Abstract
The heart is the first organ to form and function during vertebrate development and is absolutely essential for life. The left ventricle is derived from the classical primary or first heart field (FHF), while the right ventricle and outflow tract are derived from a distinct second heart field (SHF). The recent discovery of the SHF has raised several fundamental and important questions about how the two heart fields are integrated into a single organ and whether unique molecular programs control the development of the two heart fields. This review briefly highlights the contributions of the SHF to the developing and mature heart and then focuses primarily on our current understanding of the transcriptional pathways that function in the development of the SHF and its derivatives in transgenic and knockout mice.
Keywords: second heart field, anterior heart field, Isl1, MEF2C, transcription factor
The vertebrate heart forms from progenitor cells found in two bilaterally symmetrical regions of anterior lateral mesoderm, and as early heart development proceeds, the two lateral halves of this population are brought together at the midline to form a linear tube, which begins to exhibit slow peristaltic contractions [1,2]. Subsequently, the linear heart tube undergoes rightward looping and is remodeled into ventricular and atrial chambers with inflow and outflow regions and valves to control the flow of blood. In birds and mammals, the four-chambered heart is separated into two ventricles and two atria by septal divisions that keep oxygenated blood from mixing with deoxygenated blood [3].
Minor errors in cardiac development can result in profound congenital heart defects, which are the most common form of genetic birth defects and are the leading cause of infant mortality in developed countries [1,2,4]. Among the most common and serious congenital heart defects are ventricular septal defects (VSDs), valve defects, and outflow tract alignment and septation abnormalities such as persistent truncus arteriosus (PTA), overriding aorta, and pulmonary artery stenosis [2,4]. In spite of the prevalence and severity of congenital heart defects, relatively little is known about the genetic and molecular basis of these abnormalities, although some progress has been made in recent years [4,5].
The use of model systems has proven invaluable for determining the morphogenetic events, the genes, and the molecules that control normal cardiac development and has provided insight into how disruptions in normal development lead to congenital heart disease [3,5,6]. Work in the invertebrate Drosophila melanogaster has defined many of the essential regulators of cardiac specification and differentiation and has shown that the cardiac regulatory network has been remarkably conserved over the hundreds of millions of years since humans and flies last had a common ancestor [3,6]. These studies, combined with many studies in vertebrate systems, have identified a core group of transcription factors that are required for cardiac development, including members of the NK class of homeodomain proteins, GATA zinc-finger transcription factors, the MADS domain transcription factor MEF2, T-box and Forkhead transcription factors, and the Hand class of basic helix-loop-helix (bHLH) factors, as well as many other essential transcriptional regulators of cardiac development [3,6].
The complexities of vertebrate genetics and development have made the task of defining the transcriptional regulatory network controlling heart development a difficult one. It has been hypothesized that vertebrates have had two rounds of genome duplication since our last common ancestor with invertebrates, such as Drosophila and Ciona [5,7]. As a result, the majority of transcription factor families associated with cardiac development are represented by multiple members with overlapping temporal and spatial patterns of expression in the heart. In addition, morphological complexity has been added to the heart throughout vertebrate evolution, and distinct patterns of gene expression within the heart are apparent. Even the Drosophila heart is complex with distinct patterns of gene expression in different cells within each hemisegment along the dorsal vessel [8–10]. The evolution of a closed circulatory system and the presence of distinct types of chamber myocardium in the atrium and the ventricle and the presence of distinct inflow and outflow tracts have made the vertebrate heart even more intricate. This additional complexity is especially apparent in amniotes. Birds and mammals each have a four-chambered heart, and each chamber has developed a specialized function, which is revealed by distinct patterns of gene expression that mark regional differences within the heart [3].
Recently, a second heart field was discovered to give rise to the right ventricle and outflow tract in birds and mammals [11–14]. This field, referred to as the anterior heart field, the secondary heart field, or the second heart field (SHF), has provided a potential explanation for the observations that many genes and transgenes are expressed in the right ventricle and outflow tract, but not the left ventricle or atria, and that there are several mutations in mice and diseases in humans that selectively impact the right ventricle [2,15]. This has led to an exciting period of research with many new studies addressing molecular, genetic, and embryological aspects of this novel field. Indeed, many excellent reviews have been written about the SHF over the last few years, particularly with regard to the embryological origins and contributions of this field to the heart and outflow tracts [11,15–18]. In this review, I provide an overview of some of the key transcription factors involved in the development of the SHF and its derivatives. Since this issue is devoted to model systems for the study of cardiovascular development, I will focus primarily on the mouse as a model system to define the transcriptional networks involved in SHF development and gene expression. I will refer to the two heart fields as the first and second heart fields, using the nomenclature put forth by Buckingham, Meilhac, and Zaffran [11].
1. A second mesodermal lineage contributes to the mammalian and avian hearts
The notion that the arterial pole and the outflow tract might arise from an extra-cardiac population of cells in the pharyngeal mesoderm dates back to the 1970s. Cell labeling experiments performed in living chicken embryos demonstrated that portions of the outflow tract seemed to be added to the heart after its initial formation and that the source of these cells appeared to be from a position anterior to the heart and not from the bilateral regions of cardiogenic anterior lateral mesoderm [19]. More recent studies, performed independently by three groups, have provided convincing evidence that the arterial pole of the heart, most notably the outflow tract and portions of the right ventricle arise from a second heart field, which resides outside of the traditionally-defined primary or first heart field (FHF) in the cardiac crescent [12–14].
Using quail-chick chimeras and fluorescent labeling experiments in chicken embryos, Kirby and coworkers demonstrated that outflow tract myocardium is derived from splanchnic mesoderm underlying the caudal pharynx [14]. This extra-cardiac population of mesodermal progenitors was termed the secondary heart field as a distinction from the bilateral primary heart fields that contribute to the linear heart tube and inflow pole of the heart [14]. In a similar set of dye-labeling experiments, Mjaatvedt and colleagues concluded that the outflow tract of the chicken heart is derived from a novel heart-forming field that surrounds the aortic sac anterior to the linear heart tube [13]. These conclusions were further supported by ablation studies and labeling with lacZ-expressing adenoviruses, which also indicated that the outflow tract conus region of the heart was derived from an extra-cardiac field termed the anterior heart field [13]. Neither of these original studies performed in chick embryos indicated a significant contribution of this novel heart forming population to the right ventricle [13,14].
A fortuitous enhancer trap upstream of the Fgf10 locus in mouse resulted in a transgenic line that expressed lacZ in the pharyngeal mesoderm, outflow tract, and right ventricle [12]. By examining the expression directed by this transgene under the control of Fgf10 regulatory sequences, combined with fluorescent dye labeling experiments, Buckingham and coworkers identified that the outflow tract and most or all of the right ventricle in mice are derived from an extra-cardiac anterior heart field [12,20]. At mouse embryonic day (E) 7.5, which represents an early stage of cardiac development, the Fgf10-lacZ enhancer trap directed expression to a population of cells that was distinct from myocardial progenitors in the FHF of the cardiac crescent, as defined by cardiac alpha-actin expression [12] (Fig. 1). During ventral morphogenesis and foregut invagination in the mouse, these mesodermal progenitors, which reside medial and caudal to the cardiac crescent are displaced anterior and dorsal to the linear heart tube in the splanchnic and pharyngeal mesoderm [11,12,15]. These cells are then added progressively to the heart at the time of cardiac looping, when the heart dramatically elongates, to form the right ventricle and outflow tract [11,15].
Fig. 1.
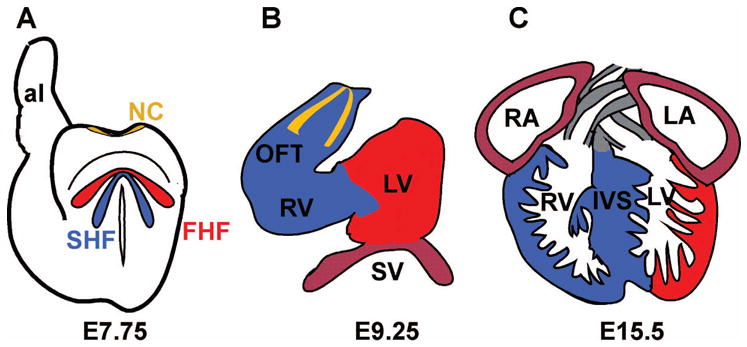
A schematic representation of three stages of heart development in the mouse. (A) A representation of a mouse embryo at embryonic day (E) 7.75 showing the location of three major populations of cells that contribute to the developing heart and outflow tract. The first heart field (FHF) is shown in red; the second heart field (SHF), which resides dorsal and medial to the FHF, is depicted in blue; and cardiac neural crest (NC) progenitors are shown at the dorsal neural tube in yellow. (B) A schematic of a mouse heart undergoing looping morphogenesis at E9 to E9.5. The FHF (red) contributes to the left ventricle. The SHF (blue) contributes primarily to the right ventricle (RV) and outflow tract (OFT). The sinus venosus (SV) or future atrial chamber is probably composed of FHF and SHF descendants. Two streams of plexinA2-positive cardiac neural crest cells entering the outflow tract are depicted in yellow. (C) A representation of a nearly mature embryonic mouse heart at E15.5. The free wall and majority of the left ventricle (LV) chamber is composed almost exclusively of cells derived from the FHF (red). The RV and interventricular septum (IVS) are composed primarily of cells descended from the SHF. The atria may be derived from a mixture of cells of FHF and SHF origins and the OFT is composed of cells of both NC and SHF origin. al, allantois; LA, left atrium; RA, right atrium.
While some minor differences were observed in the source of the second mesodermal progenitor population, in general the studies clearly agreed that the cells of the second field were located dorsal and anterior to the FHF and contributed to the heart from the splanchnic and pharyngeal mesoderm [11]. The studies also differed somewhat in their conclusions about the contribution of the SHF to the developing heart. In general, the two studies in chick concluded that the contribution of the SHF was to the outflow tract, whereas the mouse work suggested that the second lineage contributed more broadly to the heart, including the outflow tract and much or all of the right ventricle [11–14]. These different conclusions may represent differences in the experimental approaches used or may represent bona fide differences in the contribution of the second lineage to the hearts of birds compared to mammals [11]. Alternatively, the secondary/anterior heart fields described in the chick may represent a subset of a broader field that makes a more substantial contribution to the heart, as the mouse studies suggested [11,21].
The original Fgf10 enhancer trap showed lacZ expression in the outflow tract and right ventricle with a sharp boundary at the sulcus marking the future interventricular septum with no transgene expression in the left ventricle [12]. This study suggested, at least superficially, that the boundaries of SHF contribution to the mouse heart are tightly restricted to the right ventricle and outflow tract [12,15]. However, these transgene expression studies could not discriminate between the fate of cells in the SHF and de novo expression of the transgene in the heart. In addition, this type of expression study cannot rule out the possibility that SHF cells make a more extensive contribution to the heart but that the regulatory elements in the promoter and enhancer are down regulated in SHF descendants. Therefore, a variety of fate mapping and cell labeling approaches have been employed to try to define the boundaries of SHF in mice. Zaffran et al. used a combination of fluorescent labeling of embryonic tissues ex vivo and in vivo restrospective labeling to confirm by different approaches that right ventricular myocardium is derived from the SHF [20].
The retrospective labeling approach is a powerful tool to mark the fate of individual cells and their descendants in vivo [22]. The approach relies on a version of a lacZ reporter transgene, referred to as nlaacZ, which contains a stop codon preventing expression of a function β-galactosidase enzyme [22]. Rare intramolecular recombination events in nlaacZ result in restoration of a functional β-galactosidase [22,23]. Statistical analyses suggest that these events are only likely to occur once or less in any given embryo and thus provide a record of all cells descended from the cell where the single recombination event occurred [23]. These analyses demonstrated that the FHF and SHF originate from a common progenitor population that segregates prior to the cardiac crescent stage, probably just after gastrulation [11,23]. These studies also supported the notion that the right ventricle and outflow tract are derived from the SHF, although the more recent studies suggested a greater contribution of the second lineage than earlier studies [11,23]. The retrospective studies support a model in which the outflow tract is derived exclusively from the SHF and the left ventricle is derived from the FHF, whereas the atria, right ventricle, and inflow region represent a mixture of the two populations [11,23].
Other studies by our group used a genetic approach to address the contribution of the SHF to the heart [21]. A transgenic mouse line in which Cre recombinase is expressed under the control of a SHF-restricted element from the mef2c gene were crossed to the Rosa26R lacZ reporter strain, which allows a fate map of all of the cells expressing the mef2c-Cre transgene and all of the descendants of those cells [21,24]. These studies, demonstrated that the SHF, as marked by the mef2c-Cre transgene, contributed widely to the outflow tract and right ventricle, and also demonstrated that the entire interventricular septum appears to be derived from the SHF [21] (Fig. 2). These studies also showed that the endocardium in the right ventricle and outflow tract and the outflow cushions are derived from the SHF [21] (Fig. 2). Other studies by Franco and coworkers showed that the interventricular septum is derived from both right and left ventricular myocytes, suggesting that the interventricular septum originates from SHF cells that cross into the left ventricle and then interdigitate with SHF-derived cells in the right ventricle to form the septum [25].
Fig. 2.
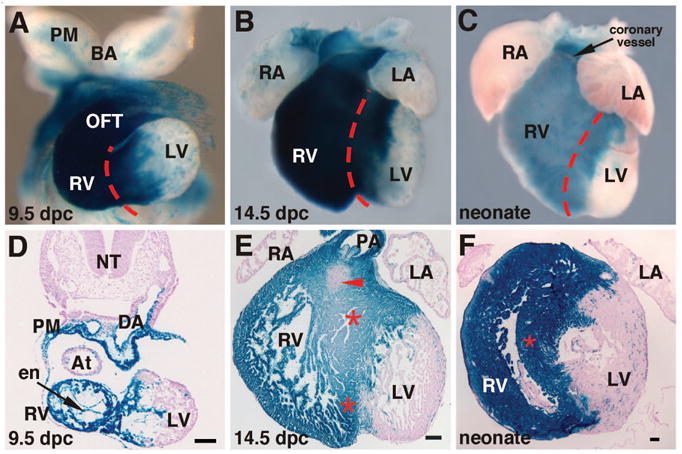
Mef2c-AHF-Cre marks the SHF and its descendants. Transgenic mice harboring the mef2c-AHF-Cre transgene were crossed to ROSA26R lacZ reporter mice and progeny were collected at 9.5 days post coitus (dpc) (A, D), 14.5 dpc (B, E) or as neonates immediately after birth (C, F) and stained with X-gal as a measure of lacZ expression. Whole mount hearts are shown in (A–C). Transverse sections are shown in (D, F). A frontal section is shown in (E). These results support the idea that the right ventricle (RV), outflow tract (OFT), and interventricular septum (red asterisks and red dashed lines) are derived from the SHF. The red arrowhead in (E) marks the AV canal, which is unmarked by mef2c-AHF-Cre-expressing cells and their descendants. BA, branchial arches; At, atrium; DA, dorsal aorta; LA, left atrium; LV, left ventricle; NT, neural tube; PA, pulmonary artery; PM, pharyngeal mesoderm; RA, right atrium. Portions of this figure were derived with permission from Verzi et al., 2005 [21].
Fate mapping studies performed using a mouse line in which Cre recombinase is expressed under the control of the murine Isl1 locus have suggested that the SHF makes much more extensive contributions to the heart than other fate mapping and labeling studies [26]. Isl1-Cre itself is not expressed in the heart, but when crossed to the Rosa26R Cre-dependent reporter strain of mice [24], Evans and coworkers found that Isl1+ progenitor cells contributed broadly to the heart, including the outflow tract, the majority of the right ventricle, and portions of the left ventricle, atria, and the inflow pole [26]. These data also suggested that the FHF contributes exclusively only to part of the left ventricle and that both fields contribute cells to the atria and the right ventricle [26], which is supported by the conclusions of the retrospective analyses as well [23]. The broader contribution of Isl1-Cre-expressing descendants to the heart than from cells marked by other methods or genes, such as mef2c-AHF-Cre-expressing cells, may indicate that Isl1 is expressed in the mesoderm prior to split of first and second heart fields [21,26]. Alternatively, mef2c and other genes, like Fgf10, may mark only a subset of the Isl1 domain and represent a separate subdomain within the SHF defined by Isl1 expression [21,26]. In any case, it is clear that the SHF makes extensive contributions to the outflow tract, right ventricle, and ventricular septum in the mouse and that significant portions of the left ventricle are not SHF-derived [11,16].
2. Transcription factors required for SHF development
The LIM-homeodomain protein Islet-1 (ISL1) is required for the development of the SHF and its derivatives [26]. The Isl1 gene is broadly expressed during development, with expression evident in the allantois, throughout the lateral and pharyngeal mesoderm in the early post-gastrula embryo, as well as in the pharyngeal endoderm and ventral neural tube [21,26–32]. ISL1 was identified as an important regulator of the Insulin1 promoter by binding to the consensus sequence element, YTAATGR [33]. Inactivation of Isl1 in mice results in defects in pancreatic development and motor neuron specification [27,31]. In addition, Evans and coworkers found that Isl1 null mice display profound defects in cardiac development [26]. The hearts of Isl1 null mice fail to undergo looping morphogenesis and appear to be missing the majority of the anterior region of the heart. Remnants of the common atrium and the left ventricle are present in Isl1 null mice, but the right ventricle and outflow tract appear to be missing [26]. This anatomical interpretation of the Isl1 null phenotype is supported by marker analyses in which left ventricular markers, such as Hand1 and Tbx5, were expressed in Isl1 knockout mice, but markers of the outflow tract and right ventricle, including Fgf10, were absent [26]. Importantly, expression analysis of Isl1 itself indicated that it is not expressed in the outflow tract and right ventricle but rather is expressed in SHF progenitor cells in the pharyngeal mesoderm [26].
Several other mouse knockouts show similar cardiac phenotypes to that of Isl1 null mice (Table 1). Inactivation of mef2c in mice results in defective cardiac looping and lethality by E10 [34]. Mice lacking MEF2C have only a single ventricular chamber and gross abnormalities of the inflow and outflow tracts, suggesting a SHF phenotype [11,34,35]. More recent studies have shown that cells expressing a transgenic marker of the second heart lineage and its derivatives are present in the single ventricular chamber in mef2c knockout mice, but the expressing cells are abnormally distributed within the ventricle [21]. Together, these data support an essential role for MEF2C in the transcriptional program for SHF development and in the normal addition and distribution of SHF progenitors to the heart [3,21,35]. These studies do not exclude a role for MEF2C in the FHF as well. Indeed, mef2c is expressed weakly in FHF and its derivatives, and the inflow tract phenotype observed in mef2c knockout mice may be due to a requirement for this transcription factor in either the first or second heart fields or both [21,34,36]. Indeed, recent studies have suggested a requirement for MEF2C function in the normal allocation of cells from the FHF to the sinoatrial region and the left ventricle [36].
Table 1.
Some of the transcription factors required for the development of the second heart field (SHF) and its derivatives. OFT, outflow tract; RV, right ventricle.
| Gene | Expression in the heart | Mutant phenotype | References |
|---|---|---|---|
| Isl1 | Primarily expressed in SHF progenitors. Weak expression in the distal OFT. No detectable expression in RV, LV or atria | Embryonic lethal at E10. Failed looping. No RV or OFT is formed. Other defects in neuronal and pancreatic development. | [26,27,31] |
| Mef2c | Expressed early in SHF progenitors. Expressed throughout the heart in myocardium and endocardium. Strongest expression at E9 to E10.5 in OFT and RV. After E9.5, mef2c is expressed broadly throughout the embryo. | Embryonic lethal by E10. Failed looping. Single ventricular chamber and defective OFT. Minor inflow defects. Profound vascular defects. | [34,36, 72,73] |
| Foxh1 | Expressed broadly throughout the embryo, including SHF progenitors at crescent stage and in pharyngeal mesoderm at E9. | Embryonic lethal by E10. Failed looping. Lack of a distinct RV. Normal inflow and atrial regions. | [37,74] |
| Foxc1/c2 | Both genes are expressed broadly in mesodermal and neural crest derivatives. Both genes are expressed in SHF progenitors at E8 and later in splanchnic and pharyngeal mesoderm. Subsequent expression in many areas of the heart. | Embryonic lethality by E10.5. Double mutant mice have severe defects in RV formation. Also have defects in coronary vessel and epicardial development and abnormal inflow tract formation. | [38,39] |
| Hand2 | Expressed in both FHF and SHF progenitors at cardiac crescent stage. Later expression in the heart becomes largely restricted to the RV and OFT. | Embryonic lethal by E10.5. Defective looping with a single ventricular chamber with left-sided identity. | [41,42] |
| Smyd1 (Bop) | Expressed in precardiac mesoderm at E7.75 and throughout the heart from the linear heart tube stage onward. Minimal expression in the pharyngeal mesoderm. | Embryonic lethal. Single ventricular chamber. Defective looping. Apparently normal atrial development. | [43] |
The forkhead transcription factor FoxH1 is also required for proper formation of the right ventricle and outflow tract [37]. Foxh1 null mice have normal inflow and atrial regions, but have defective development of anterior cardiac structures [37]. Like mef2c null mice, Foxh1 mutants form a linear heart tube and a single ventricle, but fail to undergo looping morphogenesis, and a distinct right ventricle fails to form [37]. Numerous early cardiac markers showed disrupted expression in Foxh1 mutants, including mef2c, Hand2, Fgf8, and Tbx5 [37]. Notably, Isl1 expression in the splanchnic and pharyngeal mesoderm was not affected by loss of FoxH1 expression, suggesting that ISL1 functions upstream or in parallel to FoxH1 in SHF development [37].
Mice lacking both FoxC1 and FoxC2 function have severe defects in right ventricle and outflow tract formation but also exhibit other defects, including abnormal epicardial development [38,39]. Interestingly, Foxc1/Foxc2 double mutant mice have disrupted expression of Tbx1, Fgf10 and Fgf8 suggesting a key role in the hierarchy controlling outflow tract development from SHF mesoderm [39].
The bHLH protein Hand2 is another transcription factor required for proper development of the outflow tract and right ventricle, although the cardiac phenotype of Hand2 null mice is somewhat less severe than the Isl1 null phenotype [40]. The hearts of Hand2 null mice fail to undergo rightward looping and lack a morphologically distinct right ventricle, and the outflow tract appears to make direct connection to a single left ventricle-like chamber [40]. Hand2 expression is restricted within the developing mouse heart at E9 to the right ventricle and outflow tract so the loss-of-function phenotype is consistent with its expression pattern [40–42]. Mutation of the Bop (Smyd1) gene, which encodes a SET domain-containing transcriptional repressor, results in mice with a normal common atrium, but a defective single ventricular chamber with little or no evidence of rightward looping. The common ventricular chamber in Bop null mice was interpreted as having a left ventricular phenotype, which is consistent with an observed loss of Hand2 expression in Bop mutants [43]. While it appears that BOP function is required for several aspects of cardiac growth in all parts of the heart, at least part of the phenotype appears to affect selectively the derivatives of the SHF, possibly through loss of Hand2 expression [43].
Numerous other transcription factors are also required for SHF development as part of a more general requirement for cardiogenesis or with more mild phenotypes than a nearly complete loss of the outflow tract and right ventricle as seen in Isl1, Foxh1, mef2c, and Hand2 knockout mice [1,3,11,16]. For example, inactivation of the gene encoding the T-box transcription factor Tbx1 in mice results in multiple defects in neural crest-derived tissues as well as in defective septation of the outflow tract and ventricles [44–47].
Recently, it was shown that Tbx1 function is required in the SHF for outflow tract septation [48]. The relative mildness of the SHF phenotype in Tbx1 knockout mice suggests that Tbx1 may function downstream in the hierarchy for the development of SHF-derived structures.
All of the described genetic studies, as well as numerous other knockout studies in mice, demonstrate that a number of transcription factors are required for the development of the SHF and its derivatives. A remaining challenge, if we are to understand the specification and differentiation of myocytes and the morphogenesis of the heart, is to define the connections among these transcription factors by placing them into pathways with the goal of defining the complete transcriptional network for SHF development.
3. An ISL1-dependent transcriptional network in the SHF
The very early expression of Isl1 in the anterior lateral mesoderm and subsequently in the pharyngeal mesoderm combined with the requirement for ISL1 for right ventricle and outflow tract formation established this transcription factor as a critical early regulator of SHF development [26]. Furthermore, the observation that Isl1 is not expressed in the outflow tract and right ventricle demonstrates its requirement in the transcriptional program controlling SHF development is early in the progenitor cell population rather than in differentiated cardiac myocytes themselves.
Recent studies have identified two other essential cardiac transcription factors as direct transcriptional targets of ISL1 in the SHF, establishing the initial basis for a transcriptional network controlling development of this field into the right ventricle, outflow tract, and other regions of the heart (Fig. 3). As described in Section 2, mef2c is required for outflow tract and right ventricle development in mice [34]. Several recent studies have shown that mef2c is regulated by multiple, independent modular enhancers that each govern a subset of the endogenous mef2c expression pattern [35,49–51]. One of the enhancers from the mef2c gene directs expression exclusively to the SHF and its derivatives in the right ventricle and outflow tract [35]. The function of this enhancer depends on two evolutionarily conserved, perfect consensus ISL1 binding sites. Interestingly, the activity of this enhancer requires the ISL1 sites for activity in the right ventricle and outflow tract even though ISL1 itself is not expressed in those regions [35]. This will be discussed again in Section 4 (below).
Fig. 3.
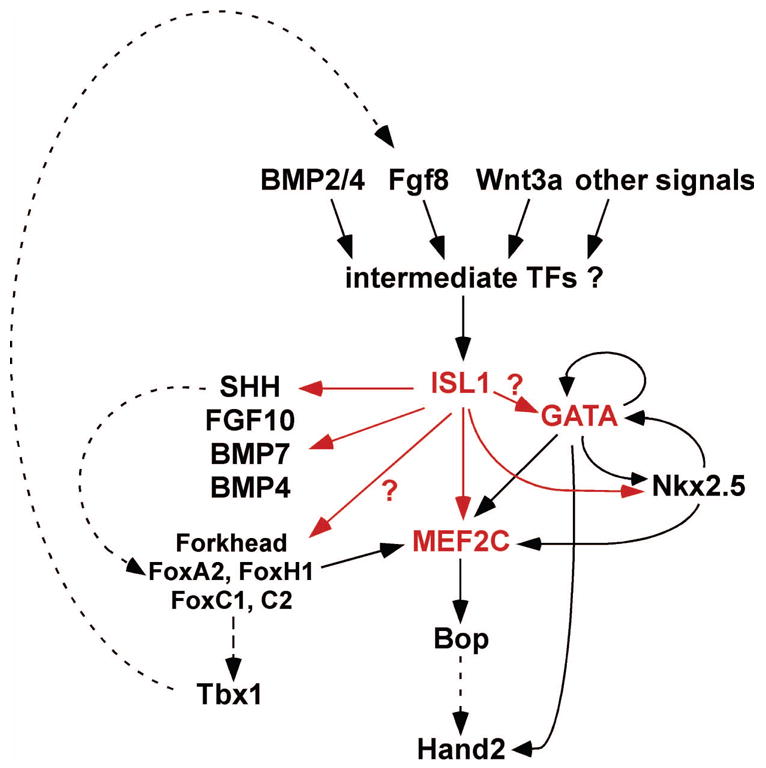
An ISL1-dependent transcriptional network for SHF development. In this model, the LIM-homeodomain transcription factor ISL1 functions as a key early regulator of SHF development by activating MEF2C and GATA transcription factors, which function as part of a core network for right ventricle and outflow tract development.
The ISL1-dependent mef2c SHF enhancer is also dependent on two conserved GATA transcription factor binding sites, suggesting that ISL1 and GATA factors may cooperate to activate a transcriptional program in the SHF [35]. Alternatively, these two classes of transcription factors may function sequentially with ISL1 serving as an initial activator of mef2c and other genes followed by subsequent activation by GATA factors. This type of model has been proposed previously for GATA transcription factors in the endoderm, where Forkhead transcription factors serve as initial activators and GATA transcription factors reinforce expression initiated by the activity of the Forkhead "pioneer factor" [52,53].
The notion of a combinatorial relationship between ISL1 and GATA transcription factors has been suggested previously for the regulation of the insulinotropic polypeptide gene in neuroendocrine cells [54]. The possibility that these factors may regulate an entire transcriptional program in the SHF is supported by our studies on mef2c regulation and also by our studies on Nkx2-5 regulation [35,55]. Like mef2c, Nkx2-5 is regulated by multiple, modular transcriptional enhancers [56]. One of the most well characterized enhancers from the Nkx2-5 gene directs expression to the pharyngeal mesoderm, outflow tract, and right ventricle during development, and this enhancer has been shown to be a direct transcriptional target of GATA factors [57,58]. At the time that this enhancer was originally identified, the SHF had not yet been described, and ISL1 was not known to be a critical regulator of heart development. However, subsequent studies have shown that this SHF enhancer from Nkx2-5 is also a direct transcriptional target of ISL1 via a conserved ISL1 binding site adjacent to the previously identified, essential GATA binding site [55]. Taken together, the analyses of the mef2c and Nkx2-5 enhancers support a central role for a transcriptional program involving ISL1 and GATA transcription factors in the SHF and suggest a possible coordinated program for gene activation in that lineage (Fig. 3).
Hand2 has also been shown to be a direct target of GATA factors in the right ventricle and outflow tract via two essential, conserved consensus GATA sites in a proximal upstream enhancer [59]. Because of the highly restricted pattern of expression directed by the Hand2 enhancer to the SHF and its derivatives, we hypothesized that this element might also be a target of ISL1 via the same ISL1-GATA dependent mechanism as the Nkx2-5 and mef2c SHF enhancers described above. However, a search of the Hand2 enhancer described in those earlier studies and other evolutionarily conserved regions in the Hand2 locus failed to reveal any conserved, consensus ISL1 sites (A. Heidt and BLB, unpublished observations). The authors of the Hand2 enhancer study proposed a model in which Hand2 may exhibit chamber-restricted expression in the right ventricle through negative regulation in the left ventricle [59], which seems plausible. Alternatively, there may be additional positive regulatory elements in the enhancer, other than ISL1 binding sites, that cooperate with GATA factors to mediate chamber-restricted expression. It has been shown previously that the transcriptional repressor Bop is required for Hand2 expression in the developing right ventricle and outflow tract, suggesting a role for Bop in the transcriptional network for SHF development [43]. Bop has been proposed as part of the transcriptional cascade for SHF development as a regulator of Hand2 [3,43,60]. Indeed, Bop has been shown to be a direct transcriptional target of MEF2 via a conserved enhancer element, which does support the possibility that Bop may function downstream of ISL1 and MEF2C in the SHF transcriptional network [60].
While it seems clear that the ISL1-GATA-MEF2C pathway is a central component to the transcriptional network in the SHF, additional pathways involving Forkhead factors also functions in the second heart lineage to regulate Tbx1 expression [61]. Importantly, the Forkhead-dependent pathways, which appear to be partially independent of the ISL1 pathway, also intersect and reinforce the ISL1-MEF2C pathway [37,62] (Fig. 3). These reinforcing intersections of distinct transcription pathways may provide robustness to myocardial differentiation in cells of SHF origin by providing multiple routes for activating and maintaining expressing of core cardiac transcription factors [3,6].
Recently, Srivastava and colleagues identified an enhancer from the Tbx1 gene that is sufficient to direct expression to the SHF [61]. This enhancer is activated and bound in vitro by FoxA2, FoxC1, and FoxC2 via a consensus Forkhead-binding site in the enhancer, suggesting that Tbx1 is dependent on the activity of one or more of these Forkhead proteins for activation in the SHF [61] (Fig. 3). In an earlier study, the same group identified an enhancer from Fgf8 that required Tbx1 binding for function in the pharyngeal mesoderm [62], and recent studies from Anne Moon and coworkers showed that loss of Fgf8 function in the SHF resulted in a reduction in Isl1 expression in the pharyngeal mesoderm and outflow tract [63]. These studies suggest an early role for Fgf8 in the hierarchy for SHF development, possibly even upstream of Isl1 [26,63]. Taken together with the observation that Tbx1, which is expressed later than either Fgf8 or Isl1, is required for subsequent Fgf8 expression, these observations suggest a reinforcing role for the Forkhead-Tbx1 pathway by maintenance or amplification of Fgf8 expression. In addition, these studies suggest an intersection of the Forkhead-Tbx1 pathway with the ISL1-GATA-MEF2C pathway (Fig. 3).
As noted above, mef2c is essential for development of SHF derivatives and is also a key direct target of ISL1 [35] (Fig. 3). In addition, mef2c appears to be a direct target of the Forkhead transcription factor FoxH1 [37]. Wrana and colleagues showed that mef2c expression in SHF derivatives is dependent on FoxH1 and identified an enhancer element from mef2c that directs expression to the SHF[37]. This enhancer is a distinct element from the ISL1-dependent enhancer described above, and the activity of this enhancer is not specific to the SHF [37]. However, this additional enhancer element appears to require FoxH1 and Nkx2-5 for activity in SHF derivatives in vivo [37]. These observations suggest that MEF2C functions as a crucial nodal point for development of the second heart lineage and its derivatives and that multiple enhancers from the mef2c gene function to integrate inputs from, which also may serve to reinforce the transcriptional program for myocyte development through activation of MEF2C (Fig. 3).
An excellent example of reinforcing transcriptional circuits in the SHF and its derivatives is found between GATA factors and Nkx2-5. As noted earlier in this review, a SHF-restricted enhancer from Nkx2-5 is dependent on GATA transcription factors [57,58]. Importantly, an analysis of a Gata6 enhancer by Molkentin and colleagues showed that the activity of the enhancer is restricted to the pharyngeal mesoderm, outflow tract, and right ventricle at E9.5 in mouse development [64]. This element contains a consensus Nkx binding site, which is bound by Nkx2-5 in vitro and is required for in vitro activation of the Gata6 enhancer by Nkx2-5 [64]. Taken together, these studies suggest a reinforcing circuit with GATA factors functioning both upstream and downstream of Nkx2-5 in the SHF [3,6,64]. As noted earlier in this review, multiple intersection points on key downstream transcription factors and multiple reinforcing circuits within the transcriptional network for SHF development are likely to provide robustness to the developmental program. This notion is supported by numerous studies that demonstrate positive feedback and feed-forward transcriptional circuits in the heart and other systems [3,65].
4. Future Challenges
It has become clear over the last decade or so that the programs controlling cardiac development contain an elaborate network of reinforcing transcriptional pathways where a core group of transcription factors activate each other's expression [3,6]. This provides a degree of robustness to the cardiogenic program and may help explain why no single factor has been identified as a "master regulator" for cardiac myogenesis. The complex regulatory relationships among the core cardiac transcription factors, such as GATA proteins, Nkx2-5, MEF2C, Forkhead, T-box, and Hand factors, as well as numerous other regulators, has also made unraveling the earliest transcriptional circuits in the heart challenging [1,3].
The recent discovery of the SHF provides an embryological basis for many of the chamber restricted patterns of expression observed for many genes and transgenes and suggests that at least two distinct transcriptional hierarchies for cardiac development almost certainly exist [3,11,15,16]. Within the SHF, ISL1 is clearly an early regulator and may even function as the earliest transcriptional regulator of the lineage. Thus, it will be essential to determine how Isl1 is activated in the SHF and whether its expression is regulated by multiple enhancers. If transcriptional enhancers from Isl1 are identified, it will be important to determine whether other cardiac transcription factors activate and maintain Isl1 expression, as is the case for other core cardiac transcription factors, or whether Isl1 may be a target of intercellular signaling molecules, such as Fgf8 and others [63]. If Isl1 is indeed activated by intercellular signals and not simply by other cardiac transcription factors, this might indicate that ISL1 serves as a type of master regulator for the SHF and may also explain why Isl1 expression is not maintained in SHF derivatives in the outflow tract and right ventricle.
Although Isl1 itself is not expressed in the right ventricle and outflow tract, its function is required there for the activation of SHF enhancers from both the Nkx2-5 and mef2c genes [26,35,55]. These observations present something of a paradox since ISL1 is required for enhancer function in tissues where ISL1 itself is not expressed, suggesting that ISL1 may leave a stable mark on these enhancers. How this may be accomplished is not known, but a model in which ISL1 remodels chromatin to a more relaxed configuration allowing for subsequent activation and expression mediated by other transcription factors seems plausible. It will be important in the future to determine how ISL1 activates gene expression and whether this LIM-homeodomain transcription factor can assemble chromatin remodeling complexes in SHF progenitor cells.
Another important challenge for the future will be to define the role of Nkx2-5 in SHF development. The phenotype of Nkx2-5 null mice has been interpreted as a loss of FHF derivatives, while a rudimentary outflow tract and right ventricle are retained in these animals [66,67]. However, even if the primary genetic requirement for Nkx2-5 is in the FHF, it still functions in SHF development. As discussed in Section 3, Nkx2-5 is implicated in activation of both Gata6 and mef2c in the SHF, and the Nkx2-5 gene itself has at least one a discrete modular enhancer for the SHF [37,56–58,64]. Thus, it will be interesting to determine if Nkx2-5 functions in an early specification role in the SHF, as it probably does in the FHF, or whether it simply represents part of the core cardiac transcription factor hierarchy and it is required for amplification and maintenance of the cardiac program in the SHF.
Nkx2-5, like other cardiac transcription factor genes, contains multiple, independent modular transcriptional enhancers that each direct a subset of the endogenous expression pattern [56]. Why some cardiac genes, such as mef2c and Nkx2-5, have multiple transcriptional enhancers that direct overlapping temporal and spatial patterns of activity is not known. One possibility is that distinct enhancer elements respond to unique upstream cues, which would allow multiple pathways to converge on core transcription factors, such as MEF2C and Nkx2-5, via different elements [3]. This type of regulatory strategy would be important if core factors for cardiac gene expression, like MEF2C, Nkx2-5, and GATA4, were activated by multiple signaling pathways in the SHF. There is evidence for this for Nkx2-5 regulation where distinct enhancers respond to different cues, including BMPs and probably others [66,68,69]. Alternatively, these genes may have separate enhancer elements that control initial activation versus maintenance of expression. This type of regulatory arrangement might function on mef2c, for example. Although there is no evidence for two-step activation and maintenance via different mef2c enhancers at this time, it is plausible that mef2c could be activated by ISL1 via one enhancer and maintained by Nkx2-5 and FoxH1 via a separate element [35,37]. In the future, it will be essential to understand this regulatory complexity mediated by multiple enhancers if we are to decipher the elaborate networks governing SHF development.
Finally, recent work by Chien and Evans and colleagues has identified the presence of Isl1+ cells in the adult myocardium [70]. While these cells are only present very sparsely throughout the rodent myocardium, they retain the ability to differentiate into fully functional cardiomyocytes ex vivo [70,71]. These progenitor cells appear to originate in the SHF, as their Isl1 expression would suggest, but rather than differentiating into myocytes during development, these cells retain Isl1 expression and exist as a resident progenitor population in the adult heart [70,71]. These studies raise the exciting and intriguing possibility that the heart may have the capacity for self-repair by reactivation and expansion of these residual SHF progenitor cells retained in the adult myocardium [71]. Likewise, it is very attractive to speculate that these Isl1+ SHF progenitors derived from either embryos or adults may be suitable for myocardial regeneration and repair of ischemic injuries as an intervention in adult cardiology and as a potential source of multipotent cardiac progenitors for repair of congenital heart defects.
Acknowledgments
I am grateful for helpful comments from members of my laboratory and other colleagues. Work in the Black lab is supported by grants HL64658 and AR52130 from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–48. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–7. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci. 2006;63:1141–8. doi: 10.1007/s00018-005-5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava D. Genetic assembly of the heart: implications for congenital heart disease. Annu Rev Physiol. 2001;63:451–69. doi: 10.1146/annurev.physiol.63.1.451. [DOI] [PubMed] [Google Scholar]
- 6.Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- 7.Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev Biol. 2007;18 doi: 10.1016/j.semcdb.2006.12.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo PC, Frasch M. Establishing A-P polarity in the embryonic heart tube: a conserved function of Hox genes in Drosophila and vertebrates? Trends Cardiovasc Med. 2003;13:182–7. doi: 10.1016/s1050-1738(03)00074-4. [DOI] [PubMed] [Google Scholar]
- 9.Lovato TL, Nguyen TP, Molina MR, Cripps RM. The Hox gene abdominal-A specifies heart cell fate in the Drosophila dorsal vessel. Development. 2002;129:5019–27. doi: 10.1242/dev.129.21.5019. [DOI] [PubMed] [Google Scholar]
- 10.Zaffran S, Reim I, Qian L, Lo PC, Bodmer R, Frasch M. Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development. 2006;133:4073–83. doi: 10.1242/dev.02586. [DOI] [PubMed] [Google Scholar]
- 11.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–35. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 12.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–40. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 13.Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 14.Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–88. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 15.Kelly RG, Buckingham ME. The anterior heart-forming field: voyage to the arterial pole of the heart. Trends Genet. 2002;18:210–6. doi: 10.1016/s0168-9525(02)02642-2. [DOI] [PubMed] [Google Scholar]
- 16.Kelly RG. Molecular inroads into the anterior heart field. Trends Cardiovasc Med. 2005;15:51–6. doi: 10.1016/j.tcm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Abu-Issa R, Waldo K, Kirby ML. Heart fields: one, two or more? Dev Biol. 2004;272:281–5. doi: 10.1016/j.ydbio.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Yutzey KE, Kirby ML. Wherefore heart thou? Embryonic origins of cardiogenic mesoderm. Dev Dyn. 2002;223:307–20. doi: 10.1002/dvdy.10068. [DOI] [PubMed] [Google Scholar]
- 19.de la Cruz MV, Sanchez Gomez C, Arteaga MM, Arguello C. Experimental study of the development of the truncus and the conus in the chick embryo. J Anat. 1977;123:661–86. [PMC free article] [PubMed] [Google Scholar]
- 20.Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA. Right ventricular myocardium derives from the anterior heart field. Circ Res. 2004;95:261–8. doi: 10.1161/01.RES.0000136815.73623.BE. [DOI] [PubMed] [Google Scholar]
- 21.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–45. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 22.Meilhac SM, Kelly RG, Rocancourt D, Eloy-Trinquet S, Nicolas JF, Buckingham ME. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development. 2003;130:3877–89. doi: 10.1242/dev.00580. [DOI] [PubMed] [Google Scholar]
- 23.Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6:685–98. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 24.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 25.Franco D, Meilhac SM, Christoffels VM, Kispert A, Buckingham M, Kelly RG. Left and right ventricular contributions to the formation of the interventricular septum in the mouse heart. Dev Biol. 2006;294:366–75. doi: 10.1016/j.ydbio.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 26.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–60. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- 28.Dong J, Asa SL, Drucker DJ. Islet cell and extrapancreatic expression of the LIM domain homeobox gene isl-1. Mol Endocrinol. 1991;5:1633–41. doi: 10.1210/mend-5-11-1633. [DOI] [PubMed] [Google Scholar]
- 29.Mitsiadis TA, Angeli I, James C, Lendahl U, Sharpe PT. Role of Islet1 in the patterning of murine dentition. Development. 2003;130:4451–60. doi: 10.1242/dev.00631. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa Y, O'Leary DD. Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci. 2001;21:2711–25. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 1996;84:309–20. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 32.Radde-Gallwitz K, Pan L, Gan L, Lin X, Segil N, Chen P. Expression of Islet1 marks the sensory and neuronal lineages in the mammalian inner ear. J Comp Neurol. 2004;477:412–21. doi: 10.1002/cne.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;344:879–82. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- 34.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–7. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 36.Vong L, Bi W, O'Connor-Halligan KE, Li C, Cserjesi P, Schwarz JJ. MEF2C is required for the normal allocation of cells between the ventricular and sinoatrial precursors of the primary heart field. Dev Dyn. 2006;235:1809–21. doi: 10.1002/dvdy.20828. [DOI] [PubMed] [Google Scholar]
- 37.von Both I, Silvestri C, Erdemir T, Lickert H, Walls JR, Henkelman RM, Rossant J, Harvey RP, Attisano L, Wrana JL. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7:331–45. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Seo S, Kume T. Forkhead transcription factors, Foxc1 and Foxc2, are required for the morphogenesis of the cardiac outflow tract. Dev Biol. 2006;296:421–36. doi: 10.1016/j.ydbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Kume T, Jiang H, Topczewska JM, Hogan BL. The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 2001;15:2470–82. doi: 10.1101/gad.907301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–60. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 41.Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–9. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 42.Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev Biol. 1998;196:228–36. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- 43.Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. doi: 10.1038/ng866. [DOI] [PubMed] [Google Scholar]
- 44.Stennard FA, Harvey RP. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development. 2005;132:4897–910. doi: 10.1242/dev.02099. [DOI] [PubMed] [Google Scholar]
- 45.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–91. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 46.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 47.Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–29. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 48.Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–27. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 49.De Val S, Anderson JP, Heidt AB, Khiem D, Xu SM, Black BL. Mef2c is activated directly by Ets transcription factors through an evolutionarily conserved endothelial cell-specific enhancer. Dev Biol. 2004;275:424–34. doi: 10.1016/j.ydbio.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 50.Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120:1021–1032. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- 51.Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128:4623–33. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- 52.Zaret K. Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev Biol. 1999;209:1–10. doi: 10.1006/dbio.1999.9228. [DOI] [PubMed] [Google Scholar]
- 53.Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–17. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- 54.Jepeal LI, Boylan MO, Wolfe MM. Cell-specific expression of the glucose-dependent insulinotropic polypeptide gene functions through a GATA and an ISL-1 motif in a mouse neuroendocrine tumor cell line. Regul Pept. 2003;113:139–47. doi: 10.1016/s0167-0115(03)00046-6. [DOI] [PubMed] [Google Scholar]
- 55.Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, Hui CC, Henkelman RM, Nemer M, Black BL, Nagy A, Bruneau BG. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–74. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz RJ, Olson EN. Building the heart piece by piece: modularity of cis-elements regulating Nkx2-5 transcription. Development. 1999;126:4187–92. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- 57.Searcy RD, Vincent EB, Liberatore CM, Yutzey KE. A GATA-dependent nkx-2.5 regulatory element activates early cardiac gene expression in transgenic mice. Development. 1998;125:4461–70. doi: 10.1242/dev.125.22.4461. [DOI] [PubMed] [Google Scholar]
- 58.Lien CL, Wu C, Mercer B, Webb R, Richardson JA, Olson EN. Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development. 1999;126:75–84. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- 59.McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–41. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- 60.Phan D, Rasmussen TL, Nakagawa O, McAnally J, Gottlieb PD, Tucker PW, Richardson JA, Bassel-Duby R, Olson EN. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development. 2005;132:2669–78. doi: 10.1242/dev.01849. [DOI] [PubMed] [Google Scholar]
- 61.Maeda J, Yamagishi H, McAnally J, Yamagishi C, Srivastava D. Tbx1 is regulated by forkhead proteins in the secondary heart field. Dev Dyn. 2006;235:701–10. doi: 10.1002/dvdy.20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development. 2004;131:5491–502. doi: 10.1242/dev.01399. [DOI] [PubMed] [Google Scholar]
- 63.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–33. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molkentin JD, Antos C, Mercer B, Taigen T, Miano JM, Olson EN. Direct activation of a GATA6 cardiac enhancer by Nkx2.5: evidence for a reinforcing regulatory network of Nkx2.5 and GATA transcription factors in the developing heart. Dev Biol. 2000;217:301–9. doi: 10.1006/dbio.1999.9544. [DOI] [PubMed] [Google Scholar]
- 65.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–95. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 66.Prall OWJ, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. A Nkx2-5/Bmp2/Smad1 negative feedback loop orchestrates cardiac progenitor cell specification and proliferation in the second heart field. Cell. 2007 doi: 10.1016/j.cell.2007.01.042. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–66. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 68.Lien CL, McAnally J, Richardson JA, Olson EN. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev Biol. 2002;244:257–66. doi: 10.1006/dbio.2002.0603. [DOI] [PubMed] [Google Scholar]
- 69.Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev Biol. 2002;244:243–56. doi: 10.1006/dbio.2002.0604. [DOI] [PubMed] [Google Scholar]
- 70.Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–53. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent Embryonic Isl1(+) Progenitor Cells Lead to Cardiac, Smooth Muscle, and Endothelial Cell Diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 72.Bi W, Drake CJ, Schwarz JJ. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol. 1999;211:255–67. doi: 10.1006/dbio.1999.9307. [DOI] [PubMed] [Google Scholar]
- 73.Lin Q, Lu J, Yanagisawa H, Webb R, Lyons GE, Richardson JA, Olson EN. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–74. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- 74.Weisberg E, Winnier GE, Chen X, Farnsworth CL, Hogan BL, Whitman M. A mouse homologue of FAST-1 transduces TGF beta superfamily signals and is expressed during early embryogenesis. Mech Dev. 1998;79:17–27. doi: 10.1016/s0925-4773(98)00160-9. [DOI] [PubMed] [Google Scholar]


