Abstract
Accurate chromosome segregation during cell division requires not only the establishment, but also the precise, regulated release of chromosome cohesion. Chromosome dynamics during meiosis are more complicated, because homologues separate at anaphase I whereas sister chromatids remain attached until anaphase II. How the selective release of chromosome cohesion is regulated during meiosis remains unclear. We show that the aurora-B kinase AIR-2 regulates the selective release of chromosome cohesion during Caenorhabditis elegans meiosis. AIR-2 localizes to subchromosomal regions corresponding to last points of contact between homologues in metaphase I and between sister chromatids in metaphase II. Depletion of AIR-2 by RNA interference (RNAi) prevents chromosome separation at both anaphases, with concomitant prevention of meiotic cohesin REC-8 release from meiotic chromosomes. We show that AIR-2 phosphorylates REC-8 at a major amino acid in vitro. Interestingly, depletion of two PP1 phosphatases, CeGLC-7α and CeGLC-7β, abolishes the restricted localization pattern of AIR-2. In Ceglc-7α/β(RNAi) embryos, AIR-2 is detected on the entire bivalent. Concurrently, chromosomal REC-8 is dramatically reduced and sister chromatids are separated precociously at anaphase I in Ceglc-7α/β(RNAi) embryos. We propose that AIR-2 promotes the release of chromosome cohesion via phosphorylation of REC-8 at specific chromosomal locations and that CeGLC-7α/β, directly or indirectly, antagonize AIR-2 activity.
Keywords: AIR-2; meiosis; C. elegans; cohesion; aurora-B
Introduction
Sister chromatid separation in mitosis is precisely controlled to ensure the equal segregation of genetic material to both daughter cells. The mechanism by which sister chromatids remain joined after DNA replication and then separate at the onset of anaphase has recently been elucidated. It has been shown that sister chromatids are held together by a proteinacious scaffold whose destruction at anaphase triggers chromosome separation (Uhlmann et al., 2000; Waizenegger et al., 2000). In budding yeast, the degradation of one cohesin subunit, Scc1p, by the protease separase is both necessary and sufficient to trigger chromosome separation (Uhlmann et al., 1999, 2000). In yeast, degradation of Scc1p is regulated via its phosphorylation by Cdc5p/Polo kinase and by a pathway leading to the activation of separase at the onset of anaphase (Uhlmann et al., 1999, 2000; Alexandru et al., 2001). The pathway leading to the activation of separase is composed of the anaphase-promoting complex (APC)* and securin (Ciosk et al., 1998; Zou et al., 1999), and appears to be evolutionarily conserved (for review see Nasmyth, 2001). In human cells, SCC1 cleavage by separase is necessary for anaphase (Hauf et al., 2001).
The APC–securin–separase pathway appears to also regulate chromosome separation in meiosis (Buonomo et al., 2000; Golden et al., 2000; Salah and Nasmyth, 2000; Siomos et al., 2001). In Saccharomyces cerevisiae, activation of separase at anaphase I and II results in the degradation of the meiotic-specific cohesin molecule, Rec8p (Buonomo et al., 2000). In addition, Caenorhabditis elegans separase mutants are defective in meiotic chromosome separation (Siomos et al., 2001). However, despite the conservation of the APC–securin–separase regulatory pathway, chromosome separation in meiosis is quite different from that in mitosis. During meiosis, two rounds of chromosome segregation follow one round of DNA replication, generating four haploid gametes from one diploid germ cell. In the first meiotic division, homologous chromosomes, but not sister chromatids, are separated. Sister chromatids remain held together until the onset of anaphase II. Several unique chromosomal behaviors during meiosis ensure the faithful segregation of homologues at anaphase I. First, recombination between homologues generates chiasmata that hold homologues together, promoting their proper alignment at metaphase I. Second, the kinetochores of sister chromatids function as a single unit during meiosis I, which facilitates their segregation to the same pole. Third, a mechanism exists in meiosis that selectively destroys the cohesion complex holding homologues together, but preserves the cohesion between sister chromatids at anaphase I.
How this selective loss of chromosome cohesion is accomplished during meiosis I remains unclear. It has been shown in yeast and worms that Rec8p/REC-8 is localized to meiotic chromosomes in both divisions and is required for chromosome cohesion in both divisions (Buonomo et al., 2000; Pasierbek et al., 2001). Therefore, the selective loss of chromosome cohesion in anaphase I could be achieved by differential degradation of the REC-8 molecules that are responsible for homologue cohesion. This differential degradation could be achieved by several mechanisms, including selectively localizing active separase to specific sites, selectively marking a subset of REC-8 linking homologous chromosomes for degradation, or selectively protecting a subset of cohesin from being degraded during anaphase I. It has been shown both in vivo and in vitro that the phosphorylated form of the mitotic cohesin, Scc1p, is degraded more efficiently than the nonphosphorylated form (Uhlmann et al., 2000). In fact, Cdc5p/Polo has been demonstrated to be the kinase regulating Scc1p phosphorylation, leading to its destruction during mitosis in S. cerevisiae (Alexandru et al., 2001). In S. pombe, the meiotic cohesin Rec8p is also phosphorylated during both meiotic divisions (Parisi et al., 1999; Watanabe and Nurse, 1999), although the same preference of separase for phosphorylated Rec8p has not been demonstrated and the kinase responsible for Rec8p phosphorylation has not been identified. An intriguing possibility is that the selective release of chromosome cohesion during meiosis I involves the differential phosphorylation, thereby differential degradation, of cohesin molecules distal to chiasmata.
One class of protein kinases that have been implicated in chromosome segregation during mitosis and meiosis are the aurora-B kinases (for reviews see Bischoff and Plowman, 1999; Adams et al., 2001a). The role of the aurora-B kinases in chromosome segregation has been illuminated by loss-of-function studies in various organisms and from their interesting cell cycle–regulated subcellular localization patterns (Francisco et al., 1994; Bischoff et al., 1998; Schumacher et al., 1998; Shindo et al., 1998; Tatsuka et al., 1998; Hsu et al., 2000). They belong to a group of proteins termed chromosomal passengers, as these proteins translocate from the chromosome to the spindle and cell cortex during anaphase and are likely to coordinate chromosome separation with cytokinesis (Adams et al., 2001a). Human aurora-B is amplified in a variety of cancers and its overexpression in cell cultures results in polyploidy (Tatsuka et al., 1998). One of the known substrates for the aurora-B kinases is histone H3 (Hsu et al., 2000). Phosphorylation of histone H3 at serine 10 has long been correlated with chromosome condensation and segregation during mitosis in a variety of organisms (Bradbury et al., 1973; Gurley et al., 1974; Hendzel et al., 1997; Wei et al., 1998). Removal of the aurora-B–like kinase from yeast, worms, and flies (Hsu et al., 2000; Adams et al., 2001b; Giet and Glover, 2001) results in severe mitotic chromosome segregation defects. These results strongly suggest that mitotic defects in aurora-B mutants are likely a result of inappropriate chromosome condensation. Depletion of the aurora-B kinase in yeast and worms also prevents proper meiosis, although the cause of the meiotic defect remains unclear (Hsu et al., 2000). In both yeast and worms, the PP1 phosphatases Glc7p and CeGLC-7 have been shown to antagonize aurora-B in vivo (Hsu et al., 2000).
Here we report that the C. elegans aurora-B kinase, AIR-2, is required for chromosome separation during meiosis. In air-2(RNAi) embryos, no separation of homologues or sister chromatids was observed. During meiosis, AIR-2 localizes specifically to chromosome arms distal to chiasmata in metaphase I and to the point of contact between sister chromatids in metaphase II. The meiotic cohesin REC-8 remains detectable in air-2(RNAi) embryos throughout meiosis. In addition, AIR-2 phosphorylates REC-8 at a specific residue in vitro. Furthermore, we show that removing the CeGLC-7α/β phosphatases by RNA interference (RNAi) results in ectopic chromosomal localization of AIR-2 both temporally and spatially. In Ceglc-7α/β(RNAi) embryos, REC-8 was barely detected on meiotic chromosomes and sister chromatids separated precociously at the onset of anaphase I. Together, our results strongly support the model that in C. elegans meiosis, AIR-2 regulates the release of chromosome cohesion via location-specific phosphorylation of the cohesin REC-8, and CeGLC-7α/β, directly or indirectly, oppose AIR-2 activity in cohesion release.
Results
Chromosome condensation appears normal in AIR-2–depleted oocytes during C. elegans meiosis
The C. elegans gonad is a tube-like syncytium consisting of thousands of germ nuclei at various developmental stages (Schedl, 1997). Nuclei at the most distal end, relative to the spermatheca, proliferate mitotically. Moving proximally, germ nuclei enter meiosis, progress through different stages of meiotic prophase I, and cellularize to become oocytes (Fig. 1 A). Full-grown oocytes are in diakinesis of prophase I and will be referred to by their positions in the gonad relative to the spermatheca, with the most proximal oocyte being −1. In the presence of sperm, −1 oocytes sequentially undergo maturation, enter the spermatheca, and become fertilized (McCarter et al., 1999). After fertilization, the oocyte-derived nucleus completes two rounds of meiotic division, each marked by the extrusion of a polar body, to become a haploid pronucleus (Fig. 1, A and B).
Figure 1.
Comparison of meiotic chromosomes in wild-type and air-2(RNAi) animals. (A) Schematic representation of one arm of the C. elegans gonad, including newly fertilized embryos. Nuclei are represented by open circles. The DNA complement in oocyte-derived nuclei is indicated by the number inside each nucleus. Ooc, oocyte-derived nucleus; pb, polar body; sp, sperm pronucleus; spm, spermatheca. (B–E) Projected fluorescence images of histone H2B–GFP in living embryos (B and C) or DAPI staining of fixed proximal gonads (D and E) derived from wild-type (B and D) or air-2(RNAi) (C and E) animals. The arrow in B points to extruded polar bodies, whereas in C, it points to the site where polar bodies should have been extruded. Arrowheads in D and E indicate single bivalents in proximal oocytes. Bars, 5 μm.
We have shown previously that in air-2(RNAi) animals, oocyte meiosis does not complete normally (Hsu et al., 2000). In air-2(RNAi) embryos, no polar bodies were extruded and the oocyte-derived nucleus retains a 4N DNA content (Fig. 1 C) as it fuses with the sperm pronucleus. The meiotic failure of air-2(RNAi) embryos was not likely due to a defect in chromosome condensation, in contrast to the mitotic failure, which was attributed primarily to the disruption of chromosome condensation. Six bivalent chromosomes in the oocytes of air-2(RNAi) animals appeared as short rods that were morphologically indistinguishable from those in wild-type oocytes (Fig. 1, D and E; Speliotes et al., 2000). This suggests that AIR-2 plays a role in meiotic chromosome segregation via a mechanism distinct from chromosome condensation.
AIR-2 is required for homologous chromosome separation during meiosis I
To better understand the defect in meiotic divisions observed in air-2(RNAi) embryos, we performed four-dimensional (4-D) imaging of meiosis in live embryos using a reporter strain carrying a histone H2B–green fluorescent protein transgene (GFP; see Materials and methods). In the wild-type embryo immediately after fertilization, the six bivalents start to line up with the long axis of each bivalent parallel to the surface of the embryo (Fig. 2, A and F; Herman et al., 1979; Albertson and Thomson, 1993). Each bivalent consists of two homologues arranged axially, or end to end, with the inner ends being the point of contact between homologues and outer ends leading toward the poles. Later, the chromosomes rotate 90° such that the long axis of each bivalent is now perpendicular to the surface of the embryo (Fig. 2, B and G). 2 min later, anaphase begins and the homologous chromosomes separate into two groups (Fig. 2, C and H). The group of chromosomes closer to the surface of the embryo is extruded as a polar body (Fig. 2, D and I). In meiosis II, the sister chromatids arrange in a similar end-to-end configuration. After the completion of meiosis I, six univalent chromosomes appear unaligned for 5 min before the whole process repeats to separate the end-to-end–joined sister chromatids (Fig. 2, E and J).
Figure 2.

Representative live H2B–GFP images of different meiotic stages from wild-type, air-2(RNAi), and Ceglc-7 α/β(RNAi) embryos. (A–E) Schematic drawing of meiotic chromosomes in the oocyte-derived nucleus in a wild-type embryo. Each bivalent is represented as two short rods joined end to end with the outer ends leading toward poles. Homologues from one bivalent are indicated by different shades of gray in A–C. After the extrusion of a polar body (pb), sister chromatids are shown as small short rods in D and E. (F–J) Wild-type; (K–O) air-2(RNAi); (P–T) Ceglc-7α/β(RNAi) meiotic embryos. (A, F, and K) Initial alignment, parallel to the cell cortex. (B and G) After a 90° rotation, the chromosomes are perpendicular to the edge of the embryo. (C and H) Anaphase I; (D and I) metaphase II; (E and J) anaphase II. Embryos in K and P, L and Q, M and R, N and S, and O are at the same stage as those in F, G, H, I, and J, respectively. All images presented here are representative slices from each time point except for that in T, which is a projection of the image stack of DAPI staining from a fixed embryo. The edge of the embryo is indicated by a black curve in A–E and a white curve in F–T. Bar, 2.5 μm.
In air-2(RNAi) embryos, the bivalents appeared morphologically normal immediately after fertilization (Fig. 2 K). However, the initial alignment on the metaphase plate was slightly disorganized, no rotation was observed, and the long axis of each bivalent remained parallel to the surface of the embryo (Fig. 2, K and L). 3 min after the initial alignment, a time point equivalent to the onset of anaphase I in wild-type embryos, all the chromosomes moved toward and were compressed against the edge of the embryo (Fig. 2 M). No separation of chromosomes was observed (n = 50). 5 min later, all the chromosomes moved back to their initial positions, appeared clearly as six bivalents, and repeated the entire process (Fig. 2, N and O). These observations suggest that AIR-2 is necessary for the separation of homologues in meiosis I and sister chromatids in meiosis II.
AIR-2 and BIR-1 localize to subchromosomal foci corresponding to the point of contact between chromosomes in metaphase I and II
The air-2(RNAi) phenotype suggests an important function for AIR-2 in chromosome separation in meiosis. A clue as to how AIR-2 might function in chromosome separation came from a detailed examination of the subchromosomal localization of AIR-2 during both meiotic divisions. The bulk of AIR-2 staining has been shown to remain cytoplasmic in oocytes until the −1 oocyte undergoes maturation (Schumacher et al., 1998). Upon oocyte maturation, AIR-2 becomes chromatin associated and remains so throughout the completion of meiosis, except during anaphase when AIR-2 temporarily relocates to the microtubule midzone (Schumacher et al., 1998).
We now show that chromatin-associated AIR-2 localizes to defined subchromosomal foci in both meiotic divisions (Fig. 3). These foci correspond to the equatorial axes of the bivalent (or the inner ends of homologues) in metaphase I (Fig. 3, D–F) and the inner ends of the end-to-end–joined sister chromatids in metaphase II (Fig. 3, J–L). At anaphase I, when homologues separate, AIR-2 is detected only in the microtubule midzone between homologues (Fig. 3, G–I). The survivin-like protein, BIR-1, has a temporal and spatial staining pattern similar to AIR-2 and has been shown previously to be required for the localization of AIR-2 to chromosomes (Speliotes et al., 2000). bir-1(RNAi) embryos are indistinguishable from air-2(RNAi) embryos (unpublished data; Speliotes et al., 2000). We show that BIR-1 also localizes to the inner ends between homologues in metaphase I and between sister chromatids in metaphase II (Fig. 3, M–P), identical to the pattern observed with AIR-2. These foci represent the last points of contact between separating chromosomes, either homologues or sister chromatids. These results suggest the intriguing possibility that AIR-2 phosphorylation of a substrate(s) at these foci triggers the release of cohesion selectively at these locations.
Figure 3.
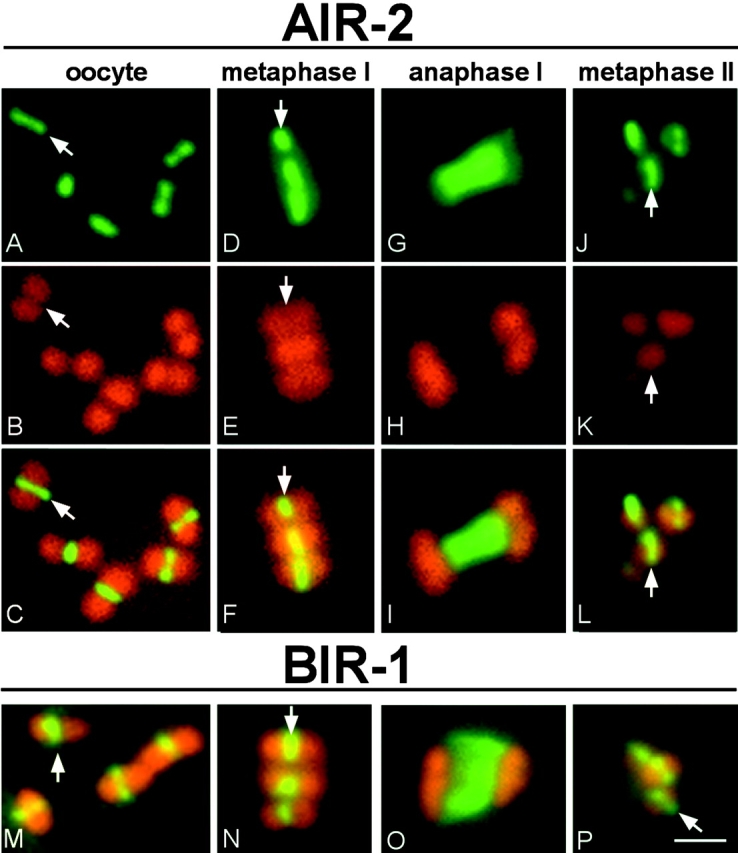
AIR-2 and BIR-1 localize to subchromosomal foci on meiotic chromosomes. Antibody staining is presented in green and DAPI staining in red. (A–L) Representative slice images of AIR-2 (A, D, G, and J), DAPI (B, E, H, and K), or merged (C, F, I, and L) staining in oocyte-derived nuclei at different meiotic stages. (M–P) Only merged images are shown for BIR-1 at corresponding stages. Arrows in A–F and M–N point to the staining between homologues in meiosis I, and in J–L and P, they point to the staining between sister chromatids in meiosis II. Bar, 2 μm.
The subchromosomal localization of AIR-2 and BIR-1 is dependent on chiasmata formation
Because there is on average one crossover per homologue pair in C. elegans meiosis (Brenner, 1974; Barnes et al., 1995), bivalents initially appear cross shaped, with the center of each cross being the chiasma, and become rod shaped as chromosomes condense further. Because centromeres are located at the ends of meiotic chromosomes in C. elegans, the equatorial axes of the rod-shaped bivalent chromosomes correspond to the chromosome arms distal to the chiasmata and the poleward (or long) axis proximal in vertebrate and yeast chromosomes (Fig. 4, A and B; Albertson and Thomson, 1993).
Figure 4.

Subchromosomal localization of AIR-2 and BIR-1 is dependent on chiasma formation. (A and B) Schematic drawing of the bivalent in vertebrates (A) and C. elegans (B). Sister chromatids are shown in the same color, whereas homologues are represented in different colors. The direction of pulling force is indicated by arrows. Green and orange bars represent cohesin molecules connecting chromosome arms distal and proximal, respectively, to chiasmata. (C–F) Merged and projected images of chromosomes from the −1 oocyte of him-5(e1490) (C and E) and her-1(e1518) XO (D and F) animals stained with either AIR-2 (C and D) or BIR-1 (E and F) antibodies. Arrows point to univalents, and arrowheads point to representative bivalents. Bar, 2 μm.
As the subchromosomal foci we observed after AIR-2 and BIR-1 staining correspond to the chromosome arms distal to the chiasmata, we therefore asked whether these foci are dependent on chiasma formation. Mutations in him-5 and him-8 cause a high frequency of nondisjoined X chromosomes, resulting in five bivalents and two nondisjoined univalents, which can be clearly observed in diakinetic oocytes (Hodgkin et al., 1979; Broverman and Meneely, 1994). In him-5(e1490) or him-8(e1489) mutants, we observed very intense staining of AIR-2 and BIR-1 on all five bivalents, but no detectable staining on either of the univalents (Fig. 4, C and E; n = 42 for AIR-2; n = 11 for BIR-1).
To demonstrate that the intense subchromosomal foci are dependent on chiasma formation instead of a direct requirement for the gene him-5 or him-8, we examined the staining pattern for these proteins in her-1(e1518) XO animals. In C. elegans, sex is determined by the ratio of X chromosomes to autosomes (Madl and Herman, 1979). Wild-type hermaphrodites have two X chromosomes, whereas males have only one X chromosome. Mutations in the sex determination gene her-1 result in XO animals developing into relatively normal, fertile hermaphrodites (Hodgkin, 1980). The single X chromosome appears as a nonpaired univalent in oocytes of her-1 XO animals. No staining of AIR-2 or BIR-1 was observed on the univalents in oocytes of her-1(e1518) XO animals, whereas the staining on bivalents appeared wild type (Fig. 4, D and F; n = 14 for AIR-2; n = 4 for BIR-1). This result demonstrates that the observed subchromosomal foci containing AIR-2 and BIR-1 are dependent on chiasma formation and not on the function of specific genes required for meiotic crossover.
REC-8 remains detectable on both axes of bivalents in air-2(RNAi) embryos
A potential candidate for AIR-2 phosphorylation at the chromosomal arms distal to chiasmata is the meiotic cohesin REC-8. The REC-8 protein was observed to localize to both poleward and equatorial axes of bivalents in metaphase I. At anaphase I, only the REC-8 on the poleward axis remained detectable, whereas the REC-8 on the equatorial axis was absent (Pasierbek et al., 2001). Comparison to the mitotic cohesin Scc1p in budding yeast, which is degraded more efficiently as a phosphoprotein, suggests that selective removal of C. elegans REC-8 at anaphase I could be regulated via differential phosphorylation by AIR-2.
If REC-8 is one of the in vivo substrates for AIR-2 and phosphorylation of REC-8 plays an important role for its degradation, one would predict that REC-8 protein would remain localized along both axes of the bivalents in air-2(RNAi) embryos. We stained meiotic chromosomes of wild-type and air-2(RNAi) embryos with antibody to REC-8 (Pasierbek et al., 2001). In wild-type embryos whose meiotic chromosomes appeared to be in metaphase I, 74% (n = 36) had detectable REC-8 clearly on both axes of bivalents (Fig. 5 A). The other 26% had detectable REC-8 only on the poleward, but not the equatorial, axis of bivalents (Fig. 5 B). We interpret that the 26% embryos lacking equatorial REC-8 staining have initiated anaphase I and the removal of REC-8 distal to the chiasmata, but have not separated their homologues. Bivalents in air-2(RNAi) embryos do not align normally at metaphase. Because no polar bodies were extruded in air-2(RNAi) embryos, we could not distinguish the first from the second meiotic attempt. However, in embryos where bivalents were clearly recognized, 100% had detectable REC-8 on both axes (n = 22, Fig. 5 D). This suggests that REC-8 is not degraded in air-2(RNAi) embryos and that its degradation requires AIR-2 activity.
Figure 5.

REC-8 localization is abnormal in air-2(RNAi) and Ceglc-7 α/β(RNAi) embryos. Representative meiotic chromosomes stained with either REC-8 (A–E) antibody or DAPI (F). (A–C) Wild-type embryos. In all embryos examined whose homologues appeared connected, 74% had cross-shaped REC-8 staining as shown in A, whereas 26% had no detectable equatorial staining similar to that shown in B. A representative REC-8 staining in anaphase is shown in C. (D) 100% of air-2(RNAi) embryos with recognizable bivalents had cross-shaped REC-8 staining. (E) 100% of Ceglc-7α/β(RNAi) embryos had very low or no detectable REC-8 staining. The corresponding DAPI staining is shown in F. The number of embryos imaged and scored is indicated above each panel. Arrows point to the poleward axes, and arrowheads point to the equatorial axes. Bar, 2 μm.
AIR-2 phosphorylates the meiotic cohesin REC-8 in vitro
Retention of REC-8 on meiotic chromosomes in air-2(RNAi) embryos suggests that AIR-2 activity is required for the release of chromosomal REC-8 protein. It is possible that AIR-2 phosphorylates REC-8 directly to promote its release. We showed that, in vitro, recombinant AIR-2 could phosphorylate bacterially expressed REC-8, but showed very low or no activity toward two other C. elegans Scc1p-/Rad21-like molecules, COH-1 and COH-2 (Fig. 6 A; Pasierbek et al., 2001). No phosphorylation of REC-8 was observed when a kinase-dead version of AIR-2 was used in the assay. These results suggest that, in vitro, AIR-2 retains specificity for distinct substrates, such as REC-8.
Figure 6.

AIR-2 phosphorylates REC-8 in vitro. (A) Kinase assays were performed with GST–AIR-2 (AIR-2; lanes 1, 3, and 5) or kinase-dead mutant GST–AIR-2 (AIR-2D; lanes 2, 4, and 6) using GST–REC-8 (lanes 1 and 2), GST–COH-1 (lanes 3 and 4), or GST–COH-2 (lanes 5 and 6) as substrates. Two bands were pulled down with glutathione beads in lanes 1 and 2. The slower migrating band is the full-length REC-8, whereas the faster migrating band is truncated REC-8. AIR-2,* GST–AIR-2 autophosphorylation; AIR-2_P, Ponceau staining of AIR-2 protein; substrate,* phosphorylation of corresponding test substrates; substrate_P, Ponceau staining of each substrate protein. (B) Kinase assays with either wild-type (lanes 1 and 5), T625A (lanes 2 and 6), S626A (lanes 3 and 7), or T625A/S626A (lanes 4 and 8) REC-8 using wild-type AIR-2 (lanes 1–4) or kinase-dead AIR-2 (lanes 5–8). The top half is phosphorimaging and the bottom half is Ponceau staining. Full-length REC-8, AIR-2, and breakdown products are as indicated to the left.
We then mapped the in vitro phosphorylation sites on REC-8 to determine if any specific site(s) was used. We predicted potential phosphorylation sites on REC-8 based on our previous experience with preferred substrate sites for AIR-2 (unpublished data). We mutated the following residues to alanines either individually or in combination: S244, S248, S244/S248, S395, S396, S395/S396, T625, S626, and T625/S626. We then assayed their phosphorylation by AIR-2 in vitro. The T625A and T625A/S626A mutations resulted in a >70% reduction of REC-8 phosphorylation, whereas the other mutations resulted in no significant change in the overall phosphorylation by AIR-2 in vitro (Fig. 6 B; unpublished data). This result indicates that the majority of phosphorylation occurs on threonine 625. Because we still detected a low level of phosphorylation in T625A and T625A/S626A mutant REC-8, we cannot rule out the possibility that AIR-2 phosphorylates additional, nonpreferred sites in our in vitro assay. Although it remains to be further investigated whether T625 is phosphorylated by AIR-2 in vivo and whether phosphorylation of REC-8 at T625 facilitates its cleavage by separase, the sequence surrounding T625 is intriguing. Threonine 625 is two amino acids away from a consensus site, EXXR, for separase cleavage, where X represents any amino acid. In yeast, phosphorylations that facilitate the cleavage of mitotic cohesion Scc1p have been shown to occur at sites a few residues away from the separase cleavage consensus site EXXR (Alexandru et al., 2001). Together with the above results, the in vitro kinase data support our hypothesis that AIR-2 promotes the release of chromosome cohesion by phosphorylating REC-8 at specific subchromosomal foci.
Depletion of CeGLC-7 results in an increase in chromosomal AIR-2 and a decrease in chromosomal REC-8
We have shown previously that two C. elegans PP1 phosphatases, CeGLC-7α and CeGLC-7β, antagonize AIR-2 meiotic activity in vivo (Hsu et al., 2000). We tested whether CeGLC-7 function is required for proper AIR-2 chromosomal localization. In wild-type gonads, AIR-2 is detected temporally in only the most proximal oocyte and spatially at the chromosome arms distal to chiasmata (Fig. 7, A, C, and D; Schumacher et al., 1998). In Ceglc-7α/β(RNAi) animals, there is a striking increase in the number of oocytes containing chromosomal AIR-2 as well as the overall intensity of chromosomal AIR-2 staining. On average, four to five proximal oocytes have detectable chromosomal AIR-2 in Ceglc-7α/β(RNAi) animals (Fig. 7, A and B). The intensity of chromosomal AIR-2 staining is always greatest in the −1 oocyte and decreases in more distal oocytes. In all Ceglc-7α/β(RNAi) gonads imaged, 100% had chromosomal AIR-2 in the −2 oocyte at a level equal to or greater than that in a wild-type −1 oocyte (n = 30). Interestingly, in 67% of individual oocytes imaged, including all −1 oocytes, AIR-2 staining was detected on both equatorial and poleward axes of at least one bivalent (n = 83; Fig. 7, E and F). We conclude from these observations that depletion of CeGLC-7α/β results in an increase in chromosomal AIR-2 both spatially and temporally in the gonad. This increase could reflect an increase in the overall level of AIR-2 in the gonad or a redistribution of cytoplasmic AIR-2 onto chromosomes. At present, we cannot distinguish between these two possibilities. Because of the large cytoplasmic volume of oocytes, redistribution of cytoplasmic AIR-2 to chromosomal foci could result in a significant increase in the intensity of chromosomal AIR-2 staining. However, the level of cytoplasmic AIR-2 is very low even in wild-type oocytes, making it difficult to determine if it is decreased in the Ceglc-7α/β(RNAi) animals.
Figure 7.
Ectopic localization of AIR-2 in Ceglc-7 α/β(RNAi) animals. Representative proximal gonads from wild-type (A) and Ceglc-7α/β(RNAi) (B) animals stained with AIR-2 (green) and DAPI (red) and shown as a merged image. Numbers above A indicate positions of oocytes with −1 being the most proximal. In >95% of wild-type gonads examined, chromosomal AIR-2 is detected only in the −1 oocytes. In 100% of Ceglc-7α/β(RNAi) animals, chromosomal AIR-2 is detected in 2–5 proximal oocytes. (C–F) High magnification of selected bivalents from wild-type (C and D) and Ceglc-7α/β(RNAi) (E and F) oocytes stained with AIR-2 (C and E) or DAPI (D and F). Arrows point to the poleward axes, and arrowheads point to the equatorial axes. (I) A schematic model for how AIR-2 regulates the release of cohesion in meiosis I. Orange bar, unphosphorylated REC-8; red triangle, AIR-2; large green bar, phosphorylated REC-8; small green bars, degraded REC-8. Bars: (B) 5 μm; (F) 1 μm.
We then asked whether the chromosomal localization of REC-8 is affected in Ceglc-7α/β(RNAi) animals. Out of 80 Ceglc-7α/β(RNAi) animals examined, we did not detect a single gonad or embryo that had wild-type REC-8 levels. In fact, 95% of meiotic embryos had no detectable REC-8 staining at all (n = 27; Fig. 5 E). Although we were unable to detect REC-8, it is likely that a low level of REC-8 is present in Ceglc-7α/β(RNAi) animals because bivalents remain intact in these oocytes. Compare this to rec-8(RNAi) oocytes where bivalents disintegrate to form single chromatids (unpublished data; Pasierbek et al., 2001). These results suggest that CeGLC-7α/β play a role in maintaining the steady-state level of chromosomal REC-8, either by increasing its chromosomal localization or by preventing its degradation.
Sister chromatids, in addition to homologues, are separated at the onset of anaphase I in Ceglc-7α/β(RNAi) embryos
We then examined if meiotic chromosome separation is affected in Ceglc-7α/β(RNAi) embryos by performing 4-D imaging. In Ceglc-7α/β(RNAi) embryos, bivalents were observed properly aligned and rotated at metaphase I (Fig. 2, P and Q). However, at a time equivalent to the onset of anaphase I and the separation of homologues in wild-type embryos (Fig. 2 H), the bivalents in Ceglc-7α/β(RNAi) embryos dissociated into as many as 24 DNA-staining structures (Fig. 2 R). We interpret these small DNA structures to be the 24 individual sister chromatids that normally comprise the six bivalents. In all embryos imaged (n = 19), these small chromosomes never segregated into two groups and no polar bodies were extruded (Fig. 2, S and T). This result indicates that CeGLC-7α/β are required for the proper regulation of cohesion release during meiosis. This is likely a result of the reduced chromosomal REC-8 protein observed on both the equatorial and poleward axes of bivalents in Ceglc-7α/β(RNAi) embryos (Fig. 5 E). This Ceglc-7α/β RNAi phenotype is dependent on AIR-2 activity because Ceglc-7α/β(RNAi);air-2(RNAi) embryos are indistinguishable from air-2(RNAi) embryos with respect to meiotic chromosome segregation (unpublished data). The dependence of Ceglc-7α/β(RNAi) phenotype on AIR-2 activity suggests that CeGLC-7α/β might regulate the level of chromosomal REC-8 by opposing AIR-2 activity toward the phosphorylation of REC-8. Alternatively, CeGLC-7α/β might regulate the level of chromosomal REC-8 simply by restricting chromosomal localization of AIR-2 (see Discussion).
Discussion
We report here that the aurora-B kinase AIR-2 promotes the release of meiotic chromosome cohesion in C. elegans likely through phosphorylation of the meiotic cohesin REC-8. Depletion of AIR-2 results in failure of chromosome separation during anaphase I and II. AIR-2 localizes to subchromosomal foci corresponding to the point of contact between separating chromosomes in metaphase I and II, and the release of REC-8 from meiotic chromosomes depends on AIR-2 activity. In vitro, AIR-2 phosphorylates REC-8 at a specific amino acid. We also show that the CeGLC-7α/β phosphatases antagonize AIR-2 activity in chromosomal cohesion release likely by restricting AIR-2 localization. In Ceglc-7α/β(RNAi) embryos, chromosomal AIR-2 is elevated, chromosomal REC-8 is decreased, and sister chromatids separate precociously at anaphase I. We propose that AIR-2 promotes the release of chromosome cohesion via phosphorylation of REC-8 at specific chromosomal locations and that CeGLC-7α/β, directly or indirectly, antagonize AIR-2 activity.
Regulation of chromosomal cohesion release in meiosis
Although mitotic and meiotic cell divisions share many features in common, they are very different with respect to chromosome behavior. Here we propose a novel mechanism for the selective release of chromosome cohesion during meiosis I and discuss how this regulation results from crossing over in C. elegans (Fig. 7 G). Our results strongly support a model whereby the selective release of cohesion during meiosis I is, in part, regulated by selective localization of AIR-2 at chromosome arms distal to chiasmata, which results in the subsequent phosphorylation of REC-8 at these sites.
Cohesion between sister chromatids distal to chiasmata has been proposed to be responsible for holding homologues together (Maguire, 1974; Carpenter, 1994). Nasmyth and colleagues have further demonstrated that release of cohesion is only required for chromosome segregation in meiosis I if homologues recombine and form chiasmata (Buonomo et al., 2000). Therefore, the observed subchromosomal location of AIR-2 in metaphase I puts it at the right place and time for a function in selective release of a subset of cohesin responsible for holding homologues together. In addition, we show that the subchromosomal AIR-2 localization in meiosis I is dependent on chiasmata formation and that release of REC-8 from meiotic chromosomes requires AIR-2 activity. REC-8 remains between chromosomal arms distal to chiasmata in air-2(RNAi) embryos. Finally, our in vitro data that AIR-2 phosphorylates REC-8 strongly suggest that AIR-2 functions through the phosphorylation of REC-8.
Possible roles of aurora-B kinases in meiosis
Because of their diverse subcellular locations, it has been proposed that the aurora-B kinases regulate multiple biological substrates (Bischoff and Plowman, 1999; Adams et al., 2001a). These proposed substrates include the kinetochore proteins Ndc10p, Sli15p, and Dam1p in budding yeast (Biggins et al., 1999; Kang et al., 2001), histone H3 in various organisms (Hsu et al., 2000; Giet and Glover, 2001; Murnion et al., 2001), and kinesin-related proteins in frogs and worms (Giet et al., 1999; unpublished data). In many organisms it has been shown that aurora-B is part of a protein complex consisting of at least aurora-B/AIR-2/Ipl1p, INCENP/ICP-1/Sli15p, and survivin/BIR-1/Bir1p (for review see Adams et al., 2001a). Loss of function in any of the components results in a similar phenotype in vivo (Kim et al., 1999; Kaitna et al., 2000; Speliotes et al., 2000; Uren et al., 2000; Adams et al., 2001b). We now show that AIR-2 likely phosphorylates REC-8 and promotes the release of chromosome cohesion during meiosis. This finding, combined with previous findings that AIR-2 functions in cytokinesis (Schumacher et al., 1998; Severson et al., 2000) and AIR-2 is part of a passenger protein complex, suggests that AIR-2 might play a critical role in coordinating chromosome separation and cytokinesis in meiosis.
Two observations suggest that AIR-2 has an additional, as yet poorly defined, role in meiotic chromosome segregation in C. elegans. First, if the sole function of AIR-2 in meiosis is to release chromosome cohesion, the chromosome segregation defect in meiosis I should be suppressible by mutations in which homologues are not recombined to form chiasmata in the first place. We have made several attempts to suppress the air-2(RNAi) meiosis I phenotype with a mutation in spo-11 to no avail. However, in spo-11;air-2(RNAi) embryos, the 12 univalents can be clearly identified in the period after anaphase I but before anaphase II (see supplemental material at http://www.jcb.org/cgi/content/full/jcb.200110045/DC1). This suggests that in addition to the release of chromosome cohesion, AIR-2 likely has another role in meiotic chromosome segregation. A similar attempt to suppress separase sep-1(RNAi) with spo-11 mutations in C. elegans was reported to be unsuccessful (Siomos et al., 2001). Second, at anaphase, all chromosomes in air-2(RNAi) embryos consistently move toward and are pressed against the edge of the embryo, as if a force from one pole is overwhelming the other. Why this happens is unclear. This is interesting in light of the monopolar spindle phenotype observed in Drosophila aurora mutant embryos and Eg2-inhibited Xenopus oocytes (Glover et al., 1995; Giet and Prigent, 2000). However, no centrosomes have been identified to associate with the C. elegans oocyte meiotic spindle (Albertson, 1984), and we did not observe overt defects in microtubule organization before metaphase I in air-2(RNAi) embryos (unpublished data). Because bivalents in air-2(RNAi) embryos do not align normally at metaphase, it is difficult to interpret the microtubule staining result at or after metaphase. The monopolar movement of chromosomes in air-2(RNAi) embryos is currently under investigation.
Possible involvement of aurora-B kinases in mitotic chromosomal cohesion release?
Whether aurora-B kinases also play a role in the release of mitotic cohesion remains unclear. However, at least in invertebrates, all studies so far suggest that aurora-B does not function in cohesion release during mitosis. First, in budding yeast, it has been reported that mitotic chromosome cohesion is unaffected in ipl1 mutants (Biggins et al., 1999), but instead, the release of cohesion requires another kinase, Cdc5p/Polo (Alexandru et al., 2001). Second, despite the polyploidy and chromosome segregation defects associated with the oocyte-derived nucleus, separation of sperm-derived chromosomes has been observed during mitosis in air-2(RNAi), bir-1(RNAi), and icp-1(RNAi) embryos (Oegema et al., 2001). Finally, we show that AIR-2 phosphorylates REC-8 but not two other likely mitotic cohesin proteins in vitro, further suggesting that AIR-2 does not regulate cohesin in mitosis. In vertebrates, the release of mitotic cohesion has been shown to involve two separate pathways (i.e., APC–separase-dependent and APC–separase-independent; Sumara et al., 2000; Waizenegger et al., 2000). The involvement of aurora-B kinases in chromosome cohesion release in either of these pathways remains to be further investigated.
Function of CeGLC-7α/β phosphatases
The involvement of CeGLC-7α/β in the regulation of meiotic chromosome cohesion release is best demonstrated by our observation that in Ceglc-7α/β(RNAi) embryos, sister chromatids separate precociously at the onset of anaphase I. In addition, CeGLC-7α/β might function in the maintenance of chromosomal REC-8 before anaphase, as we observed a dramatic decrease in the level of chromosomal REC-8 throughout the gonad in Ceglc-7α/β(RNAi) animals. However, no phenotype was observed in Ceglc-7α/β(RNAi) animals before the onset of anaphase I. It is possible that a decrease in chromosomal REC-8 is only detrimental when combined with a high separase activity in anaphase. Alternatively, this might be due to incomplete penetrance of Ceglc-7α/β(RNAi).
We believe that the function of CeGLC-7α/β in the regulation of meiotic cohesion is mediated through AIR-2. However, we cannot rule out the additional involvement of an AIR-2–independent mechanism. It is possible that CeGLC-7α/β antagonize AIR-2 by: (a) directly inhibiting its activity; (b) restricting its chromosomal localization; (c) dephosphorylating REC-8; or (d) a combination of above mechanisms. The first possibility is supported by recent work showing that a recombinant human PP1 phosphatase can directly inhibit the activity of Xenopus aurora-B kinase in embryo extracts (Murnion et al., 2001). The second possibility is supported by our observation of ectopic chromosomal AIR-2 in Ceglc-7α/β(RNAi) animals. Alternatively, the Ceglc-7α/β(RNAi) phenotype could be the result of a chromosome-wide increase in the phosphorylation of histone H3, a previously shown substrate for aurora-B and GLC-7 (Hsu et al., 2000). It is possible that an increase in phosphorylated histone H3 causes a change in chromatin organization, which either facilitates the accessibility of AIR-2 to phosphorylate REC-8 or the accessibility of separase to degrade REC-8.
Because the functions of aurora-B kinases and GLC-7 phosphatases are essential for meiotic divisions in a variety of species, it is an intriguing possibility that they might play a role in the release of meiotic chromosome cohesion across diverse species.
Materials and methods
Strains and alleles
The N2 Bristol strain was used as the wild-type strain. The genetic markers used were: LGII, him-14(it44ts); LGIV, spo-11(ok79), him-8(e1489), nT1(IV;V); LGV, her-1(e1518), him-5(e1490). The strain AZ212 carries an integrated H2B–GFP transgene under the control of the PIE-1 promoter (Praitis et al., 2001) and was provided by J. Austin (University of Chicago, Chicago, IL). All strains were cultured as described by Brenner (1974). All other strains were provided by the C. elegans Genome Consortium.
Immunofluorescence
Fixation protocols for AIR-2 and BIR-1 were as described in Lin et al. (1998), and that for REC-8 was as described in Pasierbek et al. (2001). Antibody dilutions used were: AIR-2, 1:200; BIR-1, 1:100; REC-8, 1:200; Alexa®488 secondary antibody (Molecular Probes), 1:500. BIR-1 and REC-8 antibodies were gifts from B. Horvitz (Massachusetts Institute of Technology, Boston, MA) and J. Loidl (University of Vienna, Vienna, Austria), respectively.
We also examined the staining pattern of AIR-2 and BIR-1 in him-14(it44ts) and spo-11(ok79) mutants in which crossing-over is defective in all chromosomes, resulting in 12 visible univalents in each oocyte (Dernburg et al., 1998; Zalevsky et al., 1999). In spo-11(ok79) and him-14(it44ts) mutants, ∼50% of univalents had no detectable staining, whereas the other 50% had variable amounts of staining.
RNAi
Double-stranded RNA corresponding to air-2, Ceglc-7α, and Ceglc-7β were derived from clones yk483g8, yk393h9, and yk150g8, respectively, and were injected at concentrations of 1–3 mg/ml. Injections were performed as previously described (Hsu et al., 2000). In embryos from air-2(RNAi) or bir-1(RNAi) animals that we imaged (n = 50 and n = 5, respectively), no separation of homologues was ever observed. This is contradictory to a previous report by Speliotes et al. (2000) who claimed to have observed chromosome separation in air-2(RNAi) and bir-1(RNAi) embryos. However, no data were presented in the previous report directly regarding this issue, and the reason for this discrepancy remains unclear. 100, 20, and 0% of Ceglc-7a/b(RNAi) (n = 48), Ceglc-7β(RNAi) (n = 14), and Ceglc-7α(RNAi) (n = 9) embryos, respectively, were observed to separate sister chromatids precociously. The yk-designed clones were provided by Y. Kohara (National Institute of Genetics, Shizuoka, Japan).
Imaging and time-lapse microscopy
Imaging of immunofluorescence was performed using an Olympus Bmax 60F microscope and a MicroMax-512EBFT CCD camera from Princeton Instruments as described previously (Hsu et al., 2000). Each photograph was collected as sequential images along the z-axis with 0.13 μm between adjacent images and a total of 128 image slices. All images were collected in grayscale and pseudocolored with a custom program EditView4D, and projections were done with the free software ImageJ (http://rsb.info.nih.gov/ij).
4-D microscopy was performed on live embryos mounted on agarose pads in Boyd's buffer (Boyd et al., 1996). At each time point, GFP fluorescence and differential interference contrast images were collected as described above. Consecutive time points were ∼1.5 min apart. Because of the dynamic nature of chromosomes in newly fertilized embryos, we were unable to obtain well-defined H2B–GFP images using stack projection. Representative stacks of images can be viewed at http://www.jcb.org/cgi/content/full/jcb.200110045/DC1. Whenever needed, projection was performed with DAPI-stained, fixed embryos at equivalent stages.
Plasmid construction
The entire coding regions of W02A2.6 (REC-8) and K08A8.3 (COH-1) were amplified by RT-PCR and cloned into the pGEX-6P-3 (Amersham Pharmacia Biotech) plasmid, generating plasmids pRL586 and pRL554, respectively. The full-length coding region of F10G7.4 (COH-2) was cloned by PCR, amplifying from yk632f7, and cloned into pGEX-6p-3, resulting in pRL552. PCR-based site-directed mutageneses were performed to change REC-8 S244, S248, S395, S396, T625, and S626, respectively, to alanine. All mutant clones were confirmed by sequencing. The entire coding region of AIR-2 was amplified by PCR from yk483g8 and subcloned into the pGEX-6P-1 vector (Amersham Pharmacia Biotech). The kinase-dead version of AIR-2 has a K58M change via site-directed mutagenesis. All glutathione-S-transferase (GST) fusion proteins were expressed using the Escherichia coli expression strain BL21(DE3)pLys S.
In vitro kinase assay
GST fusion constructs were expressed by overnight induction of BL21 cells with 1 mM IPTG at 30°C. Induced cultures were pelleted, resuspended in lysis buffer (PBS containing 0.2% Triton X-100) supplemented with Complete protease inhibitors (Roche), and briefly sonicated on ice. Lysates were clarified by centrifugation at 4°C, and supernatants were added to glutathione–Sepharose beads (Amersham Pharmacia Biotech) and rocked on ice for 2 h. The beads were pelleted, washed with lysis buffer, and resuspended in elution buffer (10 mM glutathione, 50 mM Tris-HCl, pH 8.0). Phosphorylation assays were conducted for 20 min at room temperature in 15 μl kinase buffer (20 mM MOPS, pH 7.2, 25 mM β-glycerol phosphate, 5 mM EGTA, 1 mM DTT, 1 mM sodium orthovanadate, 7.5 mM magnesium chloride, 10 nM ATP, 30 μCi [3 Ci/μmol] [32P]γ-ATP [NEN]) with ∼50 ng substrate and 50 ng kinase. Kinase reactions were terminated by adding SDS-PAGE sample buffer and heating to 95°C for 4 min. The reactions were separated on 10% polyacrylamide gels and blotted to nitrocellulose. The loading was assayed by Ponceau S staining and Western blotting using anti–AIR-2 or anti-GST antibodies (Upstate Biotechnology). The incorporation of 32P was detected by phosphorimaging.
Online supplemental material
Quicktime™ videos associated with Fig. 2 and mentioned in the Discussion are provided as supplemental material and can be viewed at http://www.jcb.org/cgi/content/full/jcb.200110045/DC1. Oocyte-derived meiotic chromosome dynamics visualized using a GFP–H2B reporter are presented for wild-type, air-2(RNAi), Ceglc-7a/b(RNAi), spo-11, and spo-11;air-2(RNAi).
Supplemental Material
Acknowledgments
The authors thank Scott Robertson, Leon Avery, Dennis McKearin, and Eric Olson for critical reading of the manuscript, and X. Li for technical assistance (University of Texas Southwestern Medical Center).
E. Rogers and R. Lin are supported by a grant from the National Institutes of Health (NIH) to R. Lin (HD37933). J.A. Waddle is supported by a Burroughs Wellcome Fund Career Award in the Biomedical Sciences (2550), and J.M. Schumacher is supported by a grant from the NIH (R01 GM62181-01) and a V-Foundation scholar award. J.D. Bishop is supported by the Training Program in Molecular Genetics of Cancer (T32 CA09299).
The online version of this article includes supplemental material.
Footnotes
Abbreviations used in this paper: APC, anaphase-promoting complex; 4-D, four dimensional; GFP, green fluorescent protein; GST, glutathione-S-transferase; RNAi, RNA interference.
References
- Adams, R.R., M. Carmena, and W.C. Earnshaw. 2001. a. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54. [DOI] [PubMed] [Google Scholar]
- Adams, R.R., H. Maiato, W.C. Earnshaw, and M. Carmena. 2001. b. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson, D.G. 1984. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101:61–72. [DOI] [PubMed] [Google Scholar]
- Albertson, D.G., and J.N. Thomson. 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1:15–26. [DOI] [PubMed] [Google Scholar]
- Alexandru, G., F. Uhlmann, K. Mechtler, M.A. Poupart, and K. Nasmyth. 2001. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 105:459–472. [DOI] [PubMed] [Google Scholar]
- Barnes, T.M., Y. Kohara, A. Coulson, and S. Hekimi. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics. 141:159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., F.F. Severin, N. Bhalla, I. Sassoon, A.A. Hyman, and A.W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J.R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, et al. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J.R., and G.D. Plowman. 1999. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9:454–459. [DOI] [PubMed] [Google Scholar]
- Boyd, L., S. Guo, D. Levitan, D.T. Stinchcomb, and K.J. Kemphues. 1996. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 122:3075–3084. [DOI] [PubMed] [Google Scholar]
- Bradbury, E.M., P.D. Cary, C. Crane-Robinson, and H.W. Rattle. 1973. Conformations and interactions of histones and their role in chromosome structure. Ann. NY Acad. Sci. 222:266–289. [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broverman, S.A., and P.M. Meneely. 1994. Meiotic mutants that cause a polar decrease in recombination on the X chromosome in Caenorhabditis elegans. Genetics. 136:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, S.B., R.K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann, and K. Nasmyth. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 103:387–398. [DOI] [PubMed] [Google Scholar]
- Carpenter, A.T. 1994. Chiasma function. Cell. 77:957–962. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 93:1067–1076. [DOI] [PubMed] [Google Scholar]
- Dernburg, A.F., K. McDonald, G. Moulder, R. Barstead, M. Dresser, and A.M. Villeneuve. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 94:387–398. [DOI] [PubMed] [Google Scholar]
- Francisco, L., W. Wang, and C.S. Chan. 1994. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14:4731–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., and D.M. Glover. 2001. Drosophila aurora b kinase is required for histone h3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., and C. Prigent. 2000. The Xenopus laevis aurora/Ip11p-related kinase pEg2 participates in the stability of the bipolar mitotic spindle. Exp. Cell Res. 258:145–151. [DOI] [PubMed] [Google Scholar]
- Giet, R., R. Uzbekov, F. Cubizolles, K. Le Guellec, and C. Prigent. 1999. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J. Biol. Chem. 274:15005–15013. [DOI] [PubMed] [Google Scholar]
- Glover, D.M., M.H. Leibowitz, D.A. McLean, and H. Parry. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 81:95–105. [DOI] [PubMed] [Google Scholar]
- Golden, A., P.L. Sadler, M.R. Wallenfang, J.M. Schumacher, D.R. Hamill, G. Bates, B. Bowerman, G. Seydoux, and D.C. Shakes. 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151:1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley, L.R., R.A. Walters, and R.A. Tobey. 1974. Cell cycle–specific changes in histone phosphorylation associated with cell proliferation and chromosome condensation. J. Cell Biol. 60:356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf, S., I.C. Waizenegger, and J.M. Peters. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 293:1320–1323. [DOI] [PubMed] [Google Scholar]
- Hendzel, M.J., Y. Wei, M.A. Mancini, A. Van Hooser, T. Ranalli, B.R. Brinkley, D.P. Bazett-Jones, and C.D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360. [DOI] [PubMed] [Google Scholar]
- Herman, R.K., J.E. Madl, and C.K. Kari. 1979. Duplications in Caenorhabditis elegans. Genetics. 92:419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J. 1980. More sex-determination mutants of Caenorhabditis elegans. Genetics. 96:649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J.A., H.R. Horvitz, and S. Brenner. 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 91:67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, J.Y., Z.W. Sun, X. Li, M. Reuben, K. Tatchell, D.K. Bishop, J.M. Grushcow, C.J. Brame, J.A. Caldwell, D.F. Hunt, R. Lin, M.M. Smith, and C.D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291. [DOI] [PubMed] [Google Scholar]
- Kaitna, S., M. Mendoza, V. Jantsch-Plunger, and M. Glotzer. 2000. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 10:1172–1181. [DOI] [PubMed] [Google Scholar]
- Kang, J., I.M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C.S. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)–related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., J.S. Kang, and C.S. Chan. 1999. Sli15 associates with the Ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R., R.J. Hill, and J.R. Priess. 1998. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 92:229–239. [DOI] [PubMed] [Google Scholar]
- Madl, J.E., and R.K. Herman. 1979. Polyploids and sex determination in Caenorhabditis elegans. Genetics. 93:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire, M.P. 1974. Letter: the need for a chiasma binder. J. Theor. Biol. 48:485–487. [DOI] [PubMed] [Google Scholar]
- McCarter, J., B. Bartlett, T. Dang, and T. Schedl. 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205:111–128. [DOI] [PubMed] [Google Scholar]
- Murnion, M.E., R.R. Adams, D.M. Callister, C.D. Allis, W.C. Earnshaw, and J.R. Swedlow. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656–26665. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673–745. [DOI] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, S., M.J. McKay, M. Molnar, M.A. Thompson, P.J. van der Spek, E. van Drunen-Schoenmaker, R. Kanaar, E. Lehmann, J.H. Hoeijmakers, and J. Kohli. 1999. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 19:3515–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer, and J. Loidl. 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15:1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah, S.M., and K. Nasmyth. 2000. Destruction of the securin Pds1p occurs at the onset of anaphase during both meiotic divisions in yeast. Chromosoma. 109:27–34. [DOI] [PubMed] [Google Scholar]
- Schedl, T. 1997. Developmental genetics of the germ line. C. elegans II. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess, editors. Cold Spring Harbor Laboratory Press, Plainview, NY. 241–270. [PubMed]
- Schumacher, J.M., A. Golden, and P.J. Donovan. 1998. AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson, A.F., D.R. Hamill, J.C. Carter, J. Schumacher, and B. Bowerman. 2000. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 10:1162–1171. [DOI] [PubMed] [Google Scholar]
- Shindo, M., H. Nakano, H. Kuroyanagi, T. Shirasawa, M. Mihara, D.J. Gilbert, N.A. Jenkins, N.G. Copeland, H. Yagita, and K. Okumura. 1998. cDNA cloning, expression, subcellular localization, and chromosomal assignment of mammalian aurora homologues, aurora-related kinase (ARK) 1 and 2. Biochem. Biophys. Res. Commun. 244:285–292. [DOI] [PubMed] [Google Scholar]
- Siomos, M.F., A. Badrinath, P. Pasierbek, D. Livingstone, J. White, M. Glotzer, and K. Nasmyth. 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11:1825–1835. [DOI] [PubMed] [Google Scholar]
- Speliotes, E.K., A. Uren, D. Vaux, and H.R. Horvitz. 2000. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 6:211–223. [DOI] [PubMed] [Google Scholar]
- Sumara, I., E. Vorlaufer, C. Gieffers, B.H. Peters, and J.M. Peters. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151:749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuka, M., H. Katayama, T. Ota, T. Tanaka, S. Odashima, F. Suzuki, and Y. Terada. 1998. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 58:4811–4816. [PubMed] [Google Scholar]
- Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 400:37–42. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F., D. Wernic, M.A. Poupart, E.V. Koonin, and K. Nasmyth. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 103:375–386. [DOI] [PubMed] [Google Scholar]
- Uren, A.G., L. Wong, M. Pakusch, K.J. Fowler, F.J. Burrows, D.L. Vaux, and K.H. Choo. 2000. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 10:1319–1328. [DOI] [PubMed] [Google Scholar]
- Waizenegger, I.C., S. Hauf, A. Meinke, and J.M. Peters. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 103:399–410. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., and P. Nurse. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 400:461–464. [DOI] [PubMed] [Google Scholar]
- Wei, Y., C.A. Mizzen, R.G. Cook, M.A. Gorovsky, and C.D. Allis. 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl. Acad. Sci. USA. 95:7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky, J., A.J. MacQueen, J.B. Duffy, K.J. Kemphues, and A.M. Villeneuve. 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics. 153:1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, H., T.J. McGarry, T. Bernal, and M.W. Kirschner. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 285:418–422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




