Abstract
The simple combinatorial rules for regulation of the sloppy-paired-1 (slp1) gene by the pair-rule transcription factors during early Drosophila embryogenesis offer a unique opportunity to investigate the molecular mechanisms of developmentally regulated transcription repression. We find that the initial repression of slp1 in response to Runt and Fushi-tarazu (Ftz) does not involve chromatin remodeling, or histone modification. Chromatin immunoprecipitation and in vivo footprinting experiments indicate RNA polymerase II (Pol II) initiates transcription in slp1-repressed cells and pauses downstream from the promoter in a complex that includes the negative elongation factor NELF. The finding that NELF also associates with the promoter regions of wingless (wg) and engrailed (en), two other pivotal targets of the pair-rule transcription factors, strongly suggests that developmentally regulated transcriptional elongation is central to the process of cell fate specification during this critical stage of embryonic development.
Keywords: Segmentation, ChIP, DNase I hypersensitivity, in vivo footprinting
The Drosophila embryo provides a powerful model for investigating the regulatory mechanisms of cell fate specification during development. Gradients of maternally provided positional information are translated into differential programs of gene expression at a single-cell level of resolution within the 6000 cells that comprise the cellular blastoderm stage embryo at 3 h of development (Ingham 1988). On the antero–posterior axis, this initial specification is revealed by the segmentally repeated expression of several segment-polarity genes (Akam 1987). These metameric expression patterns are generated in response to positional cues provided by the DNA-binding transcription factors encoded by the pair-rule segmentation genes (Howard 1990). One key component of segment-polarity gene regulation is context-dependent activation and repression by the pair-rule transcription factor Runt, the founding member of the Runx family of developmental regulators (Coffman 2003; Levanon and Groner 2004). Recent studies reveal that Runt activity is modulated by the Ftz homeodomain protein and further identify the slp1 gene as an attractive model for investigating the molecular mechanisms of Runt-dependent transcriptional regulation (Swantek and Gergen 2004).
The segmentally repeated expression of slp1 initiates during the late cellular blastoderm stage and becomes fully apparent by the onset of gastrulation (∼3.5 h of development). The combinatorial rules that generate this pattern differ in odd- and even-numbered parasegments. Expression in the posterior half of odd parasegments requires the pair-rule transcription factors Runt and Odd-paired (Opa). In contrast, in the adjacent cells that comprise the anterior half of the even parasegments, the presence of Ftz converts Runt from an activator to a repressor of slp1 expression (Swantek and Gergen 2004). Normally, these combinatorial rules only apply within the presegmental region of the embryo. However, ectopic coexpression of Runt and Opa in embryos that are mutant for Ftz leads to slp1 activation in all somatic blastoderm cells, whereas ectopic coexpression of both Runt and Ftz gives slp1 repression throughout the embryo (Fig. 1A). Thus, all late blastoderm stage nuclei can adopt slp1 expression states characteristic of posterior odd-numbered or anterior even-numbered parasegments in response to these pair-rule transcription factors. The uniform repression of slp1 in response to Runt and Ftz is of particular interest, as it is straightforward to generate staged populations of these embryos that can be used to investigate the in vivo biochemistry of this repression. Indeed, in matched embryo collections processed 3–4 h AED (after egg deposition), quantitative RT–PCR reveals 40-fold less slp1 mRNA in embryos that are ectopically coexpressing Runt and Ftz compared with wild-type embryos. Our goal in this study was to identify the molecular changes at the slp1 locus that account for this 40-fold difference in expression.
Figure 1.
Manipulation of slp1 expression by pair-rule transcription factors. (A) Expression of slp1 mRNA in gastrula-stage embryos as revealed by in situ hybridization. (Middle panel) The wild-type pattern shows 14 distinct stripes in the presegmental region with stronger expression in even-numbered parasegments. The embryos on the left and right demonstrate the response of slp1 to ectopic expression of different pair-rule transcription factors using the NGT (nanos-GAL4-tubulin) expression system (Tracey et al. 2000). (Left panel) NGT-driven expression of Runt and Opa in embryos that are mutant for ftz leads to activation throughout the presegmental region as well as in the anterior head region. (Right panel) In contrast, NGT-driven coexpression of Runt and Ftz leads to uniform slp1 repression. (B) Quantitative RT–PCR was used to determine the differences in slp1 expression in staged collections of wild-type (WT) and Runt + Ftz (R+F) coexpressing embryos.
Results and Discussion
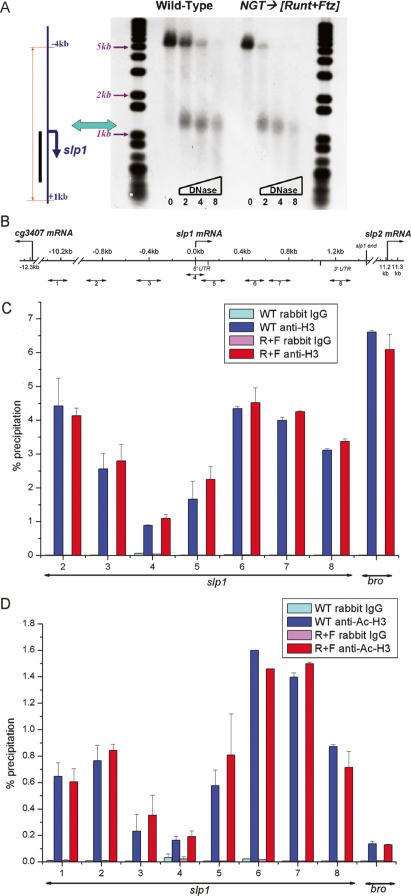
We began by using DNase I hypersensitivity to probe the chromatin structure of the slp1 locus. These assays revealed the presence of a DNase I-hypersensitive site near to the 5′-end of the slp1 transcription unit (Fig. 2A). Chromatin immunoprecipitation (ChIP) experiments with antiserum against histone H3 provide an explanation for this DNase I hypersensitivity. There is significantly reduced association of H3 with the slp1 promoter region compared with both the structural gene as well as sequences upstream of the promoter (Fig. 2B,C). These observations strongly suggest that the promoter region is nucleosome free. Importantly, matched collections of wild-type and Runt + Ftz (R+F) embryos show both the same pattern of DNase I hypersensitivity (Fig. 2A) and histone H3 association (Fig. 2C). These results indicate that the 40-fold decrease in mRNA expression in slp1-repressed embryos is not due to gross changes in the accessibility of the slp1 promoter region.
Figure 2.
slp1 repression is established without alterations in chromatin structure. (A) DNase I digestion reveals the presence of a hypersensitive site near the 5′-end of the slp1 transcription unit. The diagram on the left indicates the relationship of an ∼5-kb interval of the slp1 locus to the DNA fragments detected in the Southern blot shown on the right. The thin line on the left indicates the extent of a 5-kb fragment produced by BstXI digestion, and the adjacent thick vertical bar indicates the antisense riboprobe used to detect slp1-containing sequences. The horizontal line and downward arrow indicate the initiation site and direction of transcription, respectively. DNA samples isolated from wild-type and R+F embryos (3–4 h AED) are organized as indicated across the top, with the amounts of DNase I used on different samples indicated across the bottom. (B) The location of PCR products generated with different primer pairs used for ChIP studies is shown relative to a map of the slp1 locus. The PCR products are numbered left to right, from 1 to 8. (C) Results of quantitative PCR on ChIP samples generated using an antibody against histone H3 with chromatin isolated from wild-type (blue bars) and R+F (red bars) embryos. The nonspecific background was determined using a control rabbit IgG antiserum with these same chromatin preparations (cyan and magenta bars, respectively). Results obtained for different intervals of the slp1 locus are as labeled across the bottom. ChIP results using a primer pair that amplifies a segment of the Bro structural gene are given on the right. Error bars indicate the standard error in the percent precipitation values for each interval. (D) Results of quantitative PCR with ChIP samples generated using an antibody that detects acetylated H3, labeled as in C.
Histone acetylation and deacetylation are important for transcriptional regulation with a general correlation between histone acetylation and active transcription (Roth et al. 2001; Schubeler et al. 2004). Indeed, prior work from our laboratory demonstrated that the Rpd3 histone deacetylase is important for maintaining the Runt-dependent repression of the segment-polarity gene en (Wheeler et al. 2002). ChIP experiments reveal no significant difference in the H3 acetylation pattern of slp1 chromatin from wild-type versus R+F embryos (Fig. 2D). Although we detect no differences in H3 or Ac-H3 association that correlate with slp1 repression, there are interesting differences in the H3 acetylation levels at different genomic locations. The slp1 structural gene (primer pairs 6 and 7) shows stronger Ac-H3 association than the upstream region (primer pair 2) (Fig. 2D). This difference is not observed for the association of H3 with these same intervals (Fig. 2C), suggesting that H3 acetylation marks genomic regions that are permissive for transcription. The relative levels of H3 and Ac-H3 association with Brother (Bro), a gene that is not transcribed in the early embryo (as measured by RT–PCR) (data not shown), provide additional evidence for this trend. Although H3 association with the Bro gene is greater than for any region of the slp1 locus, the level of Ac-H3 association with Bro is lower than for any region of slp1 (Fig. 2C,D). Based on the observation that we can detect differences in H3 and Ac-H3 association that correlate with transcriptional potential and yet detect no differences between wild-type and slp1-repressed embryos, we conclude that H3 acetylation plays a negligible role in the establishment of slp1 repression.
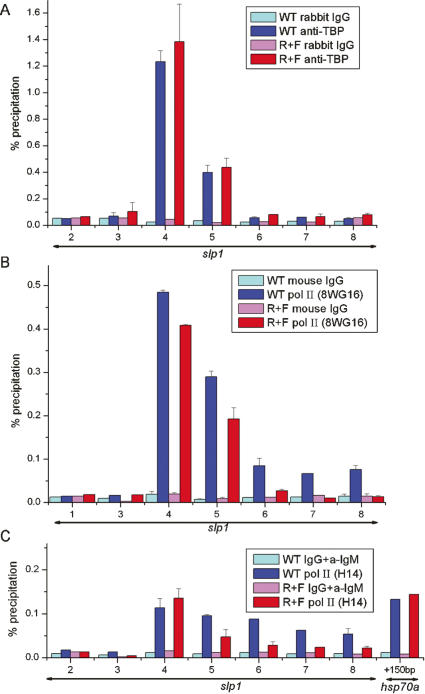
The above observations led us to characterize the interactions of the transcriptional machinery with slp1. Association of the TATA-box-binding protein (TBP) is a first step in assembly of the transcriptional machinery on a promoter (Green 2000). As expected, TBP association is detected with a promoter-proximal interval centered 6 base pairs (bp) upstream of the slp1 transcript initiation site in chromatin from wild-type embryos (Fig. 3A). A weaker signal is detected for an interval within the 5′ untranslated region (UTR), centered 124 bp downstream from the start site, whereas all other intervals give background level signals. We find very similar levels of TBP association in chromatin from R+F embryos (Fig. 3A). More surprising is the finding that there is almost no difference in the level of Pol II association with the slp1 promoter-proximal interval in chromatin from wild-type and R+F embryos (Fig. 3B). Pol II is also associated with the slp1 structural gene in wild-type embryos, but at lower levels than at the promoter (Fig. 3B). In contrast, Pol II association with the slp1 structural gene is markedly reduced in R+F embryos and near to background levels for regions downstream from the 5′-UTR (Fig. 3B). Based on these results, we conclude that promoter recruitment of Pol II is not blocked in slp1-repressed embryos. We further characterized slp1-associated Pol II using an antibody that recognizes the Phospho-Ser-5 form of the heptad repeats that comprise the C-terminal domain (CTD) of the largest Pol II subunit. Phospho-Ser-5 modification of the CTD is associated with transcription initiation (Komarnitsky et al. 2000; Sims et al. 2004; Morris et al. 2005). This antiserum also gives the strongest signals with the slp1 promoter-proximal interval in wild-type chromatin, and this signal is not reduced in chromatin from R+F embryos (Fig. 3C). This result indicates that slp1 repression occurs at a step downstream from transcription initiation.
Figure 3.
Initiation of transcription in slp1-repressed embryos. ChIP assays on chromatin from wild-type and R+F embryos using anti-TBP antiserum (A), the monoclonal antibody 8WG16 (B), which recognizes the heptad repeats of the CTD, irrespective of their phosphorylation status (Patturajan et al. 1999; Jones et al. 2004; Morris et al. 2005), and the monoclonal antibody H14 (C), which specifically recognizes phosphorylation of serine residue 5 within the heptad repeats that comprise the CTD (Patturajan et al. 1999; Jones et al. 2004; Morris et al. 2005). Background controls for each antibody were determined using nonimmune serum from the appropriate species as indicated. The labeling scheme is the same as used in Figure. 2.
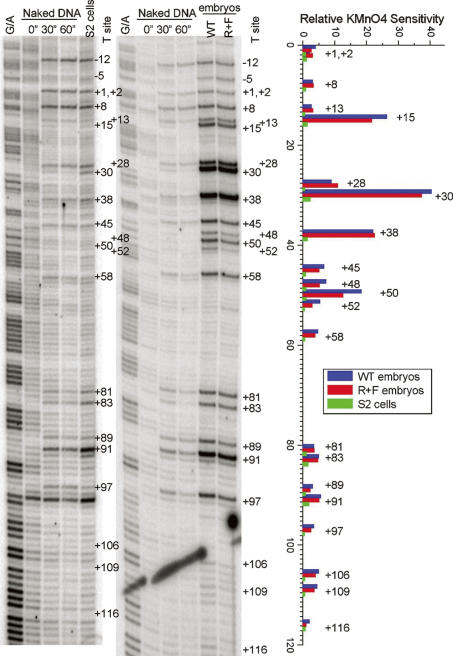
The Drosophila hsp70a promoter is an extensively studied example of regulated transcriptional elongation (Rougvie and Lis 1988; Boehm et al. 2003; Lebedeva et al. 2005). Pol II initiates transcription at the hsp70a promoter, and then, in the absence of a heat shock, pauses immediately downstream from the promoter. All somatic cells in 3–4-h-AED embryos are capable of activating the hsp70a gene, and as expected, Phospho-Ser-5-modified Pol II is readily detected on the hsp70a promoter in our chromatin preparations from non-heat-shocked embryos (Fig. 3C). The paused Pol II complex on the hsp70a promoter is also readily detected using permanganate footprinting due to the increased sensitivity of thymine residues in single-stranded regions (Weber et al. 1997). We used this same technique to carry out footprinting studies on the slp1 promoter region. The results reveal strong hyperreactivity of thymine residues at +15, +28, +30, +38, and +50 downstream from the transcription start site in blastoderm stage embryos (Fig. 4). This interval is similar, though perhaps somewhat larger than the interval detected for hsp70a, within which the most prominent increases in reactivity are at residues +22 and +30 (Weber et al. 1997). The pattern of reactivity on slp1 is extremely similar in both wild-type and slp1-repressed embryos, indicating that the hyperreactivity is not due to active transcription of the slp1 gene. Importantly, this pattern is not observed in nuclei from Drosophila tissue culture cells (Fig. 4). Thus, unlike hsp70a, the footprint on the slp1 5′-UTR is developmentally regulated.
Figure 4.
Developmental regulation of slp1 promoter-proximal pausing. The two left panels are autoradiographs revealing the permanganate sensitivity of thymine residues in an interval extending from 12 nt upstream of to 116 nt downstream from the slp1 transcription start site. Each of these panels includes a G/A ladder and three lanes showing results obtained by treating naked DNA with 10 mM KMnO4 for 0, 30, and 60 sec. In addition, the autoradiograph on the left shows in vivo results obtained from treatment of Drosophila S2 cells with 10 mM KMnO4 for 60 sec, whereas the autoradiograph on the right shows results obtained with similarly treated wild-type and R+F 3–4-h-AED Drosophila embryos. The graph on the far right presents a quantification of the raw PhosphorImager data for the S2, wild-type, and R+F samples using the Semi-Automated Footprinting Analysis software package SAFA (Das et al. 2005). The relative sensitivity of each thymine residue is the ratio of the signal for each experimental sample relative to the signal obtained from the 30-sec naked DNA control. A 10-fold or greater increase in KMnO4 sensitivity is observed for thymine residues at +15, +28, +30, +38, and +50 in both embryo samples, whereas the sensitivity of these residues in S2 cells is nearly indistinguishable from that observed on naked DNA.
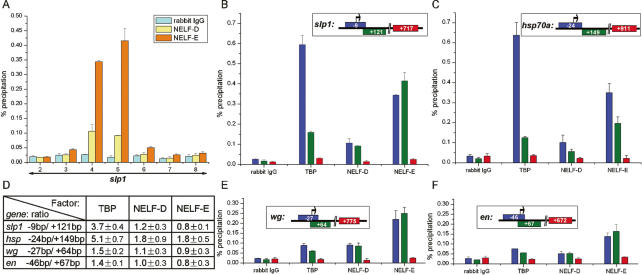
The negative elongation factor NELF is thought to play a key role in establishing the paused Pol II complex on the hsp70a promoter (Wu et al. 2003). Indeed, NELF association provides a marker for the paused complex as heat-shock-induced transcriptional elongation involves release of NELF (Wu et al. 2003, 2005). In agreement with the results of our footprinting studies, ChIP experiments reveal the NELF-D and NELF-E subunits are associated with the slp1 promoter region in chromatin from wild-type embryos (Fig. 5A), but not in chromatin from Drosophila tissue culture cells (data not shown). Strong signals are obtained in chromatin from embryos with both the promoter-proximal and 5′-UTR intervals, whereas background level signals are obtained with other intervals of the slp1 locus. It is notable that the promoter-proximal signal is less than or equal to the signal detected for the 5′-UTR interval. This pattern of association contrasts that obtained with TBP, which shows a threefold stronger signal with the promoter-proximal primer pair (Fig. 5B,D). Our interpretation of these association patterns is that NELF is bound downstream from the slp1 transcription start site, presumably as a component of the paused Pol II complex. Consistent with this interpretation, we find a similar differential pattern of TBP and NELF association with promoter-proximal and 5′-UTR intervals of hsp70a (Fig. 5C,D). These results strongly suggest that NELF plays a key role in regulating slp1 elongation in the blastoderm-stage Drosophila embryo.
Figure 5.
NELF associates with promoter-proximal 5′-UTR regions of segment-polarity genes in the blastoderm embryo. (A) ChIP assays on chromatin from wild-type 3–4-h-AED embryos using antisera against the NELF-D (yellow bars) and NELF-E (orange bars) subunits of NELF. The primer pairs used to detect NELF association with different intervals of the slp1 locus are as described for Figure 2. Background signals obtained using control IgG are shown by cyan bars. (B) Comparison of the pattern of TBP, NELF-D, and NELF-E association with intervals of slp1 detected by primer pairs 4, 5, and 7, centered 9 bp upstream of (blue bars), and 121 bp (green bars) and 717 bp (red bars) downstream from the transcription start site, respectively. (C) Pattern of TBP, NELF-D, and NELF-E association with hsp70a using a similar representation as in B. In this case, the ChIP assays were for intervals centered 24 bp upstream of (blue bars) and 149 bp (green bars) and 911 bp (red bars) downstream from the transcription start site. The preferential association of TBP that is detected with the promoter-proximal versus 5′-UTR hsp70a intervals is greater than that observed for slp1, presumably due to differences in the exact location of the primer pairs used for these two genes. The slightly stronger signal obtained with both NELF subunits and the hsp70a upstream promoter interval indicates NELF association maps closer to −24 bp than to +149 bp, relative to the transcription start site. This is consistent with in vivo data placing the paused Pol II complex 21–35 bp downstream from the hsp70a transcription start site (Rasmussen and Lis 1995). (D) A table providing the ratios of ChIP signals obtained with promoter-proximal, and 5′-UTR intervals of four different genes (slp1, hsp70a, wg, and en) with TBP, NELF-D, and NELF-E antisera. Raw data showing the association of TBP, NELF-D, and NELF-E with different regions of wg and en are shown in E and F, respectively. The intervals detected from the wg locus centered 27 bp upstream of (blue bars), and 64 bp (green bars) and 775 bp (red bars) downstream from the transcription start site. Similarly for en, the PCR-amplified intervals centered 46 bp upstream of (blue bars) and 67 bp (green bars) and 672 bp (red bars) downstream from the transcription start site.
The initial indications that slp1 expression was regulated at a step downstream from transcription initiation came from ChIP experiments on chromatin from a homogeneous population of embryos that uniformly repress slp1. Localized association of NELF in a region downstream from the transcription start site is a hallmark of promoter-proximal pausing. Importantly, this association provides a method for detecting paused Pol II complexes in chromatin from embryos that contain a mixture of cells, some of which are expressing full-length mRNA transcripts. We used ChIP assays to determine whether NELF associates with the promoter regions of wg and en, two pivotal segment-polarity gene targets of the pair-rule transcription factors. The results reveal specific association of NELF with the promoter-proximal and 5′-UTR regions of both genes in 3–4-h-AED embryos (Fig. 5E,F). Furthermore, the differential association pattern of TBP and NELF with these two intervals indicates that NELF is localized to a region immediately downstream from the initiation sites for both genes (Fig. 5D). These findings indicate that regulation of transcriptional elongation is likely to be central in generating the initial patterns of segment-polarity gene expression in the Drosophila embryo.
Regulation of transcriptional elongation has been described for several genes in addition to the Drosophila heat-shock genes, including human c-myc, c-myb, c-fos, junB, and p21 (Bender et al. 1987; Pinaud and Mirkovitch 1998; Chen and Sytkowski 2001; Aida et al. 2006; Gomes et al. 2006). A feature shared by these previously characterized examples is rapid induction of gene expression in response to external stimuli. The initial establishment of segment-polarity gene-expression patterns in response to the pair-rule transcription factors occurs within a relatively brief developmental window of ∼30 min, spanning the completion of cellularization and the beginning of germ band extension. The temporal advantages offered by regulating these genes at a transcriptional elongation step as compared with chromatin remodeling and/or Pol II initiation complex assembly may be essential for the timely establishment of differing gene expression programs during cell fate specification in the Drosophila blastoderm embryo. The observations that Pol II molecules are enriched at the 5′-ends of a number of genes (Gariglio et al. 1981; Rougvie and Lis 1990), coupled with findings that defects in transcriptional elongation factors produce specific developmental defects (Guo et al. 2000; Jennings et al. 2004), strongly suggest that regulation of transcriptional elongation is a hitherto overlooked, but potentially widespread strategy for controlling gene expression during development.
Materials and methods
Drosophila strains and transgenes
The Drosophila strain y w[67c23] that was used to develop different transgenic lines was used as the wild-type control for all in situ hybridization and biochemical experiments. Uniform ectopic expression of different pair-rule genes was obtained using the NGT (nanos-GAL4-tubulin) maternal GAL4 expression system (Tracey et al. 2000). The genetic cross to express Runt and Opa in embryos mutant for ftz was between females homozygous for the NGT[40] GAL4-driver and heterozygous for the ftz[11] mutation and males homozygous for both the UAS-runt[15]and UAS-opa[14] transgenes and heterozygous for ftz[11]. Ectopic coexpression of Runt and Ftz was obtained by crossing females homozygous for both the NGT[40] and NGT[A] GAL4 drivers with males homozygous for both the UAS-runt[15] and UAS-ftz[263] transgenes. These different NGT, UAS-runt, UAS-opa, and UAS-ftz transgenes have all been described previously (Swantek and Gergen 2004).
In situ hybridization and RT–PCR
In situ hybridization was carried out as described previously (Swantek and Gergen 2004) using the digoxigenin-labeled riboprobe for slp1 described in Wheeler et al. (2002). RNA used for PCR after reverse-transcription was isolated from homogenates of ∼200 embryos from appropriately staged collections. RNA was extracted using the High Pure RNA isolation Kit (Roche). cDNA was synthesized using the 1st Strand cDNA synthesis Kit (Roche) programmed with 1 ug of RNA. Quantitative PCR was conducted using a Light Cycler (Roche) with a primer pair centered 504 bp downstream from the slp1 promoter (primer pair 6 in Fig. 2B). The slp1 RT–PCR signal obtained with different RNA samples was normalized using the RT–PCR signal for rp49.
DNase I-hypersensitive site assay
Nuclei isolated from ∼250 mg of staged Drosophila embryos (Cartwright et al. 1999) were resuspended in DNase I digestion buffer, aliquoted into microfuge tubes, and incubated with 0, 2, 4, or 8 U of DNase I for 3 min on ice. Digestion was stopped by addition of EDTA to 15 mM and SDS to 0.5%. DNA was purified by phenol/chloroform extraction and ethanol precipitation, and resuspended in water. Approximately 5 ug of DNA samples digested with BstXI were run on 1% agarose gels and transferred to nylon membranes for Southern blot hybridization. The slp1 riboprobe used for these experiments is similar to the one used for in situ hybridization, although it was not subjected to partial hydrolysis prior to use for hybridization. DNA size standards were detected on Southern blots using a digoxigenin-labeled DNA probe.
ChIP
Staged embryo collections were dechorionated, fixed with 2% formaldehyde for 15 min at room temperature, washed, and snap-frozen at −80°C for storage (Orlando et al. 1998). Approximately 100 mg of embryos were homogenized for 1 min in 10 mM EDTA and 50 mM Tris (pH 8.1). After addition of SDS to a final concentration of 1% and incubation on ice for 10 min, glass beads (150–200 μm) were added and the homogenates were sonicated to give sheared chromatin preparations with an average DNA size of 300–400 bp. Chromatin preparations from the equivalent of ∼25 mg of embryos were used for each immunoprecipitation using the Chromatin Immunoprecipitation Assay Kit (Upstate Biotechnology) with the following antisera: anti-rabbit IgG (Sigma), anti-mouse IgG (Sigma), anti-mouse IgM (Sigma), anti-Histone H3 (Abcam), anti-acetyl-Histone H3 (Upstate Biotechnology), anti-TBP (Lebedeva et al. 2005), anti-RNA pol II 8WG16 (Covance), anti-RNA pol II H14 (Covance), and anti-NELF-D and anti-NELF-E (Wu et al. 2003). The relative amount of immunoprecipitated DNA was quantified using real-time PCR (Roche Light-Cycler) using primer pairs with a Tm in the range of 59°C–66°C that generate products between 150 and 211 bp. Primer sequences are available on request. The percent precipitation values that are reported were calculated using a dilution series of input chromatin to determine the relative efficiency for each primer pair. PCR was done multiple times on samples from at least two independent immunoprecipitation assays using at least two different chromatin preparations.
Permanganate footprinting
Permanganate footprinting on Drosophila S2 cells was performed as described previously using 10 mM KMnO4 for a 1-min incubation (Weber et al. 1997). For embryos, collections of 3–4-h-AED embryos (10 μL, corresponding to ∼103 embryos or 107 genomes) were dechorionated, transferred into 150 μL of ice-cold dissection buffer (130 mM NaCl, 5 mM NaCl, 5 mM KCl, 1.5 mM CaCl2), and briefly homogenized prior to addition of 50 μL of 40 mM KMnO4. Reactions were stopped after 1 min by addition of 200 μL of stop solution (20 mM Tris at pH 7.5, 20 mM NaCl, 40 mM EDTA, 1% SDS, 0.8 M β-mercaptoethanol) to the lysate. Nested primers for detecting cleavage within the slp1 promoter and 5′-UTR interval by Ligation-Mediated PCR span the interval from 161 to 199 nucleotides (nt) downstream from the transcription start site. The sequences and annealing temperature for PCR are as follows: slp1 primer-1, 5′-GTTTTGATGGGTTGAGTTG-3′ (51°C); slp1 primer-2 5′-GGGT TGAGTTGCGGTGT-3′ (55°C); slp1 primer-3, 5′-GCGGTGTTGAT GGGTTTCTT-3′ (58°C).
Acknowledgments
We thank Steven Naymagon and Jocelin Abraham for technical assistance, and Achim Breiling, Ping Hu, Nouria Hernandez, Ann Sutton, and Rolf Sternglanz for technical advice in developing ChIP protocols for early Drosophila embryos. We thank James Kadonaga for providing antisera to TBP. Comments on the manuscript were provided by Carl Anderson, Michael Frohman, Maurice Kernan, and Rolf Sternglanz. This work was supported by research grants from the NSF (MCB 0344486, to J.P.G) and the NIH (GM 47477 to D.S.G).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1521207
References
- Aida M., Chen Y., Nakajima K., Yamaguchi Y., Wada T., Handa H., Chen Y., Nakajima K., Yamaguchi Y., Wada T., Handa H., Nakajima K., Yamaguchi Y., Wada T., Handa H., Yamaguchi Y., Wada T., Handa H., Wada T., Handa H., Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Bender T.P., Thompson C.B., Kuehl W.M., Thompson C.B., Kuehl W.M., Kuehl W.M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Boehm A.K., Saunders A., Werner J., Lis J.T., Saunders A., Werner J., Lis J.T., Werner J., Lis J.T., Lis J.T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I.L., Cryderman D.E., Gilmour D.S., Pile L.A., Wallrath L.L., Weber J.A., Elgin S.C., Cryderman D.E., Gilmour D.S., Pile L.A., Wallrath L.L., Weber J.A., Elgin S.C., Gilmour D.S., Pile L.A., Wallrath L.L., Weber J.A., Elgin S.C., Pile L.A., Wallrath L.L., Weber J.A., Elgin S.C., Wallrath L.L., Weber J.A., Elgin S.C., Weber J.A., Elgin S.C., Elgin S.C. Analysis of Drosophilachromatin structure in vivo. Methods Enzymol. 1999;304:462–496. doi: 10.1016/s0076-6879(99)04028-8. [DOI] [PubMed] [Google Scholar]
- Chen C., Sytkowski A.J., Sytkowski A.J. Erythropoietin activates two distinct signaling pathways required for the initiation and the elongation of c-myc. J. Biol. Chem. 2001;276:38518–38526. doi: 10.1074/jbc.M105702200. [DOI] [PubMed] [Google Scholar]
- Coffman J.A. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 2003;27:315–324. doi: 10.1016/s1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- Das R., Laederach A., Pearlman S.M., Herschlag D., Altman R.B., Laederach A., Pearlman S.M., Herschlag D., Altman R.B., Pearlman S.M., Herschlag D., Altman R.B., Herschlag D., Altman R.B., Altman R.B. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariglio P., Bellard M., Chambon P., Bellard M., Chambon P., Chambon P. Clustering of RNA polymerase B molecules in the 5′ moiety of the adult β-globin gene of hen erythrocytes. Nucleic Acids Res. 1981;9:2589–2598. doi: 10.1093/nar/9.11.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes N.P., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M., Llorente B., Szostek S.A., Emerson B.M., Espinosa J.M., Szostek S.A., Emerson B.M., Espinosa J.M., Emerson B.M., Espinosa J.M., Espinosa J.M. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes & Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R. TBP-associated factors (TAFIIs): Multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- Guo S., Yamaguchi Y., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H., Rosenthal A., Yamaguchi Y., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H., Rosenthal A., Schilbach S., Wada T., Lee J., Goddard A., French D., Handa H., Rosenthal A., Wada T., Lee J., Goddard A., French D., Handa H., Rosenthal A., Lee J., Goddard A., French D., Handa H., Rosenthal A., Goddard A., French D., Handa H., Rosenthal A., French D., Handa H., Rosenthal A., Handa H., Rosenthal A., Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- Howard K. The blastoderm prepattern. Semin. Cell Biol. 1990;1:161–172. [PubMed] [Google Scholar]
- Ingham P.W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Jennings B.H., Shah S., Yamaguchi Y., Seki M., Phillips R.G., Handa H., Ish-Horowicz D., Shah S., Yamaguchi Y., Seki M., Phillips R.G., Handa H., Ish-Horowicz D., Yamaguchi Y., Seki M., Phillips R.G., Handa H., Ish-Horowicz D., Seki M., Phillips R.G., Handa H., Ish-Horowicz D., Phillips R.G., Handa H., Ish-Horowicz D., Handa H., Ish-Horowicz D., Ish-Horowicz D. Locus-specific requirements for Spt5 in transcriptional activation and repression in Drosophila. Curr. Biol. 2004;14:1680–1684. doi: 10.1016/j.cub.2004.08.066. [DOI] [PubMed] [Google Scholar]
- Jones J.C., Phatnani H.P., Haystead T.A., MacDonald J.A., Alam S.M., Greenleaf A.L., Phatnani H.P., Haystead T.A., MacDonald J.A., Alam S.M., Greenleaf A.L., Haystead T.A., MacDonald J.A., Alam S.M., Greenleaf A.L., MacDonald J.A., Alam S.M., Greenleaf A.L., Alam S.M., Greenleaf A.L., Greenleaf A.L. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho E.J., Buratowski S., Cho E.J., Buratowski S., Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva L.A., Nabirochkina E.N., Kurshakova M.M., Robert F., Krasnov A.N., Evgen’ev M.B., Kadonaga J.T., Georgieva S.G., Tora L., Nabirochkina E.N., Kurshakova M.M., Robert F., Krasnov A.N., Evgen’ev M.B., Kadonaga J.T., Georgieva S.G., Tora L., Kurshakova M.M., Robert F., Krasnov A.N., Evgen’ev M.B., Kadonaga J.T., Georgieva S.G., Tora L., Robert F., Krasnov A.N., Evgen’ev M.B., Kadonaga J.T., Georgieva S.G., Tora L., Krasnov A.N., Evgen’ev M.B., Kadonaga J.T., Georgieva S.G., Tora L., Evgen’ev M.B., Kadonaga J.T., Georgieva S.G., Tora L., Kadonaga J.T., Georgieva S.G., Tora L., Georgieva S.G., Tora L., Tora L. Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc. Natl. Acad. Sci. 2005;102:18087–18092. doi: 10.1073/pnas.0509063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Groner Y., Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- Morris D.P., Michelotti G.A., Schwinn D.A., Michelotti G.A., Schwinn D.A., Schwinn D.A. Evidence that phosphorylation of the RNA polymerase II carboxyl-terminal repeats is similar in yeast and humans. J. Biol. Chem. 2005;280:31368–31377. doi: 10.1074/jbc.M501546200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V., Jane E.P., Chinwalla V., Harte P.J., Paro R., Jane E.P., Chinwalla V., Harte P.J., Paro R., Chinwalla V., Harte P.J., Paro R., Harte P.J., Paro R., Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: Dynamic changes during early Drosophilaembryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M., Conrad N.K., Bregman D.B., Corden J.L., Conrad N.K., Bregman D.B., Corden J.L., Bregman D.B., Corden J.L., Corden J.L. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 1999;274:27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
- Pinaud S., Mirkovitch J., Mirkovitch J. Regulation of c-fos expression by RNA polymerase elongation competence. J. Mol. Biol. 1998;280:785–798. doi: 10.1006/jmbi.1998.1905. [DOI] [PubMed] [Google Scholar]
- Rasmussen E.B., Lis J.T., Lis J.T. Short transcripts of the ternary complex provide insight into RNA polymerase II elongational pausing. J. Mol. Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Denu J.M., Allis C.D., Denu J.M., Allis C.D., Allis C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Rougvie A.E., Lis J.T., Lis J.T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Rougvie A.E., Lis J.T., Lis J.T. Postinitiation transcriptional control in Drosophila melanogaster. Mol. Cell. Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Kooperberg C., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., van Leeuwen F., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., Gottschling D.E., O’Neill L.P., Turner B.M., Delrow J., O’Neill L.P., Turner B.M., Delrow J., Turner B.M., Delrow J., Delrow J., et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R.J., III, Belotserkovskaya R., Reinberg D., Belotserkovskaya R., Reinberg D., Reinberg D. Elongation by RNA polymerase II: The short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Swantek D., Gergen J.P., Gergen J.P. Ftz modulates Runt-dependent activation and repression of segment-polarity gene transcription. Development. 2004;131:2281–2290. doi: 10.1242/dev.01109. [DOI] [PubMed] [Google Scholar]
- Tracey W.D., Jr., Ning X., Klingler M., Kramer S.G., Gergen J.P., Ning X., Klingler M., Kramer S.G., Gergen J.P., Klingler M., Kramer S.G., Gergen J.P., Kramer S.G., Gergen J.P., Gergen J.P. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154:273–284. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.A., Taxman D.J., Lu Q., Gilmour D.S., Taxman D.J., Lu Q., Gilmour D.S., Lu Q., Gilmour D.S., Gilmour D.S. Molecular architecture of the hsp70 promoter after deletion of the TATA box or the upstream regulation region. Mol. Cell. Biol. 1997;17:3799–3808. doi: 10.1128/mcb.17.7.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J.C., VanderZwan C., Xu X., Swantek D., Tracey W.D., Gergen J.P., VanderZwan C., Xu X., Swantek D., Tracey W.D., Gergen J.P., Xu X., Swantek D., Tracey W.D., Gergen J.P., Swantek D., Tracey W.D., Gergen J.P., Tracey W.D., Gergen J.P., Gergen J.P. Distinct in vivo requirements for establishment versus maintenance of transcriptional repression. Nat. Genet. 2002;32:206–210. doi: 10.1038/ng942. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Yamaguchi Y., Benjamin L.R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D., Yamaguchi Y., Benjamin L.R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D., Benjamin L.R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D., Larsson J., Lambertsson A., Handa H., Gilmour D., Lambertsson A., Handa H., Gilmour D., Handa H., Gilmour D., Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes & Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Lee C., Fan R., Smith M.J., Yamaguchi Y., Handa H., Gilmour D.S., Lee C., Fan R., Smith M.J., Yamaguchi Y., Handa H., Gilmour D.S., Fan R., Smith M.J., Yamaguchi Y., Handa H., Gilmour D.S., Smith M.J., Yamaguchi Y., Handa H., Gilmour D.S., Yamaguchi Y., Handa H., Gilmour D.S., Handa H., Gilmour D.S., Gilmour D.S. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 2005;33:1269–1279. doi: 10.1093/nar/gki274. [DOI] [PMC free article] [PubMed] [Google Scholar]