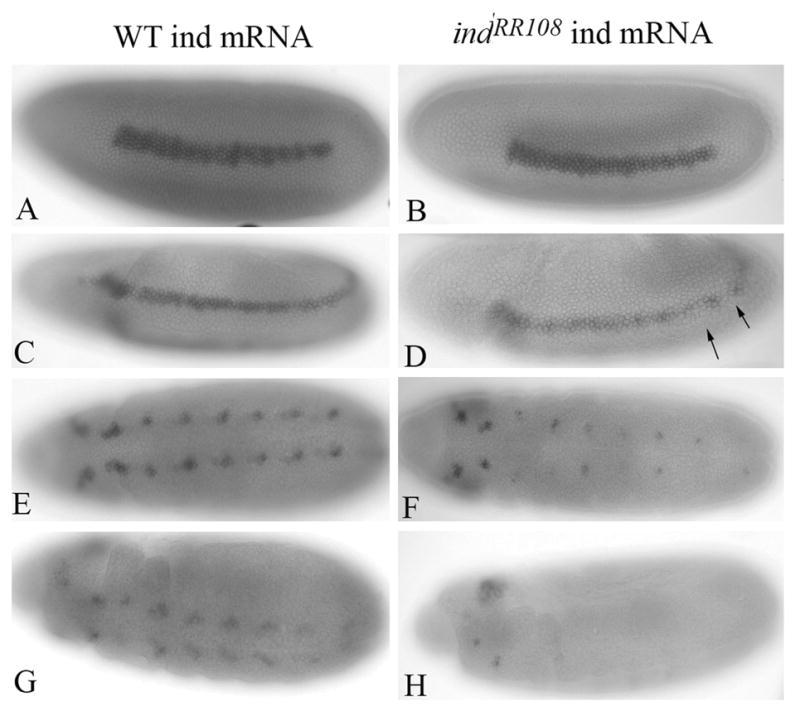
Ind activity is required to maintain expression of ind mRNA. A, C, E, G) Wild type ind mRNA expression. (B, D, F, H) expression of ind mRNA in indRR108 homozygous mutant embryos. Expression is initiated normally (compare A and B). At stage 8 expression of ind mRNA begins to deteriorate (compare C and D) Arrow indicates a gap in ind pattern beginning to form at stage 8. ind message is gone from the trunk regions by late stage 11.
2.3 Putative ind binding sites are located upstream of the Ind coding sequence in a novel regulatory element
If Ind is capable of positively regulating its own expression, there should be an enhancer element that Ind is acting through. In addition to other groups, we have independently identified an ind regulatory element located 3’ to the ind coding sequence. This regulatory element drives expression of the lacZ reporter gene in a pattern consistent with the ind expression pattern (Figure 3A). Expression from this regulatory element responds to the DV signaling pathways in a manner consistent with regulation of ind (Stathopolous and Levine, 2005; Weiss et al., 1998) TVO unpublished results). However, we failed to see expression of the lacZ transcript from this element in the neuroblasts derived from the ind domain (Figure 3). This, in combination with our observation that Ind activity is required to maintain expression of ind transcripts (Figure 2), led us to speculate that an additional enhancer element might be required for expression of ind at later stages of development. Expression from this element should be dependent on the activity of Ind. We predicted that such an enhancer element should contain sequences to which Ind will bind. Sequence analysis failed to reveal Gsh1/Ind binding sites located within the previously described downstream regulatory element. We used the cis-analyst search program available at the Berkeley Drosophila Genome Project as well as the seqseek program developed by Eric Johnson, (University of Oregon) to search genomic sequences around the ind gene for Gsh1/Ind-like binding sites (see Materials and Methods).
Figure 3.
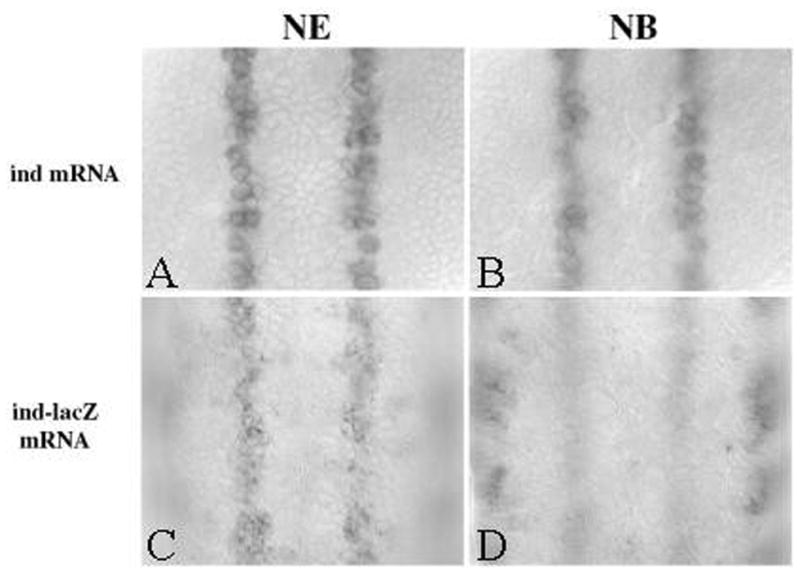
Enhancer element located downstream of ind coding region drives expression in neurectoderm but not neuroblasts. Top panels show expression of endogenous ind mRNA in A) neurectoderm and B) neuroblasts. Bottom panels show expression of ind enhancer driven lacZ expression in C) neuroectoderm, but expression in D) neuroblasts is undetectable.
Interestingly, both search programs identified four potential Gsh1/Ind binding sites (Figure 4A). Whether they identified the same sites was unclear, because only the seekseq program provided detailed sequence of the region. Based on the seekseq program these sites were all located within approximately 2.8 kb upstream of the ind coding sequence. Therefore, we generated new reporter constructs with the upstream genomic fragment driving expression of the lacZ reporter gene. An approximately 4.2 kb fragment of upstream genomic sequences, designated GN4 (Genomic N-terminal 4 kb), drives expression in a pattern in the embryo that replicated the expression of endogenous ind (Fig 4). Specifically, lacZ transcription was initiated as early as stage 6 (data not shown), shortly after endogenous ind mRNA was initiated and was maintained until after stage 11. Multiple independent insertions gave identical expression patterns. The observation that lacZ message initiated from this element at stages earlier than the indRR108 mutants predict, is not entirely surprising; because Ind may be sufficient to activate its own expression before it is necessary. These results do not exclude the possibility that the upstream regulatory element, like the downstream element, is capable of responding to the early DV signaling pathways that initiate ind expression. However, this element does not contain consensus-binding sites for the known transcription factor components of the Dorsal, EGFR or Dpp pathways (data not shown).
Figure 4.
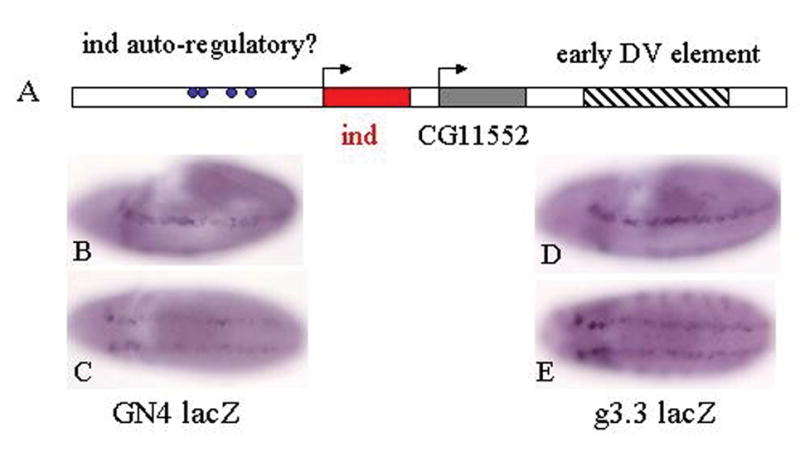
Identification of a previously uncharacterized Ind regulatory element located upstream of ind coding sequences. A) Illustration of genomic region around ind gene. The hatched region encompasses the early DV enhancer. The ind gene is shown in red and the CG11552 gene is gray. Blue dots indicate the relative positions of putative Gsh1/Ind binding sites. B) lacZ mRNA from GN4 upstream regulatory element in a stage 9 embryo. C) GN4LacZ mRNA expression in a stage 10/11 embryo. D) lacZ expression from downstream (g3.3lacZ) regulatory element in a stage 9 embryo. E) lacZ expression from downstream regulatory element in a stage 10/11 embryo.
Because the identified fragment was so large, 4.2 kb, we wanted to further narrow down the region responsible for maintenance of ind expression. To do this we built two additional reporter constructs. GN1.2LacZ contains the proximal 1.2 kb of genomic DNA upstream of ind coding sequences (Figure 5C). This element does not contain any of the potential Ind binding sites. GN1.6lacZ contains the next 1.6 kb of genomic DNA upstream of Ind and includes all four putative binding sites. When we examined expression from these reporter constructs in transgenic embryos we found that GN1.2 lacZ transgene always failed to express in an ind-like pattern (Figure 5C). However, the GN1.6lacZ transgene did drive lacZ expression in an ind-like pattern (Figure 5D). This provided further evidence that a region of genomic DNA containing Ind-like binding sites is important for conferring ind-like reporter gene expression. Finally, we performed site directed mutagenesis of the putative Ind binding sites within the GN1.6 element. We find that expression from the mutant GN1.6LacZ reporter construct is reduced relative to wild type but not entirely absent (Figure 5E). These results suggest that there are either additional Ind binding sites not identified by computational means or that other factors in addition to Ind are capable of regulating expression from this element. It is also possible that Ind binds to sites different from that of the Gsh consensus sequence. Nevertheless, the result that expression is reduced when the putative binding sites are mutated further supports our hypothesis that Ind binding to this enhancer is important for expression.
Figure 5.
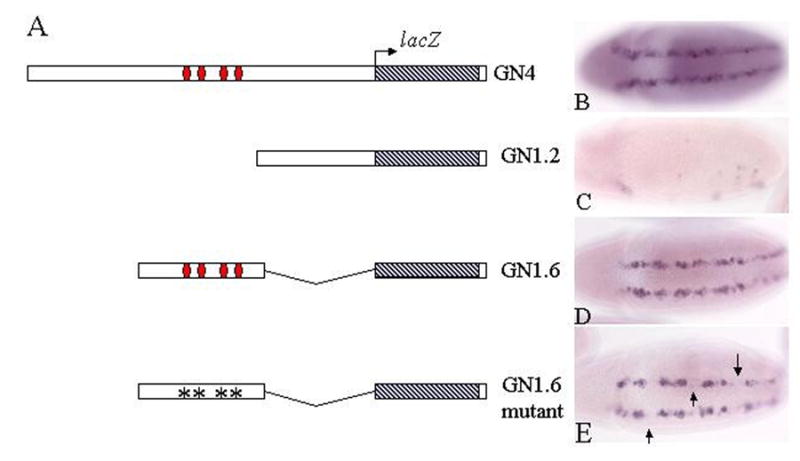
Deletion analysis of GN4 enhancer reveals regulatory element is contained in 1.6 kb fragment containing potential Ind binding site. A) Schematic of reporter constructs built and tested in embryos. B–E) Expression of lacZ mRNA in stage 10 embryos from corresponding reporter constructs to the left of photo. B) GN4lacZ. C) GN1.2 lacZ. D) GN1.6 lacZ. E) Mutant GN1.6 lacZ, arrows indicate the presence of gaps in the pattern not observed with the wild type reporter.
2.4 Expression of GN4lacZ requires Ind activity
If this novel upstream regulatory element responds to positive regulation by Ind, we predicted it would not be expressed in ind mutant embryos. Conversely, the downstream element that responds to the early DV patterning signals should be initiated normally in ind mutant embryos. To test this hypothesis we built recombinant fly lines in which the upstream regulatory element or the downstream regulatory elements were recombined onto the ind mutant chromosomes. For these experiments we used two different ind alleles; ind 16.2 and ind RR108(Weiss et al., 1998). We found that in both mutant backgrounds, the absence of ind activity resulted in a loss lacZ mRNA expression from the upstream regulatory element (GN4lacZ; Figure 6B&C). However, we were able to detect normal initiation of expression from the downstream element (g3.3lacZ; Figure 6E&F). These results were consistent with our hypothesis that the downstream element responds to the early dorsoventral patterning cues and the upstream element responds directly to Ind activity. In addition, when we examined expression from the GN4lacZ construct in a Paired Gal4-UAS Ind gain of function background we were able to detect ectopic lacZ expression in the Prd pattern (Figure 6G). This suggests that not only is Ind activity necessary for reporter expression from this element but Ind is also sufficient to activate expression of lacZ when Ind is ectopically expressed outside the endogenous Ind domain. These results further support our hypothesis that Ind positively regulates its own expression via this newly identified enhancer element located upstream of the coding region.
Figure 6.
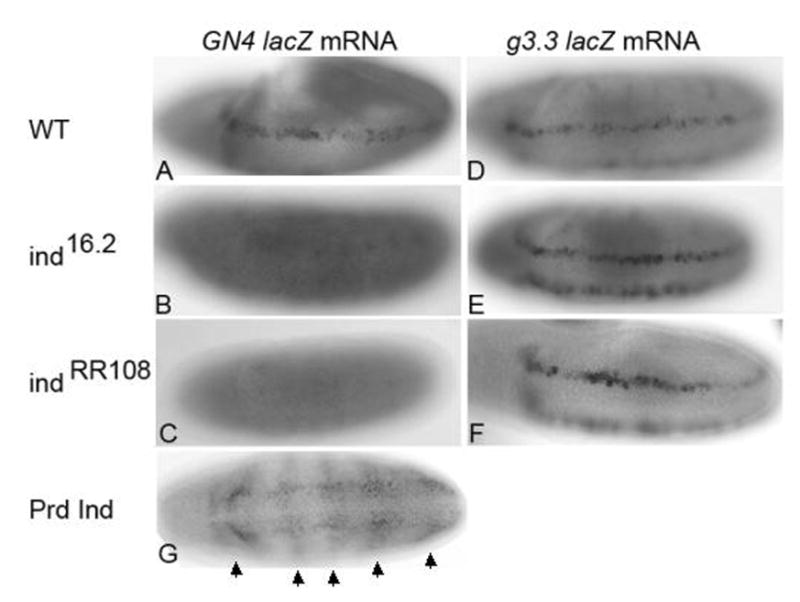
Ind activity is necessary and sufficient for GN4lacZ expression. A) Expression of GN4lacZ in WT embryo. B–C) Expression of GN4lacZ is absent in ind mutant embryos either ind16.2 (B) or ind RR108 mutants. D–F) Expression from the downstream regulatory element is unaffected in ind mutant embryos. G) Prd Gal4 UAS Ind embryo expressing GN4lacZ in Prd pattern. Arrows indicate the positions of pair rule-like stripes of lacZ expression observed in Prd Gal4 UAS Ind embryos. All embryos are stage 9–10 and anterior is to the left.
3. Discussion
Patterning of the CNS in Drosophila involves regulation of gene expression by three DV restricted homeodomain containing proteins. Previous data suggest that this is accomplished by a cascade of transcriptional repression (Cowden and Levine, 2003; Stathopolous and Levine, 2005). It is in fact well documented that the DV restricted homeodomain proteins act as transcriptional repressors. Recent papers have suggested that at least Vnd can act as a transcriptional activator (Yu et al., 2005) (Uhler et al., 2002). Here we present in vivo data suggesting that the Ind homeodomain protein can also act as a transcriptional activator. Specifically, the data show that Ind activity is required to maintain ind expression and that this autoregulation takes place through a previously uncharacterized regulatory element located upstream of the ind coding sequence. It is entirely possible that an additional as yet unidentified positive regulator is required for the maintenance of Ind expression.
Our results suggest that Ind can act as a transcriptional activator. However, we cannot rule out the possibility that loss of ind function causes derepression of a repressor. One possible repressor of ind could be Msh. We do not think this is the case for three reasons; first I have previously demonstrated that expression of Msh is expanded ventrally in ind mutant embryos (Weiss et al., 1998). In ind mutant embryos Msh expression is expanded ventrally into the Ind domain at the time of initiation, around stage 6. However, ind mRNA in the RR108 mutant appears largely normal until stage seven and eight. Second, Cowden and Levine, (2003) state that Msh over-expression does not repress ind expression (Cowden and Levine, 2003). It is possible there could be another as yet unidentified repressor as suggested in (Stathopolous and Levine, 2005). Third, expression of ind does not expand dorsally in msh mutant embryos (data not shown).
Regulation of ind expression appears to involve to separable regulatory elements. The previously described element that is located downstream of the ind coding sequence and is required for initiation (Cowden and Levine, 2003; Stathopolous and Levine, 2005; Weiss et al., 1998). An additional element located upstream of the coding sequence appears to be dependent on Ind activity. A parallel regulation might also be possible for Vnd where the early neurectodermal enhancer is located within the first intron (Markstein et al., 2004; Stathopolous et al., 2002). However, elements controlling expression in neuroblasts are located upstream of the coding sequence (Shao et al., 2002). Moreover, Vnd can bind to the upstream element and regulate reporter gene expression from it (Saunders et al., 1998; Wang et al., 2005; Yu et al., 2005). It should be noted, that these results are based on tissue culture reporter assays and not in vivo results. Nevertheless, these data do support the idea that similar to ind, vnd expression might be regulated by separable enhancer elements which control initiation and maintenance independently.
The ability of Vnd to act as a transcriptional activator appears to be in part regulated by interaction with the HMG domain-containing protein Dichaete (Yu et al., 2005). Genetic data suggest that Ind and Vnd interact with Dichaete in a similar manner (Buescher et al., 2002; Zhao and Skeath, 2002). Ind expression is normal in dichaete mutant embryos (Zhao and Skeath, 2002). Thus, we hypothesized that expression of Ind is most likely initiated normally in dichaete mutants and despite the apparently normal expression of Ind protein in dichaete mutants we might see an effect on expression from our upstream regulatory element. However, following recombination of the GN4lacZ transgene onto the dichaete chromosome we failed to see a loss of lacZ expression (data not shown). Therefore we are not convinced that Dichaete is involved in this aspect of Ind function.
Our data provides evidence that following initiation by global patterning signals, expression of the Ind homeodomain is maintained by the activity of Ind itself. This occurs through a newly identified regulatory element that is positioned upstream of the coding sequence and away from the element controlling initiation. A parallel type of regulation might occur for the Vnd homeodomain protein. However, additional work is required to confirm this hypothesis.
4. Materials and Methods
4.1 Fly stocks and In situ hybridizations
All fly stocks were maintained on standard media, yw flies were used for wildtype. The ind mutant fly lines used were indRR108/TM3 ftz lacZ; ind 16.2/TM3 ftz lacZ (Weiss et al., 1998). Recombination crosses were done as described in; Greenspan, 1997). The Prd Gal4 line was obtained from Chris Q. Doe (Oregon). All other fly lines are transgenics of constructs described in sections 4.2.
In situ hybridizations were done according to standard procedures (Jiang et al., 1991; Tautz and Pfeiffle, 1989)
4.2 Constructs
To build the UASind construct, PCR primers were designed to amplify the coding sequence from ind cDNA in NB40 were as follows: IndF cccgctcgagcggggaaataccccagaaacccaagatg and IndR ggggtaccccacgcctcaaccttcaattcgtg (Weiss et al., 1998). This product was cloned into pUAST (Brand and Perrimon, 1993) at the XhoI/KpnI sites.
GN4 lacZ
Phage DNA containing the genomic sequences surrounding Ind coding region were kindly provided by Joe Weiss (Stanford). A 4.5 kb Eco RI to Xho I fragment was cloned into the CaSper Hs43 lacZ reporter vector. Restriction mapping of phage clones was done via Southern blotting using the Chemiluminscent Dig labeling kit (Roche). Multiple single and double digests were performed, and blots were probed with Dig-labeled probes to either the ind cDNA or the previously described domain containing Vnd binding sites (Weiss et al., 1998). The g3.3 lacZ construct was built with a 2.5 kb SalI/EcoRI fragment located approximately 1.5 kb downstream of the stop codon of ind. This fragment was also ligated into the CaSper Hs43 LacZ reporter vector.
GN1.2 lacZ
The 1.2 kb PCR product was amplified from GN4 using the following primers: GN1.2 F tggatgatccttgccgc and GN1.2 R cgatcgctgactgtgcg. GN1.2 PCR product was cloned in to the TOPO TA cloning vector (Invitrogen) and subsequently moved into the pCaSpeR HS43 lacZ reporter vector at the EcoRI site.
GN1.6 LacZ
1.6 Kb ClaI fragment from GN4 was cloned into Bluescript KS at the ClaI site, then moved in to the pCaSpeR HS43 lacZ reporter vector with the NotI and XbaI sites. The Non-Mammalian Model Systems unit at Duke University injected all transgenes.
4.3 His Tagged ind homeodomain and DNA binding experiments
The coding region for the C-terminal of the Ind protein was cloned into the pET28c vector (Novagen) between the Not I and HindIII sites. His6IndHD was expressed and purified by Abgent Corporation (San Diego CA). Oligomers containing the Gsh1 binding site are tgaccagctaattagagacacatt and aatgtgtctctaattagctggtca. Oligos containing the mutant site are tgaccagctaggggagacacatt and aatgtgtctccccctagctggtca these serve as a control for non-specific binding. WT and mutant oligos were annealed to form dsDNA molecules. EMSA assay was performed using the Dig non-radioactive Gel shift kit (second generation, Roche). The DNA/protein complexes were resolved on a 5% non-denaturing polyacrylamide gel.
4.4 Sequence analysis by Cis analyst and seq seek searches
We used two separate search programs to search for Gsh1/Ind-like binding sites in the genomic regions around Ind. We used the cis-analyst search program available at the Berkeley Drosophila Genome Project (BDGP; http://rana.lbl.gov/cis-analyst/cgi/viewer.php; (Berman et al., 2001) website as well as the seqseek program developed by Eric Johnson, (University of Oregon; http://flycompute.uoregon.edu/cgi-bin/indiv_gene.pl; (Freeman et al., 2003) to search genomic sequences around the ind gene for Gsh1 like binding sites. In the cis-analyst program we entered up to 10 of the sites identified in (Valerius et al., 1995). In the seekseq program we use the consensus sequence gcymattaa and degeneracy model of gcnnattan to identify potential sites. Both programs identified multiple potential sites within approximately the same upstream region. Whether they identified the same sites in unclear, as only the seqseek program gave a detailed output.
4.5 Site directed mutagenesis
Putative Ind binding sites in the GN4 enhancer element we mutagenized using the Quick Change Multi-site directed mutagenesis kit (Stratagene). The following mutagenic primers were used to convert the core ATTA sequence to GGGG or CCCC.
Gsh site 1:tc aaa atg caa atg tca caa cca ccc cat gtc aat aaa tga tta acc caa tc
Gsh site 2:gaa ttt ttt ttg aaa cac ccc ttg ccc cgg caa atc cgg ttt gat gtt ctc
Gsh site 3: gtt ttt ttt tgc aca gcc cgc cac ccc agc aga agc caa ata ctt aaa aac
Gsh site 4:cta cgt aga agt cgg tcg aca ccc cac aat taa cgt cag caa ttg tga tcg g
Acknowledgments
We would like to thank Drs Chris Q. Doe, Keith Chapes and Rob Denell for critically reading the manuscript. Eric Spana and Jamie Roebuck at the Non-Mammalian model systems unit at Duke for all the help with injections. This publication was made possible by Grant Number P20 RR016475 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH."
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. PNAS. 2001;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Dev. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Buescher M, Hing FS, Chia W. Formation of Neuroblasts in the embryonic central nervuos system of Drosophila melanogaster is controlled by SoxNeuro. Development. 2002;129:4193–4203. doi: 10.1242/dev.129.18.4193. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer-Verlag; Berlin Heidelberg: 1985. p. 227. [Google Scholar]
- Cowden J, Levine M. Ventral dominance governs sequential patterns of gene expression across the dorsal-ventral axis of the neurectoderm in the Drosophila embryo. Dev Bio. 2003;262:335–349. doi: 10.1016/s0012-1606(03)00395-6. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: GCM target genes regulating Glial Development, Diversification and Function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Greenspan RJ. Fly Pushing, The theory and practice of Drosophila genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 1997. p. 155. [Google Scholar]
- Jiang J, Hoey T, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes Dev. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopolous A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2364–2387. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Mc Donald JA, Holbrook S, Isshiki T, Weiss J, Doe CQ, Mellerick DM. Dorso-ventral patterning in the Drosophila CNS: the vnd homeobox gene specifies ventral column identity. Genes Dev. 1998;12:3606–3612. doi: 10.1101/gad.12.22.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerick DM, Nirenberg M. Dorsal-Ventral patterning genes restrict NK-2 homeobox gene expression to the ventral half of the central nervous system of Drosophila embryos. Developmental Biology. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- Saunders HMH, Koizumi K, Odenwald W, Nirenberg M. Neuroblast pattern formation: regulatory DNA that confers the vnd/NK2 homobox gene pattern on a reporter gene in transgenic lines of Drosophila. PNAS. 1998;95:8316–8321. doi: 10.1073/pnas.95.14.8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Koizumi K, Nosworthy N, Tan DP, Odenwald W, Nirenberg M. Regulatory DNA required for vnd/NK-2 homeobox gene expression pattern in neuroblasts. Proc Natl Acad Sci U S A. 2002;99:113–7. doi: 10.1073/pnas.012584599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopolous A, Levine M. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev Bio. 2005;280:482–493. doi: 10.1016/j.ydbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Stathopolous A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of Dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeiffle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Uhler J, Garbern J, Yang L, Kamholz J, Mellerick DM. Nk6, a novel Drosophila homeobox gene regulated by vnd. Mech Dev. 2002;116:105–116. doi: 10.1016/s0925-4773(02)00155-7. [DOI] [PubMed] [Google Scholar]
- Valerius MT, Li H, Stock JL, Weinstein M, Kaur S, Singh G, Potter SS. Gsh-1: A novel murine homeobox gene expressed in the central nervous system. Dev Dyn. 1995;203:337–351. doi: 10.1002/aja.1002030306. [DOI] [PubMed] [Google Scholar]
- Von Ohlen T, Doe CQ. Convergence of Dorsal, Dpp and Egfr signaling pathways subdivides the Drosophila neuroectoderm into three dorsal-ventral columns. Dev Bio. 2000;224:362–372. doi: 10.1006/dbio.2000.9789. [DOI] [PubMed] [Google Scholar]
- Wang LH, Chmelik R, Tang D, Nirenberg M. Identification and analysis of vnd/NK-2 homeodomain binding sites in genomic DNA. PNAS. 2005;102:7097–7102. doi: 10.1073/pnas.0502261102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JB, Von Ohlen T, Mellerick D, Dressler G, Doe CQ, Scott MP. Dorsoventral patterning in the Drosophila Central Nervous System: The intermediate neuroblasts defective Homeobox gene specifies intermediate column identity. Genes Dev. 1998;12:3591–3602. doi: 10.1101/gad.12.22.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Syu LJ, Mellerick DM. Contextual interaction determine whether the Drosophila homeodomain protein, Vnd acts as a repressor or activator. Nucleic Acids Research. 2005;33:1–12. doi: 10.1093/nar/gki140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Skeath JB. The Sox-domain containing gene Diachaete/fish-hook acts in concert with vnd and ind to regulate cell fate in the Drosophila neuroectoderm. Development. 2002;129:1165–1174. doi: 10.1242/dev.129.5.1165. [DOI] [PubMed] [Google Scholar]


