Abstract
HMGA2, a high-mobility group protein, is oncogenic in a variety of tumors, including benign mesenchymal tumors and lung cancers. Knockdown of Dicer in HeLa cells revealed that the HMGA2 gene is transcriptionally active, but its mRNA is destabilized in the cytoplasm through the microRNA (miRNA) pathway. HMGA2 was derepressed upon inhibition of let-7 in cells with high levels of the miRNA. Ectopic expression of let-7 reduced HMGA2 and cell proliferation in a lung cancer cell. The effect of let-7 on HMGA2 was dependent on multiple target sites in the 3′ untranslated region (UTR), and the growth-suppressive effect of let-7 on lung cancer cells was rescued by overexpression of the HMGA2 ORF without a 3′UTR. Our results provide a novel example of suppression of an oncogene by a tumor-suppressive miRNA and suggest that some tumors activate the oncogene through chromosomal translocations that eliminate the oncogene’s 3′UTR with the let-7 target sites.
Keywords: let-7, microRNA, HMGA2, lung cancer
HMGA2 (also called HMGI-C), a member of the high-mobility group AT-hook (HMGA) family of nonhistone chromatin proteins, is an architectural transcription factor (for review, see Reeves 2001). HMGA2 protein plays a critical role in growth during embryonic development, as evidenced by the pigmy phenotype of mutant mice deficient in hmga2 and by its high expression in embryos (Zhou et al. 1995; Rogalla et al. 1996). It is normally expressed at low levels in adult tissues, but disruption of the gene by chromosomal rearrangements at chr12q13-15 and attendant overexpression of the protein result in benign mesenchymal tumors such as lipoma, uterine leiomyoma, pulmonary chondroid hamartoma (for review, see Fedele et al. 2001), pleomorphic salivary gland adenoma (Geurts et al. 1997), and endometrial polyps (Bol et al. 1996). The chromosomal break separates the ORF of HMGA2 from the 3′ untranslated region (UTR) and leads to overexpression of HMGA2 protein (Fig. 3A, below). As most of the breakpoints are within the 140-kb third intron, the rearrangement results in the expression of a chimeric HMGA2 protein that lacks a C-terminal acidic domain but is fused to another protein from the translocated region. Although the deletion and/or fusion partners were initially thought to be important for tumorigenesis, overexpression of full-length HMGA2 protein was found to be sufficient to cause a neoplastic transformation of mesenchymal cells (Zaidi et al. 2006). Consistent with this, at least in a couple of cases the translocation site was mapped in the 3′UTR, leaving the ORF intact (Schoenmakers et al. 1995; Geurts et al. 1997; Quade et al. 2003).
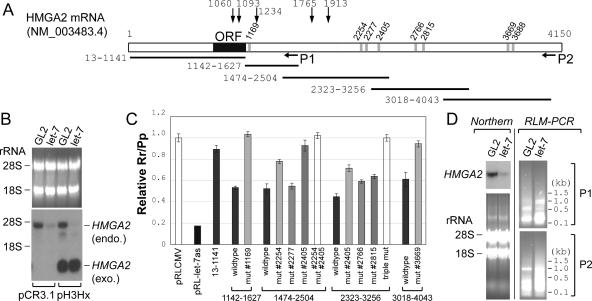
Figure 3.
The destabilization of HMGA2 mRNA by let-7 is dependent on the predicted target sites in the 3′UTR. (A) A schematic of HMGA2 mRNA. All numbers are based on the nucleotide sequence of NM_ 003483.4. Vertical gray bars indicate the potential binding sites of let-7, predicted by miRanda, PICTAR, or TargetScan (for details, see Supplementary Table S2). The target sites are named according to the nucleotide coordinate of target sequence that pairs with the 5′ end of let-7. P1 and P2 indicate approximate locations of each set of primers for the RLM-PCR (shown in D) (for details, see also Supplementary Fig. S3 and Supplementary Table S2). Horizontal lines represent segments of HMGA2 used in the luciferase reporter assays in C. Vertical arrows with the nucleotide coordinates indicate locations of chromosomal breaks in human tumors (Schoenmakers et al. 1995; Geurts et al. 1997; Ashar et al. 2003; Quade et al. 2003; Nilsson et al. 2005; Odero et al. 2005). (B) Northern hybridization was performed with the H1299-derived cell line stably expressing HMGA2 from the plasmid pH3Hx or the control cell line containing the parental vector pCR3.1, as indicated at the bottom. Endogenous HMGA2 mRNA (endo.) and the HMGA2 mRNA lacking the 3′UTR (from pH3Hx; exo.) are indicated. The rest are as in the Northern blot in Figure 2B. (C) Repression of luciferase reporter by let-7 in H1299 cells. pRL-CMV derivatives contained the indicated segments of HMGA2 mRNA inserted downstream from the Renilla luciferase ORF (the numbers at the bottom match the horizontal lines in A). pRL-let-7as is a control plasmid containing a sequence perfectly complementary to let-7 downstream from the luciferase and is repressed by the let-7 duplex by siRNA mechanisms. The relative Rr/Pp value (Y-axis) indicates the extent to which let-7 represses a given reporter. The average and standard deviation of triplicate samples are indicated. (D) RLM-PCR was performed with RNA from H1299 cells at 24 h after transfection of GL2 or let-7 duplex. (Left two panels) Northern hybridization as described in Figure 2B. (Right two panels) Agarose gel electrophoresis of RLM-RACE products from primer set P1 and P2 (shown in A) (Supplementary Fig. S3; Supplementary Table S1), with molecular size markers (in kilobases) indicated.
HMGA2 protein is also elevated in several mouse and human neoplasias such as non-small-cell carcinoma of the lung (Giancotti et al. 1989; Sarhadi et al. 2006). HMGA2 was overexpressed in 90% of lung cancers, and the presence of the protein in the nucleus correlated with high cell proliferation and poor survival. The HMGA2 gene is selectively amplified and overexpressed in 10% of liposarcomas (Berner et al. 1997). Chromosomal rearrangements at the HMGA2 locus and overexpression of the protein have also been reported in myeloid neoplasias with dysplastic features (Odero et al. 2005). Supporting a role of HMGA2 in oncogenesis, the inhibition of HMGA2 blocked retrovirally induced neoplastic transformation (Berlingieri et al. 1995). These observations, together with the proposed importance of HMGA2 in embryonic growth, suggest that overexpression of HMGA2 is tumorigenic in differentiated tissues where HMGA2 expression is normally undetectable.
MicroRNAs (miRNAs) are small noncoding regulatory RNAs that are processed by Dicer from precursors with a characteristic hairpin secondary structure. Among hundreds of miRNAs, a fraction have been shown to play a role in a variety of biological processes including development, differentiation, proliferation, and cell death. Since tumorigenesis often involves the dysregulation of these very same processes, miRNAs are potential oncogenes or tumor suppressors (for review, see Lee and Dutta 2006). A significant fraction of miRNAs are mapped to cancer-associated genomic regions or to fragile sites and many miRNAs are up- or down-regulated in cancers, suggesting that more miRNAs can potentially be involved in tumorigenesis.
let-7, one of the founding members of the miRNA family, was first identified in Caenorhabditis elegans (Reinhart et al. 2000). In human and mouse, like C. elegans, the expression of let-7 is barely detectable in embryonic stages but increases after differentiation and in mature tissues (Lee et al. 2005). Several studies proposed let-7 as a putative tumor suppressor, especially in lung cancer. The expression of let-7 is reduced in lung cancer cell lines or tumor samples relative to normal lung tissue (Takamizawa et al. 2004), and the reduction has been correlated with the overexpression of RAS, an oncogene that is a regulatory target of let-7 (Johnson et al. 2005).
In this study, we show that HMGA2 is negatively regulated by let-7, which might explain the suppression of HMGA2 during the later stages of differentiation and development. The direct linkage between let-7 and HMGA2 is another example of a miRNA being implicated in growth suppression because of its capacity to repress an oncogenic target. As suppression of HMGA2 by let-7 is mediated through its 3′UTR, our results suggest an intriguing model of oncogene activation where chromosomal breaks that separate the ORF from the 3′UTR enable the oncogene to escape repression by a miRNA.
Results and Discussion
Destabilization of HMGA2 mRNA through a miRNA-dependent pathway in the cytoplasm of HeLa cells
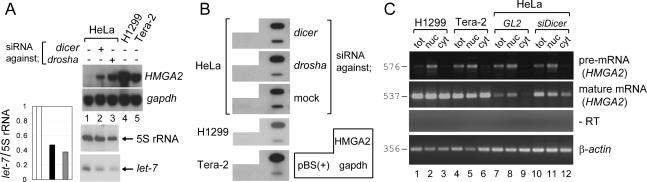
In mammalian cells, miRNAs often promote the degradation of the target mRNAs (Eulalio et al. 2007). To identify putative target genes of any miRNA, we performed a microarray hybridization of cellular mRNA after small interfering RNA (siRNA)-mediated knockdown of Dicer, an enzyme essential for miRNA biogenesis (Hutvagner et al. 2001). HMGA2 was the most up-regulated gene in the microarrays upon Dicer knockdown in HeLa cells (data not shown), suggesting that HMGA2 may be regulated by miRNAs. Consistent with previous results, HMGA2 mRNA is barely detectable in untreated HeLa cells (Fig. 1A, lane 1). In contrast, the embryonic carcinoma cell line Tera-2 and the lung cancer cell line H1299 expressed abundant levels of HMGA2 (Fig. 1A, lanes 4,5). Despite the dissimilarity in the steady-state mRNA levels, the transcriptional rates in the three cell lines were comparable in nuclear run-on assays (Fig. 1B), suggesting a destabilization of HMGA2 after transcription in HeLa cells.
Figure 1.
Destabilization of HMGA2 mRNA in the cytoplasm of HeLa cells through the miRNA pathway. (A, top two panels) Northern hybridization to detect HMGA2 and gapdh as a control. HeLa cells were transfected with GL2 (lane 1) or siRNA against Dicer (target sequence: AACUCUGCAAACCAGGUUGCU) or Drosha (target sequence: AACGAGUAGGCUUCGUGACUU) (lanes 2,3). (Lanes 4,5) The results from H1299 and Tera-2 cells are included for comparison. (Bottom two panels and the graph on the left) The levels of let-7 miRNA and 5S rRNA were measured by small RNA Northern. The blot was exposed to PhosphorImager Storm 860 (Molecular Dynamics), and each band was quantified with ImageQuant 5.2 software. The intensity of let-7 was normalized to that of 5S rRNA and plotted on the left of the blot (plain bar, lane 1; black bar, lane 2; gray bar, lane 3). (Lane 1) The value of the GL2 transfected sample was set as 1. (B) Nuclear run-on experiments were performed with the nuclei corresponding to the samples in A. The identity of each band is marked in a separate box on the right. pBS(+) indicates the pBluescript vector negative control. (C) To detect the pre-mRNA and mature mRNA of HMGA2, RT–PCR was performed with the RNAs from total (tot), nuclear (nuc), and cytoplasmic (cyt) fractions. Each amplified fragment with its expected size in base pairs is indicated. To ensure PCR products are clearly visible but are not saturated, we adjusted the amount of input cDNA and the number of cycles in each PCR amplification (35 cycles with fourfold more cDNA to detect the pre-mRNA of HMGA2, 30 cycles for mature mRNA of HMGA2, and 22 cycles for β-actin control). A representative result from the reaction without reverse transcriptase (−RT) is shown.
To determine the site in the cell of the post-transcriptional disappearance of HMGA2 in HeLa cells, we isolated RNA from nuclear and cytoplasmic fractions. The fractionation was validated by nuclear marker snoU38b (a small nucleolar RNA), which was detected exclusively in the nuclear fraction (Supplementary Fig. S1B, top panel). The spliced, mature form of HMGA2 mRNA was detectable in the nuclear fraction but undetectable in the cytoplasmic fraction of HeLa cells (Fig. 1C, lane 9; Supplementary Fig. S1B, bottom panel), indicating that the HMGA2 mRNA is destabilized in the cytoplasm. To measure the pre-mRNA for comparison, we designed a pair of primers that amplified across an exon–intron junction. The 576-base-pair (bp) PCR product from the pre-mRNA is detected only in the nuclear RNA (Fig. 1C, top panel), also validating the quality of our fractionation. Whereas the level of mature mRNA in HeLa differs significantly from those of H1299 and Tera-2 (Fig. 1A,C, second panel), the level of pre-mRNA of HMGA2 was comparable between the three cell lines (Fig. 1C, top panel), consistent with the nuclear run-on result in Figure 1B.
RNA interference (RNAi) knockdown of Dicer in HeLa cells increased HMGA2 mRNA (Fig. 1A,C). HMGA2 induction is also evident after knockdown of Drosha (Fig. 1A), another critical enzyme for miRNA biogenesis (Lee et al. 2003). RNAi of both Dicer and Drosha effectively reduced Dicer and Drosha mRNAs (Supplementary Fig. S1A) and decreased a miRNA, let-7 (Fig. 1A, bottom two panels and the quantification on the left). Neither treatment affected the expression of HMGA2 at the transcriptional stage (Fig. 1B), as evidenced by the similar intensities in the nuclear run-on assay. Also, neither treatment increased HMGA2 mRNA in Tera-2 or H1299 cells (data not shown). The mature HMGA2 mRNA was increased in the cytoplasmic fraction of HeLa cells after the RNAi of Dicer, with relatively little effect on HMGA2 mRNA in the nuclear fraction (Fig. 1C, cf. lanes 8,9 and 11,12, respectively). These results demonstrate that HMGA2 mRNA is destabilized by a miRNA(s) in the cytoplasm of HeLa, but not in H1299 or Tera-2 cells.
let-7 repressed HMGA2 expression
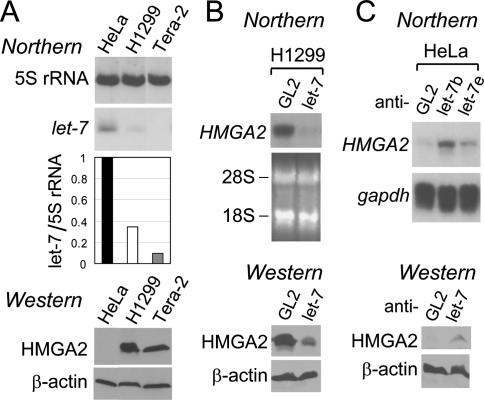
To identify a miRNA(s) that could destabilize HMGA2 mRNA, miRanda, Pictar, and TargetScan were used to search for potential miRNAs that can interact with HMGA2 mRNA. The programs predicted eight putative target sites of let-7 (Fig. 3A, below; Supplementary Table S2) in the 3′UTR of HMGA2. We focused on let-7, inspired not only by the computational prediction but also by the anti-correlation of let-7 and HMGA2 levels during in vitro differentiation of Tera-2. let-7 is induced as Tera-2 cells differentiate (Lee et al. 2005), while the converse is seen with HMGA2 (Ayoubi et al. 1999; data not shown). Consistent with the previous results, proliferating Tera-2 cells express HMGA2 mRNA and protein (Figs. 1A,C, 2A) but not let-7 (Fig. 2A). This inverse correlation between HMGA2 and let-7 is also evident in the two other cells lines, H1299 and HeLa (Figs. 1A,C, 2A).
Figure 2.
The effect of let-7 miRNA on HMGA2 expression. (A, top two panels and graph) let-7 and 5S rRNA from the indicated cell lines were measured as described in Figure 1A (black bar, HeLa; plain bar, H1299; gray bar, Tera-2). (Bottom two panels) Immunoblots for HMGA2 protein (using an antibody from Oxis International, Inc.) and β-actin as a loading control. (B, top two panels) After transfection of GL2 and let-7 duplex (a mixture of let-7b and let-7e) into H1299 lung cancer cells, HMGA2 mRNA was measured by Northern hybridization, with ethidium bromide staining of two ribosomal RNA bands shown as a loading control. (Bottom two panels) Immunoblots for HMGA2 and β-actin proteins. (C) HMGA2 mRNA (top two panels) and protein (bottom two panels) were detected after transfection of the indicated 2′-O-methyl antisense oligonucleotides into HeLa cells, with gapdh mRNA and β-actin protein as controls, respectively.
There are many variants of let-7 miRNA, let-7a to let-7i and miR-98. All the variants share an identical seed sequence that is critical for target recognition (Doench and Sharp 2004). Among them, let-7b and let-7e are most divergent with differences in four nucleotides. Since both were predicted and shown to target HMGA2 (Supplementary Table S2; Supplementary Fig. S2B [bottom panel], D), we used the two together as representative of all let-7 variants for the subsequent experiments.
First, we introduced let-7 into H1299 cells by transfecting a short RNA duplex containing let-7 sequence on one strand. Such duplexes are known to mimic the miRNA function (Hutvagner and Zamore 2002). Indeed, luciferase reporters containing artificial let-7 target sites in the 3′UTR were repressed by such duplexes (Supplementary Fig. S2A,B, top panel). Transfection of let-7 duplex to H1299 reduced the HMGA2 mRNA and protein (Fig. 2B; Supplementary Fig. S2D). Down-regulation of HMGA2 mRNA by let-7 was also evident in Tera-2 and retinoblastoma cell lines Y79 and WERI-Rb-1 (Supplementary Fig. S2D,E).
Next, we inhibited let-7 in HeLa cells by using 2′-O-methyl antisense oligonucleotide against let-7 (anti-let-7 hereafter). The inhibition of let-7 was effective, as anti-let-7 derepressed the luciferase activity from the cognate reporter plasmids (Supplementary Fig. S2C). Inhibition of let-7 in HeLa cells induced HMGA2 mRNA and protein (Fig. 2C). A similar result was obtained in another cell line, SaOS-2 (data not shown). Thus, let-7 contributes to the post-transcriptional destabilization of HMGA2 mRNA in HeLa and SaOS-2 cells.
Multiple redundant target sites for let-7 in the 3′UTR of HMGA2
Previous results indicated that HMGA2 expression is controlled by negative regulatory element(s) in its 3′UTR (Ayoubi et al. 1999; Borrmann et al. 2001). The putative target sites of let-7 are also located in the 3′UTR (Fig. 3A; Supplementary Table S2). To test whether the 3′UTR is responsible for the reduction of HMGA2 mRNA by let-7, we constructed a derivative of H1299 cells stably expressing a HMGA2 ORF without the 3′UTR. This cell line expresses a shorter HMGA2 mRNA lacking the 3′UTR (Fig. 3B, right two lanes), which is not visible in a control cell line with the parental vector pCR3.1 (Fig. 3B, left two lanes). Transfection of let-7 decreased the expression of only full-length HMGA2 mRNA, but not the truncated HMGA2 mRNA without the 3′UTR.
The cis-element for the repression by let-7 was narrowed down further by reporter assays in H1299 cells with plasmids expressing luciferase fused to various fragments of HMGA2 mRNA with let-7 duplex. Consistent with the result in Figure 3B, the segment containing 5′UTR and ORF (Fig. 3C, 13–1141) barely responded to the cotransfected let-7. The 3′UTR was divided into four fragments, each of which contains one or more target sites for let-7. All four fragments conferred repression by let-7 when inserted downstream from luciferase ORF (Fig. 3C; Supplementary Fig. S2B, bottom panel). Mutagenesis of individual target sites revealed that six out of eight predicted target sites in the HMGA2 3′UTR respond to let-7, the exceptions being sites #2277 and #3688.
let-7 promotes degradation of HMGA2 mRNA
According to current models of miRNA action in mammalian cells, a miRNA recognizes its target sites and recruits the target mRNA to cytoplasmic P-bodies or to other nuclease complexes, leading to mRNA decapping, deadenylation, and eventual degradation by exonucleases (for review, see Eulalio et al. 2007). To trap any intermediate products during the degradation, RNA was harvested at 24 h post-transfection of let-7, a time when HMGA2 mRNA decrease begins but is not completed (Fig. 3D, top left panel). RNA ligase-mediated (RLM)-PCR was used to map the 5′ ends of any HMGA2 degradation fragments. Despite the significant decrease in total HMGA2 RNA, more RLM-PCR products are visible in the let-7 transfected cells (Fig. 3D, right two panels), indicating that degradation fragments of HMGA2 mRNA increased significantly after the introduction of let-7. Nonspecific degradation of all RNA is ruled out by the detection of total RNA by ethidium bromide (Fig. 3D, bottom left panel). Cloning and sequencing of the RLM-PCR products revealed that the 5′ ends of degradation fragments are not confined to the let-7 target sites (Supplementary Fig. S3). The distributed pattern of the 5′ ends of degradation products is similar to our previous results with another miRNA, miR-206 (Kim et al. 2006), and is consistent with the notion that the mRNA is degraded by exonucleases (with pause sites) instead of by a miRNA directed site-specific endonuclease.
let-7-resistant HMGA2 rescues growth inhibition by let-7
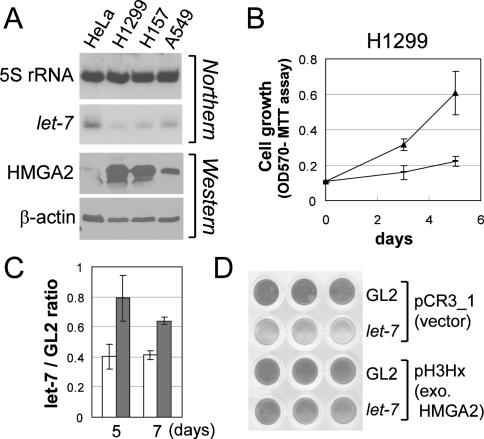
let-7 has been reported to be down-regulated in lung cancers (Takamizawa et al. 2004; Johnson et al. 2005). The decrease of let-7 and the corresponding increase of HMGA2 were evident in all three lung cancer cell lines studied (Fig. 4A). The growth of H1299 lung cancer cells was inhibited by transfection of let-7 duplex (Fig. 4B). The growth inhibition was significantly attenuated by expressing HMGA2 ORF (without the let-7-responsive 3′UTR) in these cells (Fig. 4C,D), suggesting that repression of HMGA2 is an important component of the growth inhibition by let-7. Note that the level of ectopically expressed HMGA2 is approximately fivefold that of endogenous HMGA2 in H1299 and Tera-2 (Figs. 3B, 1A; data not shown) and that the failure to completely rescue the growth inhibition suggests that other significant targets of let-7 remain to be discovered.
Figure 4.
Inhibition of proliferation of H1299 by let-7 is rescued by let-7-resistant HMGA2. (A) Decrease in let-7 and increase in HMGA2 in three lung cancer cell lines—H1299, H157, and A549. The rest are as described in Figure 2A. (B) The growth of H1299 under 2% serum was measured after transfection of GL2 (filled triangle) or let-7 duplex (dash). X-axis shows days after the first transfection; Y-axis shows OD at 570 nm in MTT assays. (C,D) Same experiment as in B was performed in H1299 stably expressing HMGA2 ORF (H1299 pH3Hx; gray bar) or containing the vector control (H1299 pCR3.1; plain bar). (Y-axis) The MTT value of cells transfected with let-7 relative to those transfected with the control duplex GL2. (D) A representative scan of the wells after the MTT assay at day 7.
Concluding remarks
miR-15a/miR-16-1 have been suggested to act as tumor suppressors by virtue of their ability to repress the anti-apoptotic oncogene Bcl2 (Cimmino et al. 2005). Chronic lymphocytic leukemias eliminate the tumor suppressor miRNAs by deleting the miRNA gene locus. The results reported here add a new twist to the story by suggesting that chromosomal translocations that activate the HMGA2 oncogene do so by deleting the 3′UTR, which is normally repressed by the action of let-7. Because multiple genes distributed throughout the genome encode for let-7, chromosomal deletions are unlikely to eliminate all the let-7 genes. Likewise, the presence of multiple target sites in the 3′UTR of HMGA2 make it unlikely that tumors can select for multiple point mutations inactivating all the target sites. In this context, a deletion eliminating the entire 3′UTR seems to be the most efficient mechanism by which the HMGA2 oncogene can escape repression by the let-7 tumor suppressor miRNA.
Although it is clear that let-7 contributes to the destabilization of HMGA2 mRNA, we cannot completely rule out the possibility that miRNA-independent or let-7-independent pathways are also involved in destabilizing HMGA2 mRNA. Indeed, HMGA2 gene contains target sites for miRNAs besides let-7 (Smalheiser and Torvik 2004). In addition to miRNA target sites, there are 10 AUUUA sequence motifs of AU-rich element (ARE) in the 3′UTR of HMGA2 that have been assumed to play a role in the destabilization of HMGA2 mRNA, (for review, see Eulalio et al. 2007). The miR-16 miRNA may be required for ARE-mediated mRNA decay (Jing et al. 2005), so that inhibition of miRNA biogenesis might additionally induce HMGA2 mRNA through the inactivation of the ARE pathway.
In lung cancers, HMGA2 is reported to be up-regulated while let-7 is down-regulated. Currently it is not known why let-7 is reduced in lung cancers. The high level of HMGA2 and low level of let-7 in lung cancer cell lines, together with the inhibition of cell proliferation after ectopic expression of let-7 and the rescue of the growth inhibition by overexpressed HMGA2, strongly suggests a causal link between let-7 and HMGA2 during lung cancer development. The rescue is significant but not complete, suggesting a role(s) of other let-7 targets. The full-length HMGA2 mRNA was overexpressed in many lung cancers (Sarhadi et al. 2006), raising the possibility that a decrease in let-7 might be the cause of HMGA2 overexpression.
In summary, our results reveal a reciprocal relation between the let-7 growth suppressor and the HMGA2 oncogene. We hypothesize that induction of let-7 during development contributes to the absence of HMGA2 in differentiated tissues. In addition, HMGA2 overexpression during tumorigenesis may arise through two let-7-dependent pathways: (1) a loss of let-7-responsive target sites through truncation of the 3′UTR, as in several benign tumors with chromosomal rearrangements, or (2) a reduction of let-7 expression as in lung cancers. Three studies have recently appeared from other groups that confirm our findings and extend the role of the let-7 family of miRNAs in suppression of HMGA2 in a mouse model system (Mayr et al. 2007), in head and neck cancers (Hebert et al. 2007), and in larger uterine leiomyomas (Wang et al. 2007).
Materials and methods
Cell lines and plasmid DNAs
Plasmid pH3Hx, a gift from Drs. B.J. Quade and C.C. Morton, contains HMGA2 ORF in the mammalian expression vector pCR3.1 (Invitrogen Corp.). After transfection of pH3Hx or pCR3.1, H1299 cells were cultured with 0.4 mg/mL of G418 for selection and maintenance of the stably expressing cells. Proliferation of cells was measured with CellTiter 96 nonradioactive cell proliferation assay kit (Promega Corp.).
RNA oligonucleotides, transfection, analysis of RNA, and luciferase assay
siRNA transfection was performed with Oligofectamine reagent or Lipofectamine 2000 reagent (Invitrogen Corp.), using 120 nM siRNA duplex or 100 nM 2′-O-methyl antisense oligonucleotide. During a long-term experiment with multiple transfections, we first performed two transfections daily, while the third transfection onward was performed on every other day. Unless otherwise indicated, the level of HMGA2 was measured on the third or the fourth day, after two transfections on days 1 and 2.
RNA isolation and analysis, mutagenesis, and luciferase assays were performed as described in the Supplemental Material.
Acknowledgments
We thank Dr. K.L. Gross for helpful discussion and Drs. B.J. Quade and C.C. Morton for kindly providing the plasmid pH3Hx.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1540407
References
- Ashar H.R., Tkachenko A., Shah P., Chada K., Tkachenko A., Shah P., Chada K., Shah P., Chada K., Chada K. HMGA2 is expressed in an allele-specific manner in human lipomas. Cancer Genet. Cytogenet. 2003;143:160–168. doi: 10.1016/s0165-4608(03)00037-2. [DOI] [PubMed] [Google Scholar]
- Ayoubi T.A., Jansen E., Meulemans S.M., de Van Ven W.J., Jansen E., Meulemans S.M., de Van Ven W.J., Meulemans S.M., de Van Ven W.J., de Van Ven W.J. Regulation of HMGIC expression: An architectural transcription factor involved in growth control and development. Oncogene. 1999;18:5076–5087. doi: 10.1038/sj.onc.1202881. [DOI] [PubMed] [Google Scholar]
- Berlingieri M.T., Manfioletti G., Santoro M., Bandiera A., Visconti R., Giancotti V., Fusco A., Manfioletti G., Santoro M., Bandiera A., Visconti R., Giancotti V., Fusco A., Santoro M., Bandiera A., Visconti R., Giancotti V., Fusco A., Bandiera A., Visconti R., Giancotti V., Fusco A., Visconti R., Giancotti V., Fusco A., Giancotti V., Fusco A., Fusco A. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol. Cell. Biol. 1995;15:1545–1553. doi: 10.1128/mcb.15.3.1545. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Berner J.M., Meza-Zepeda L.A., Kools P.F., Forus A., Schoenmakers E.F., de Van Ven W.J., Fodstad O., Myklebost O., Meza-Zepeda L.A., Kools P.F., Forus A., Schoenmakers E.F., de Van Ven W.J., Fodstad O., Myklebost O., Kools P.F., Forus A., Schoenmakers E.F., de Van Ven W.J., Fodstad O., Myklebost O., Forus A., Schoenmakers E.F., de Van Ven W.J., Fodstad O., Myklebost O., Schoenmakers E.F., de Van Ven W.J., Fodstad O., Myklebost O., de Van Ven W.J., Fodstad O., Myklebost O., Fodstad O., Myklebost O., Myklebost O. HMGIC, the gene for an architectural transcription factor, is amplified and rearranged in a subset of human sarcomas. Oncogene. 1997;14:2935–2941. doi: 10.1038/sj.onc.1201135. [DOI] [PubMed] [Google Scholar]
- Bol S., Wanschura S., Thode B., Deichert U., de Van Ven W.J., Bartnitzke S., Bullerdiek J., Wanschura S., Thode B., Deichert U., de Van Ven W.J., Bartnitzke S., Bullerdiek J., Thode B., Deichert U., de Van Ven W.J., Bartnitzke S., Bullerdiek J., Deichert U., de Van Ven W.J., Bartnitzke S., Bullerdiek J., de Van Ven W.J., Bartnitzke S., Bullerdiek J., Bartnitzke S., Bullerdiek J., Bullerdiek J. An endometrial polyp with a rearrangement of HMGI-C underlying a complex cytogenetic rearrangement involving chromosomes 2 and 12. Cancer Genet. Cytogenet. 1996;90:88–90. doi: 10.1016/0165-4608(96)00062-3. [DOI] [PubMed] [Google Scholar]
- Borrmann L., Wilkening S., Bullerdiek J., Wilkening S., Bullerdiek J., Bullerdiek J. The expression of HMGA genes is regulated by their 3′UTR. Oncogene. 2001;20:4537–4541. doi: 10.1038/sj.onc.1204577. [DOI] [PubMed] [Google Scholar]
- Cimmino A., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Calin G.A., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Fabbri M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Iorio M.V., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Ferracin M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Shimizu M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Wojcik S.E., Aqeilan R.I., Zupo S., Dono M., Aqeilan R.I., Zupo S., Dono M., Zupo S., Dono M., Dono M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E., Behm-Ansmant I., Izaurralde E., Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Fedele M., Battista S., Manfioletti G., Croce C.M., Giancotti V., Fusco A., Battista S., Manfioletti G., Croce C.M., Giancotti V., Fusco A., Manfioletti G., Croce C.M., Giancotti V., Fusco A., Croce C.M., Giancotti V., Fusco A., Giancotti V., Fusco A., Fusco A. Role of the high mobility group A proteins in human lipomas. Carcinogenesis. 2001;22:1583–1591. doi: 10.1093/carcin/22.10.1583. [DOI] [PubMed] [Google Scholar]
- Geurts J.M., Schoenmakers E.F., de Van Ven W.J., Schoenmakers E.F., de Van Ven W.J., de Van Ven W.J. Molecular characterization of a complex chromosomal rearrangement in a pleomorphic salivary gland adenoma involving the 3′-UTR of HMGIC. Cancer Genet. Cytogenet. 1997;95:198–205. doi: 10.1016/s0165-4608(96)00411-6. [DOI] [PubMed] [Google Scholar]
- Giancotti V., Buratti E., Perissin L., Zorzet S., Balmain A., Portella G., Fusco A., Goodwin G.H., Buratti E., Perissin L., Zorzet S., Balmain A., Portella G., Fusco A., Goodwin G.H., Perissin L., Zorzet S., Balmain A., Portella G., Fusco A., Goodwin G.H., Zorzet S., Balmain A., Portella G., Fusco A., Goodwin G.H., Balmain A., Portella G., Fusco A., Goodwin G.H., Portella G., Fusco A., Goodwin G.H., Fusco A., Goodwin G.H., Goodwin G.H. Analysis of the HMGI nuclear proteins in mouse neoplastic cells induced by different procedures. Exp. Cell Res. 1989;184:538–545. doi: 10.1016/0014-4827(89)90352-2. [DOI] [PubMed] [Google Scholar]
- Hebert C., Norris K., Scheper M.A., Nikitakis N., Sauk J.J., Norris K., Scheper M.A., Nikitakis N., Sauk J.J., Scheper M.A., Nikitakis N., Sauk J.J., Nikitakis N., Sauk J.J., Sauk J.J. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol. Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Zamore P.D., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D., Balint E., Tuschl T., Zamore P.D., Tuschl T., Zamore P.D., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Chen J., Di Padova F., Lin S.C., Gram H., Han J., Di Padova F., Lin S.C., Gram H., Han J., Lin S.C., Gram H., Han J., Gram H., Han J., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J., Labourier E., Reinert K.L., Brown D., Slack F.J., Reinert K.L., Brown D., Slack F.J., Brown D., Slack F.J., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kim H.K., Lee Y.S., Sivaprasad U., Malhotra A., Dutta A., Lee Y.S., Sivaprasad U., Malhotra A., Dutta A., Sivaprasad U., Malhotra A., Dutta A., Malhotra A., Dutta A., Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Dutta A., Dutta A. MicroRNAs: Small but potent oncogenes or tumor suppressors. Curr. Opin. Investig. Drugs. 2006;7:560–564. [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Yim J., Lee J., Provost P., Radmark O., Kim S., Lee J., Provost P., Radmark O., Kim S., Provost P., Radmark O., Kim S., Radmark O., Kim S., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Kim H.K., Chung S., Kim K.S., Dutta A., Kim H.K., Chung S., Kim K.S., Dutta A., Chung S., Kim K.S., Dutta A., Kim K.S., Dutta A., Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J. Biol. Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- Mayr C., Hemann M.T., Bartel D.P., Hemann M.T., Bartel D.P., Bartel D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M., Panagopoulos I., Mertens F., Mandahl N., Panagopoulos I., Mertens F., Mandahl N., Mertens F., Mandahl N., Mandahl N. Fusion of the HMGA2 and NFIB genes in lipoma. Virchows Arch. 2005;447:855–858. doi: 10.1007/s00428-005-0037-9. [DOI] [PubMed] [Google Scholar]
- Odero M.D., Grand F.H., Iqbal S., Ross F., Roman J.P., Vizmanos J.L., Andrieux J., Lai J.L., Calasanz M.J., Cross N.C., Grand F.H., Iqbal S., Ross F., Roman J.P., Vizmanos J.L., Andrieux J., Lai J.L., Calasanz M.J., Cross N.C., Iqbal S., Ross F., Roman J.P., Vizmanos J.L., Andrieux J., Lai J.L., Calasanz M.J., Cross N.C., Ross F., Roman J.P., Vizmanos J.L., Andrieux J., Lai J.L., Calasanz M.J., Cross N.C., Roman J.P., Vizmanos J.L., Andrieux J., Lai J.L., Calasanz M.J., Cross N.C., Vizmanos J.L., Andrieux J., Lai J.L., Calasanz M.J., Cross N.C., Andrieux J., Lai J.L., Calasanz M.J., Cross N.C., Lai J.L., Calasanz M.J., Cross N.C., Calasanz M.J., Cross N.C., Cross N.C. Disruption and aberrant expression of HMGA2 as a consequence of diverse chromosomal translocations in myeloid malignancies. Leukemia. 2005;19:245–252. doi: 10.1038/sj.leu.2403605. [DOI] [PubMed] [Google Scholar]
- Quade B.J., Weremowicz S., Neskey D.M., Vanni R., Ladd C., Dal Cin P., Morton C.C., Weremowicz S., Neskey D.M., Vanni R., Ladd C., Dal Cin P., Morton C.C., Neskey D.M., Vanni R., Ladd C., Dal Cin P., Morton C.C., Vanni R., Ladd C., Dal Cin P., Morton C.C., Ladd C., Dal Cin P., Morton C.C., Dal Cin P., Morton C.C., Morton C.C. Fusion transcripts involving HMGA2 are not a common molecular mechanism in uterine leiomyomata with rearrangements in 12q15. Cancer Res. 2003;63:1351–1358. [PubMed] [Google Scholar]
- Reeves R. Molecular biology of HMGA proteins: Hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Rougvie A.E., Horvitz H.R., Ruvkun G., Horvitz H.R., Ruvkun G., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rogalla P., Drechsler K., Frey G., Hennig Y., Helmke B., Bonk U., Bullerdiek J., Drechsler K., Frey G., Hennig Y., Helmke B., Bonk U., Bullerdiek J., Frey G., Hennig Y., Helmke B., Bonk U., Bullerdiek J., Hennig Y., Helmke B., Bonk U., Bullerdiek J., Helmke B., Bonk U., Bullerdiek J., Bonk U., Bullerdiek J., Bullerdiek J. HMGI-C expression patterns in human tissues. Implications for the genesis of frequent mesenchymal tumors. Am. J. Pathol. 1996;149:775–779. [PMC free article] [PubMed] [Google Scholar]
- Sarhadi V.K., Wikman H., Salmenkivi K., Kuosma E., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S., Wikman H., Salmenkivi K., Kuosma E., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S., Salmenkivi K., Kuosma E., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S., Kuosma E., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S., Salo J., Karjalainen A., Knuutila S., Anttila S., Karjalainen A., Knuutila S., Anttila S., Knuutila S., Anttila S., Anttila S. Increased expression of high mobility group A proteins in lung cancer. J. Pathol. 2006;209:206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- Schoenmakers E.F., Wanschura S., Mols R., Bullerdiek J., Van den Berghe H., de Van Ven W.J., Wanschura S., Mols R., Bullerdiek J., Van den Berghe H., de Van Ven W.J., Mols R., Bullerdiek J., Van den Berghe H., de Van Ven W.J., Bullerdiek J., Van den Berghe H., de Van Ven W.J., Van den Berghe H., de Van Ven W.J., de Van Ven W.J. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat. Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- Smalheiser N.R., Torvik V.I., Torvik V.I. A population-based statistical approach identifies parameters characteristic of human microRNA–mRNA interactions. BMC Bioinformatics. 2004;5:139. doi: 10.1186/1471-2105-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Harano T., Yatabe Y., Nagino M., Nimura Y., Yatabe Y., Nagino M., Nimura Y., Nagino M., Nimura Y., Nimura Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Wang T., Zhang X., Obijuru L., Laser J., Aris V., Lee P., Mittal K., Soteropoulos P., Wei J.J., Zhang X., Obijuru L., Laser J., Aris V., Lee P., Mittal K., Soteropoulos P., Wei J.J., Obijuru L., Laser J., Aris V., Lee P., Mittal K., Soteropoulos P., Wei J.J., Laser J., Aris V., Lee P., Mittal K., Soteropoulos P., Wei J.J., Aris V., Lee P., Mittal K., Soteropoulos P., Wei J.J., Lee P., Mittal K., Soteropoulos P., Wei J.J., Mittal K., Soteropoulos P., Wei J.J., Soteropoulos P., Wei J.J., Wei J.J. A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer. 2007;46:336–347. doi: 10.1002/gcc.20415. [DOI] [PubMed] [Google Scholar]
- Zaidi M.R., Okada Y., Chada K.K., Okada Y., Chada K.K., Chada K.K. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–7459. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- Zhou X., Benson K.F., Ashar H.R., Chada K., Benson K.F., Ashar H.R., Chada K., Ashar H.R., Chada K., Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]