Abstract
The nonsense-mediated mRNA decay (NMD) pathway selectively degrades mRNAs harboring premature termination codons (PTCs). Seven genes (smg-1–7, for suppressor with morphological effect on genitalia) that are essential for NMD were originally identified in the nematode Caenorhabditis elegans, and orthologs of these genes have been found in several species. Whereas in humans NMD is linked to splicing, PTC definition occurs independently of exon boundaries in Drosophila. Here, we have conducted an analysis of the cis-acting sequences and trans-acting factors that are required for NMD in C. elegans. We show that a PTC codon is defined independently of introns in C. elegans and, consequently, components of the exon junction complex (EJC) are dispensable for NMD. We also show a distance-dependent effect, whereby PTCs that are closer to the 3′ end of the mRNA are less sensitive to NMD. We also provide evidence for the existence of previously unidentified components of the NMD pathway that, unlike known smg genes, are essential for viability in C. elegans. A genome-wide RNA interference (RNAi) screen resulted in the identification of two such novel NMD genes, which are essential for proper embryonic development, and as such represent a new class of essential NMD genes in C. elegans that we have termed smgl (for smg lethal). We show that the encoded proteins are conserved throughout evolution and are required for NMD in C. elegans and also in human cells.
Keywords: Nonsense-mediated decay, smg genes, RNAi screens, exon junction complex, C. elegans
Nonsense-mediated mRNA decay (NMD) is a highly conserved surveillance mechanism present in all eukaryotes examined that prevents the synthesis of truncated proteins. It does so by promoting the degradation of mRNAs containing premature translation termination codons (PTCs) (for a recent review, see Maquat 2004). The functional importance of NMD is highlighted by the fact that this pathway targets for degradation a wide array of endogenous transcripts, as well as erroneously processed transcripts and transcripts carrying spontaneous mutations (for review, see Rehwinkel et al. 2006). It is proposed that NMD modulates the phenotypic outcome of many diseases, since ∼30% of inherited genetic disorders are caused by frameshift or nonsense mutations that generate premature termination codons (PTCs). Therefore, effectors of NMD are potential targets for therapeutic intervention (Frischmeyer and Dietz 1999; Holbrook et al. 2004).
Genetic screens in the nematode Caenorhabditis elegans identified seven genes that are required for the degradation of nonsense mutant mRNAs of the unc-54 myosin heavy chain gene (Hodgkin et al. 1989; Pulak and Anderson 1993; Cali et al. 1999). In addition to their suppression-of-unc phenotype, these mutations cause abnormal morphogenesis of the male bursa and the hermaphrodite vulva, and accordingly these genes were termed smg-1–7 (for suppressor with morphological effect on genitalia). In the yeast Saccharomyces cerevisae, three genes, termed UPF1–3, that are necessary for enhanced turnover of mRNAs containing a premature stop codon were identified and are orthologs of the C. elegans smg-2, smg-3, and smg-4 (Leeds et al. 1991, 1992). Subsequently, orthologs for the smg genes have been identified in several species. The smg-1, smg-5, smg-6, and smg-7 genes, which are absent in yeast cells, are required for NMD in C. elegans, Drosophila melanogaster, and in mammalian cells (for review, see Conti and Izaurralde 2005).
The key component of the NMD pathway is the protein SMG-2/UPF1, which is an ATP-dependent RNA helicase that undergoes cycles of phosphorylation/dephosphorylation that are required for NMD. Indeed, all known smg genes other than smg-2 affect the state of SMG-2 phosphorylation. It has been shown that SMG-1, SMG-3, and SMG-4 are needed to phosphorylate SMG-2, whereas SMG-5, SMG-6, and SMG-7 are needed for its dephosphorylation (Page et al. 1999). The smg-1 gene encodes a protein kinase of the phosphatidylinositol kinase superfamily of protein kinases, which is required for SMG-2/UPF1 phosphorylation (Yamashita et al. 2001; Grimson et al. 2004). SMG-5 associates with SMG-7 and also with the catalytic and structural subunits of the protein phosphatase 2A (PP2A), suggesting that the role of this complex is to direct PP2A to its SMG-2/UPF1 substrate (Anders et al. 2003). It was revealed that the SMG-5–7 proteins have a 14–3–3-like phosphoserine-binding domain, which is essential for binding to phosphorylated SMG-2/UPF1 (Fukuhara et al. 2005). The SMG-7 protein is a key factor that links mRNA surveillance and mRNA decay by forming a complex with SMG-5 and UPF1 and targeting bound mRNAs for mRNA decay (Unterholzner and Izaurralde 2004). Whereas NMD is not essential in yeast or nematodes, targeted disruption of Rent1/UPF1 in mice resulted in embryonic lethality (Medghalchi et al. 2001).
A critical feature of the NMD pathway is how a normal termination codon is distinguished from a PTC that would trigger mRNA decay. In mammalian cells, NMD is linked to splicing since the position of a termination codon relative to the last intron will determine whether it is interpreted as premature. Termination codons that are followed by an intron positioned >50–55 nucleotides (nt) downstream elicit NMD (Nagy and Maquat 1998). Consistent with this rule, for the most part, intronless genes containing PTCs are immune to NMD (Maquat and Li 2001), with a few exceptions discussed below. The link between splicing and NMD is mediated by the exon junction complex (EJC), a complex that is deposited on every exon junction as a consequence of the splicing process and serves as a binding platform for factors involved in nonsense-mediated decay, mRNA export, and mRNA localization (Le Hir et al. 2001; Lykke-Andersen et al. 2001; for review, see Tange et al. 2004). The EJC functions as a molecular signal to trigger NMD, since the presence of a PTC causes the ribosome to terminate prematurely, leaving downstream EJC marks that are not removed from the mRNA, which in turn recruit the NMD machinery and trigger mRNA degradation. Recently, it was suggested that, in the presence of a premature translation termination event, UPF1 is recruited to PTC-containing mRNAs via interactions with the release factors eRF1 and eRF3, leading to the assembly of the SURF complex comprising SMG1, UPF1, eRF1, and eRF3. Subsequently, the SURF complex interacts with UPF2, UPF3, and EJC components bound to a downstream exon–exon boundary, resulting in the formation of the DECID (decay-inducing complex) complex that triggers UPF1 phosphorylation and the dissociation of eRF1 and eRF3 (Kashima et al. 2006; for review, see Behm-Ansmant and Izaurralde 2006). Despite the conservation of the smg genes and of EJC components, PTC definition occurs independently of exon boundaries in Drosophila, and accordingly EJC components are dispensable for NMD in this organism (Gatfield et al. 2003). Furthermore, exon boundaries are not used to define PTCs in yeast either. This is probably related to the fact that most yeast genes lack introns and, with the exception of the REF/Aly proteins, there are no identifiable homologs for the EJC components (for review, see Culbertson and Leeds 2003). One model suggests that a loosely defined downstream element in the 3′ untranslated region (UTR), termed DSE, is associated with RNA-binding proteins such as yeast hnRNP-like protein Hrp1/Nab4, and that this complex triggers NMD (Gonzalez et al. 2000). Alternatively, the “faux 3′UTR” model proposes that translation termination at a nonsense codon is intrinsically aberrant, due to the fact that a PTC is not in an appropriate context. In support of this hypothesis, tethering of poly(A)-binding protein in the proximity of a PTC mimicked a normal 3′UTR and suppressed NMD (Amrani et al. 2004, 2006). Thus, despite the conservation of the NMD machinery, different mechanisms have evolved to define PTCs and discriminate them from natural stop codons in metazoa (for review, see Conti and Izaurralde 2005).
Here, we have conducted a systematic analysis of the cis-acting sequences and trans-acting factors that are required for PTC recognition in the nematode C. elegans. We have used transgenic C. elegans strains expressing GFP/LacZ reporter constructs either with a natural stop codon or harboring PTCs. We show that the introduction of a PTC in this reporter results in a robust NMD response, as determined by the lack of GFP expression and a decrease of the corresponding mRNA abundance. Use of intronless reporters established that introns are not required to define a PTC, and we show that, accordingly, EJC components are not essential for NMD in this organism. Also, we demonstrate a distance effect whereby PTCs that are closer to the 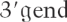 of the mRNA are less sensitive to NMD. These results underscore that despite the high conservation of the NMD machinery, different mechanisms to define a PTC and elicit NMD are found throughout evolution. We also provide evidence for the existence of several novel factors that are essential for organismal viability and that also participate in the NMD pathway in C. elegans. Use of a genome-wide RNA interference (RNAi) screen resulted in the identification of two such novel factors that are highly conserved throughout evolution. We show that these two factors are required for NMD both in C. elegans and also in human cells.
of the mRNA are less sensitive to NMD. These results underscore that despite the high conservation of the NMD machinery, different mechanisms to define a PTC and elicit NMD are found throughout evolution. We also provide evidence for the existence of several novel factors that are essential for organismal viability and that also participate in the NMD pathway in C. elegans. Use of a genome-wide RNA interference (RNAi) screen resulted in the identification of two such novel factors that are highly conserved throughout evolution. We show that these two factors are required for NMD both in C. elegans and also in human cells.
Results
NMD reporters in C. elegans
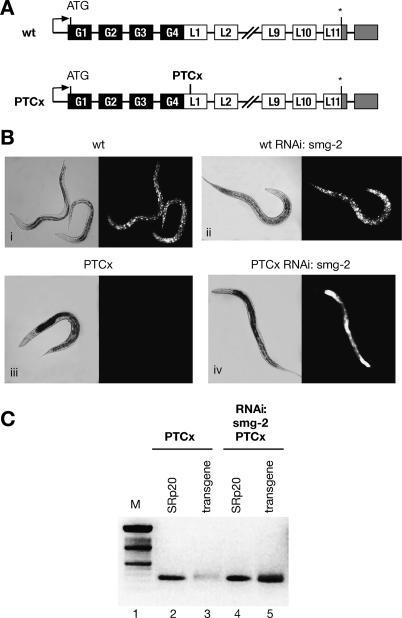
Although the requirement of smg genes for NMD in C. elegans is well documented, little is known about the mechanism of PTC definition in this organism. Here, we have created a series of NMD reporters based on the multiple synthetic-intron-containing lacZ∷GFP reporter vector pDP96.04 driven by a ubiquitously expressed promoter (Fire et al. 1990; Roberts et al. 2003). We tested transgenic C. elegans strains expressing GFP/LacZ fusions either containing a natural stop codon (referred to as “wild type”) or harboring a PTC (PTCx, placed 3513 nt upstream of the natural stop codon) (Fig. 1). As predicted, a transgenic strain expressing a wild-type reporter exhibits ubiquitous GFP expression throughout all developmental stages (Fig. 1B). Introduction of a PTC in the first exon of the LacZ gene (PTCx) in this reporter resulted in the lack of GFP expression (Fig. 1B, panel iii). In order to determine whether the observed results were indeed due to NMD, we deactivated SMG-2/UPF1, a key regulator of NMD, by RNAi. Although knockdown of SMG-2 had no effect on the wild-type reporter expression, its depletion led to the stabilization of a PTCx-containing mRNA and rescued GFP expression (Fig. 1B, cf. panels ii and iv). We confirmed that the lack of GFP expression was indeed due to NMD-induced mRNA degradation by monitoring the PTCx mRNA level before and after RNAi-mediated depletion of SMG-2/UPF1 by semiquantitative RT–PCR. As seen in Figure 1C, the level of transgenic reporter mRNA is very low in transgenic worms carrying the PTCx reporter (lane 3); however, a significant increase in reporter mRNA level was seen upon SMG-2 inactivation (lane 5). These experiments demonstrate that introduction of a PTC into the GFP/Lacz reporter induces a robust NMD response. Thus, it constitutes an excellent experimental system to address cis-acting sequences that are important to define a PTC.
Figure 1.
A PTCx transgene harboring a nonsense codon is subject to NMD in C. elegans. (A) Schematic representation of GFP/LacZ reporters. Black boxes represent GFP exons, white boxes represent LacZ exons, and gray boxes represent the 3′UTR. Intervening black lines correspond to introns. The natural stop codon is indicated by an asterisk. In the PTCx reporter, the position where a nonsense codon was generated by site-directed mutagenesis is indicated. (B, panel i) Transgenic worms carrying a wild-type reporter show ubiquitous GFP expression. (Panel ii) This expression is not affected by the depletion of SMG-2. Introduction of a nonsense codon (PTCx) resulted in lack of GFP expression (panel iii), whereas inactivation of NMD by SMG-2 RNAi led to strong GFP expression (panel iv). (C) The level of the PTCx reporter mRNA was monitored by semiquantitative RT–PCR. In the PTCx strain, the level of reporter mRNA is very low (lane 3); however, the level of transgene mRNA is significantly increased upon depletion of the smg-2 transcript (lane 5).
The position of the PTC within the transcript is critical for NMD
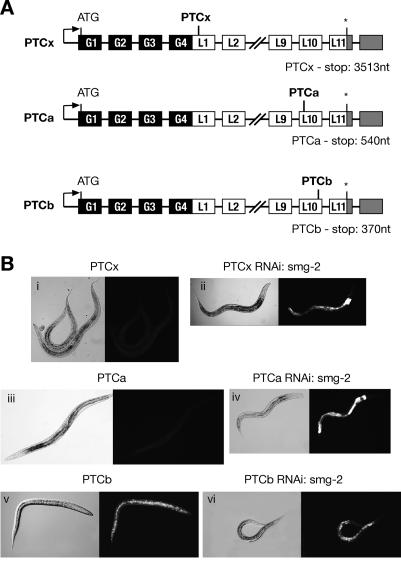
In order to investigate whether the position of the nonsense codon has an effect on its ability to elicit NMD we created two further reporters. In the PTCa reporter, a C-to-T change at position 5172 resulted in a TAA stop codon, whereas in PTCb, a G-to-A change at position 5338 resulted in a TAG stop codon. This placed PTCa and PTCb at 540 and 370 nt upstream of the natural stop codons, respectively. Thus, in both reporters the nonsense codon lies within the penultimate exon, just one exon upstream of the natural stop codon (Fig. 2A). Transgenic lines carrying the PTCa reporter showed transgene GFP expression only upon SMG-2 depletion, indicating that the PTCa transgene is subject to NMD (Fig. 2B, panels iii,iv). In contrast, we found that the PTCb reporter harboring the nonsense codon closer to the natural stop codon (170 nt downstream from PTCa) did not undergo NMD, as shown by the robust GFP expression that was not altered by inactivation of NMD by SMG-2 depletion (Fig. 2B, panels v,vi). Thus, introduction of a PTC in the penultimate exon resulted in different outcomes, depending on the distance from the natural stop or poly(A) site. This experiment strongly suggests a distance-dependent effect, whereby PTCs that are upstream of a certain boundary are not sensitive to NMD. This observation is consistent with the “faux 3′UTR” model proposed in yeast, where a stop codon is recognized as premature if placed outside signals from the normal 3′UTR (Amrani et al. 2004).
Figure 2.
The position of the PTC within the transgene influences NMD, demonstrating a polarity effect. (A) Schematic representation of PTCx, PTCa, and PTCb reporters. Black boxes represent GFP exons, white boxes represent LacZ exons, and gray boxes represent the 3′UTR. Intervening black lines correspond to introns. The natural stop codon is indicated by an asterisk. The positions of nonsense codons within the penultimate exon of the LacZ gene are indicated. (B) Transgenic worms carrying the PTCx and PTCa reporter lack GFP expression (panels i,iii, respectively); however, GFP expression is restored upon SMG-2 depletion (panels ii,iv, respectively). Transgenic worms carrying the PTCb reporter show robust GFP expression (panel v), which is not altered by the inactivation of NMD by SMG-2 depletion (panel vi).
Splicing is not required for PTC recognition in C. elegans
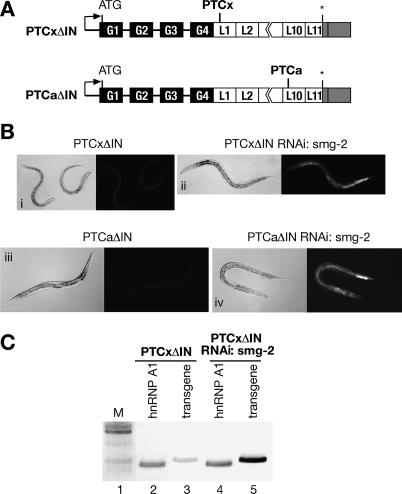
In human cells, it has been clearly established that pre-mRNA splicing plays a critical role in defining PTCs, and this is mediated by components of the EJC. In contrast, in Drosophila, despite the conservation of the splicing and NMD machineries, definition of a PTC occurs independently of exon boundaries and, accordingly, the components of the EJC are dispensable for NMD (Gatfield et al. 2003). It is not clear, however, how a PTC is defined in the nematode C. elegans. In order to address this issue, we removed the introns within the LacZ gene from the PTCx and PTCa reporters to create the PTCxΔIN and PTCaΔIN reporters, respectively (Fig. 3A). This placed the nonsense codons in the context of the last exon. Thus, if PTC definition in C. elegans is governed by the same rules as in the mammalian system, neither of the reporters should be subject to NMD, as there is no splicing and consequently no EJC complex being deposited between the PTC and the natural stop.
Figure 3.
Splicing is not required for PTC recognition in C. elegans. (A) Schematic representation of PTCxΔIN and PTCaΔIN reporters. Black boxes represent GFP exons, white boxes represent LacZ exons, and gray boxes represent the 3′UTR. Intervening black lines correspond to introns. The natural stop codon is indicated by an asterisk. Introns within the LacZ gene were removed, placing the nonsense codons within the context of the last exon. Nonsense mutations are the same as in the PTCx and PTCa reporters, respectively. (B) A transgene with no introns downstream from the PTC is subject to NMD. Transgenic worms carrying the PTCxΔIN reporter lack GFP expression (panel i); however, GFP expression is restored upon the inactivation of NMD by SMG-2 depletion (panel ii). Similarly, transgenic worms carrying the PTCaΔIN reporter lack GFP expression (panel iii), which is restored upon the inactivation of NMD by SMG-2 depletion (panel iv). (C) The level of PTCxΔIN reporter mRNA was monitored by semiquantitative RT–PCR. In the PTCxΔIN strain, the level of reporter mRNA is very low (lane 3); however, it is significantly increased upon depletion of SMG-2 (lane 5).
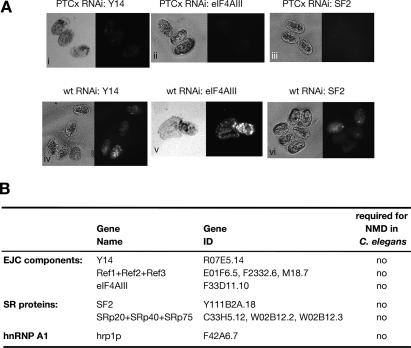
We found that transgenic strains expressing the PTCxΔIN or PTCaΔIN reporters did not show GFP expression (Fig. 3B); however, depletion of SMG-2 led to the stabilization of PTCxΔIN- and PTCaΔIN-containing mRNAs, as seen by a significant increase in reporter mRNA levels (Fig. 3C; data not shown), and consequently rescued GFP expression (Fig. 3B). This experiment clearly demonstrates that introns are not required to define a PTC in C. elegans. Accordingly, we next showed that microinjection-induced RNAi depletion of EJC components such as Y14, RNPS1, eIF4AIII, and the REF proteins did not rescue GFP expression of the PTCx construct, strongly suggesting that EJC factors are not essential for NMD in nematodes (Fig. 4; data not shown). Furthermore, depletion of SR proteins, which have been shown to activate NMD in human cells (Zhang and Krainer 2004), did not restore GFP expression, suggesting that SR proteins are not essential for NMD in nematodes (Fig. 4).
Figure 4.
EJC components and SR proteins are dispensable for NMD in C. elegans. (A) Microinjection-induced RNAi depletion of either Y14 (panel i), eIF4AIII (panel ii), or SF2/ASF (panel iii) in the PTCx strain does not rescue GFP expression in the affected embryos. As a control, depletion of Y14 (panel iv), eIF4AIII (panel v), and SF2/ASF (panel vi) does not prevent the GFP expression in the embryos of the wild-type strain. The RNAi-mediated depletion resulted in the expected phenotypes, as previously described (Longman et al. 2000, 2003). (B) Table summarizing the role of EJC components and splicing factors in NMD.
A genome-wide RNAi-based screen to identify new genes required for NMD
Previous extensive genetic screens have led to the identification of seven smg genes that are essential for NMD in C. elegans, and most of the mutations were found to be alleles of smg-1, smg-2, or smg-5 (Cali et al. 1999). As these genetic screens selected viable and relatively healthy mutants, only viable smg mutants were identified. In order to assess whether there are additional essential genes in the nematode genome that are required for NMD, we performed a preliminary genetic screen using EMS-mediated mutagenesis of the PTCx strain. We searched for the presence of GFP expression in the F2 (second-generation progeny). We observed the presence of viable mutants that were most likely alleles of the existing smg genes. Interestingly, we also observed GFP expression in dead embryos or dead/sick larvae that were most likely the result of mutations in genes required for both organismal viability and NMD. Although we could not rule out that some of the GFP-expressing nonviable mutants might carry background lethal mutations unconnected with NMD, this suggested the existence of previously uncharacterized essential NMD factors.
Technical advances in RNAi methodology and the availability of the complete genome sequence have enabled high-throughput, genome-wide RNAi analysis in C. elegans, Drosophila, and mammalian tissue culture cells. This experimental approach has been extensively used in C. elegans to identify genes involved in processes as diverse as genome stability (Pothof et al. 2003), transposon silencing (Vastenhouw et al. 2003), polyglutamine aggregation (Nollen et al. 2004), and longevity (Hamilton et al. 2005).
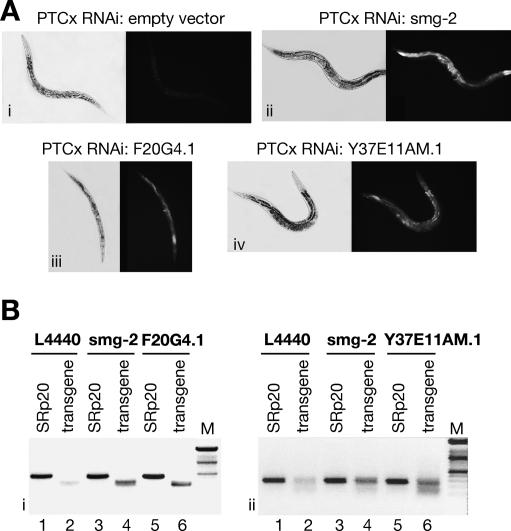
We used a commercially available RNAi bacterial feeding library containing 16,757 clones that target ∼86% of the total C. elegans predicted genes (Fraser et al. 2000; Kamath et al. 2003). In order to identify genes involved in nonsense-mediated decay, we used the C. elegans strain expressing a GPF-based reporter harboring a PTC that was described above (PTCx construct) (Fig. 1B). The reporter transgene was integrated in the C. elegans genome. Animals carrying the PTCx reporter lack GFP expression, since this reporter is subject to NMD as described above (Fig. 1B,C). The genes were identified by the criterion that their silencing restores GFP expression, as was demonstrated in controls by inactivation of the bona fide NMD factor, SMG-2/UPF1 (Fig. 1). Thus, in a high-throughput assay we screened for the appearance of green worms, dead or alive, in the PTCx strain. As a negative control we fed PTCx animals empty RNAi vector, which had no effect on the level of GFP expression (Fig. 5A, panel i). As a positive control we used a clone expressing SMG-2/UPF1, which induced strong GFP expression by abrogating the NMD process (Fig. 5A, panel ii). By screening the whole library we identified two RNAi clones that scored positive by increased GFP expression, as compared with the empty vector, or positive control (Fig. 5A, panels iii,iv). These clones were validated by RNAi mediated by injection of double-stranded RNA (dsRNA) derived from the respective genes, F20G4.1 and Y37E11AM.1 (data not shown). Semiquantitative RT–PCR analysis showed that depletion of both genes led to the increase of reporter mRNA level that was comparable with the increase induced by the depletion of SMG-2, a bona fide NMD factor (Fig. 5B). Furthermore, depletion of F20G4.1 and Y37E11AM.1 by dsRNA injections resulted in early embryonic lethality prior to morphogenesis, indicating that these newly identified NMD factors are essential for proper embryonic development (data not shown). As such, they represent a new class of essential NMD genes in C. elegans that we have termed smgl (for smg lethal; F20G4.1 and Y37E11AM.1 have been named smgl-1 and smgl-2, respectively).
Figure 5.
An RNAi screen led to the identification of two novel NMD factors in C. elegans. (A, panel i) As a negative control, RNAi was induced with empty vector. (Panel ii) As a positive control, RNAi was induced with the smg-2 clone, which led to strong GFP expression within the PTCx strain. Depletion of the F20G4.1 clone (panel iii) and the Y37E11AM.1 clone (panel iv) resulted in increased GFP expression. (B, panels i,ii) The level of PTCx reporter mRNA was monitored by semiquantitative RT–PCR. In the PTCx strain, the level of reporter mRNA is very low (lane 2); however, it is significantly increased upon depletion of SMG-2. (Panels i,ii) Depletion of F20G4.1 or Y37E11AM.1 by RNAi led to an increase in PTCx mRNA level that is comparable with the depletion of SMG-2, a bona fide NMD factor (cf. lanes 4 and 6). A control mRNA encoding the splicing factor SRp20 is not altered by any of these treatments.
Newly identified NMD factors are functionally conserved in human
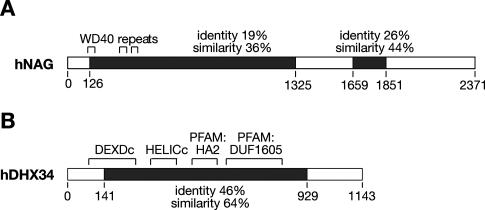
Both newly identified genes are highly conserved throughout evolution, based on sequence homology (http://www.ensembl.org), and have clear orthologs in mouse, human, and fugu, among others, but are absent in yeast (Supplementary Figs. S1, S2). The C. elegans SMGL-1 corresponds to the human NAG, with two regions of increased homology between the species (36% and 44% similarity, respectively) (Fig. 6A; Supplementary Fig. S1). The human homolog of the C. elegans smgl-2 gene is the DHX34 gene (DEAH box protein 34). SMGL-2 and DHX34 share a large central portion with high sequence conservation (46% identity and 64% similarity) (Fig. 6B; Supplementary Fig. S2). The analysis of hNAG identified three WD40 (β-propeller domain) repeats in the N-terminal part of the protein (C. Ponting, pers. comm.). This gene was previously found to be amplified in human neuroblastomas (Wimmer et al. 1999; Scott et al. 2003), but its function is not currently known. All WD repeat-containing proteins share a common function of coordinating multiprotein complex assemblies, forming a scaffold for protein–protein interactions (for review, see Smith et al. 1999). The human DHX34 gene is a member of ATP-dependent RNA helicase family, as it contains four domains associated with this family of proteins (Fig. 6B). Although the role of this helicase is not described, DEAD/DEAH-box helicases are commonly involved in many aspects of RNA metabolism including transcription, pre-mRNA splicing, translation, and mRNA decay (for recent reviews, see Rocak and Linder 2004; Fuller-Pace 2006; Linder 2006).
Figure 6.
Sequence conservation and functional domains of the newly characterized NMD factors. (A) Schematic representation of the human NAG, which is the homolog of C. elegans SMGL-1. Black boxes represent the regions of sequence conservation within proteins; the level of conservation is indicated. WD40 domains within the N terminus of the protein are indicated by brackets above the protein. (B) Schematic representation of the human DHX34, which is the homolog of C. elegans SMGL-2. Black boxes represent the regions of sequence conservation within proteins; the level of conservation is indicated. Functional domains associated with ATP-dependent RNA helicases (DEXDc, HELICc, PFAM:HA2, and PFAM:DUF 1605) are indicated by brackets above the protein.
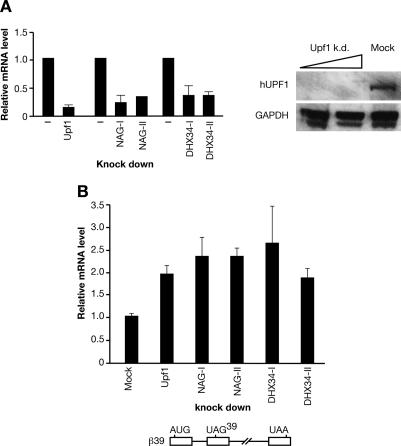
Next, we analyzed whether the human homologs of these two novel NMD factors identified in C. elegans, hNAG, and hDHX34 also had a role in NMD in human cells. First, HeLa cells were individually depleted of either hNAG or hDHX34 using two different nonoverlapping small interfering RNA (siRNA) pools (Fig. 7A). Then, depleted cells were transfected with a β-globin NMD reporter (β39), or with a wild-type β-globin reporter (βwt) as a control (Lykke-Andersen et al. 2000). Total RNA was isolated 24 h post-transfection and the steady-state level of β-globin reporter mRNA was analyzed by real-time RT–PCR. As expected, the level of wild-type β-globin mRNA remained unchanged regardless of UPF1, NAG, or DHX34 depletion (Supplementary Fig. S3). In contrast, depletion of UPF1 resulted in an increased level of the β-globin NMD reporter (β39) mRNA, compared with mock-depleted cells (Fig. 7B). Importantly, depletion of both hNAG and hDHX34 also led to increased levels of β-globin NMD reporter mRNA (Fig. 7B). These results show that hNAG and hDHX34 genes identified by sequence homology with the newly described NMD factors in C. elegans are also functionally conserved NMD factors in humans.
Figure 7.
The human homologs of the novel C. elegans factors are required for NMD in human cells. (A) HeLa cells were mock-depleted, or depleted of UPF1, NAG, and DHX34 using two nonoverlapping siRNA pools. The level of knockdown of individual depletions was measured by real-time RT–PCR. The graph represents data of at least three independent experiments for each siRNA pool. For UPF1, the level of depletion was also assessed by Western blotting. (B) Depletion of both NAG and DHX34 genes, accomplished by two nonoverlapping siRNA pools, led to the increase of the β-globin reporter (β39) mRNA level. Depleted HeLa cells were transiently transfected with NMD β-globin reporter (β39), together with an EGFP vector as a transfection control. Total RNA was isolated 24 h post-transfection and the steady-state level of the reporter mRNA was measured by real-time RT–PCR. A schematic representation of the NMD β-globin reporter (β39) is below the graph.
Discussion
Seven smg genes were identified as molecular effectors of nonsense-mediated decay in C. elegans; however, it is still not clear how a nonsense codon is interpreted as a PTC triggering NMD. Most C. elegans genes have introns, suggesting that exon boundaries might play a role in defining a PTC, as it happens in human cells, and circumstantial evidence has been accumulated supporting this notion (for review, see Mango 2001). For instance, analysis of nonsense alleles of the unc-54 gene showed that the unc-54(e1328) and unc-54(e1300) alleles, which harbor a PTC in the penultimate or in the ultimate exon, respectively, only displayed modest mRNA degradation (Dibb et al. 1985; Bejsovec and Anderson 1988). Six additional mutant RNAs harboring a stop codon in the last or penultimate exon were shown to be only intermediate targets of mRNA surveillance (Mango 2001). This led to the proposition that, if exon boundaries play a role in PTC definition in C. elegans, at least two introns are needed (Mango 2001). It should be noted that these examples are not incompatible with a boundary-dependent or polarity effect whereby distance from the natural stop codon or the poly(A) site influences the strength of NMD. Here, we were able to show that PTCs can be defined independently of exon boundaries, as shown by the strong NMD elicited by a GFP reporter lacking introns downstream from the PTC (PTCxΔIn and PTCaΔIn). This is in agreement with our observation that EJC components are not required for eliciting NMD in C. elegans (Fig. 4). Accordingly, the dpy-5(e61) allele undergoes NMD despite the fact that the dpy-5 gene has only one exon (Hodgkin et al. 1989). In mammalian cells the −55-nt boundary rule is sometimes overruled, as shown in a T-cell receptor (TCR)-β gene that did not conform to this rule (Wang et al. 2002). This illustrates that, at least in some cases, there seems to be a distance effect, with nonsense codons distant from the last intron being more effective in eliciting NMD (Wang et al. 2002). Moreover, in the case of an immunoglobulin-μ mRNA harboring a PTC, it was recently demonstrated that NMD occurs independently of introns and that the distance between the termination codon and the poly(A) tail is a critical determinant for EJC-independent NMD (Buhler et al. 2006).
Nonsense-mediated decay has a prominent role in gene expression since it not only regulates transcripts harboring nonsense mutations, but also regulates an important fraction of the transcriptome (Rehwinkel et al. 2006). For instance, global changes associated with UPF loss of function affect the abundance of hundreds of mRNAs in yeast, Drosophila, and humans (Lelivelt and Culbertson 1999; Mendell et al. 2004; Rehwinkel et al. 2005).
Use of an RNAi screen strategy resulted in the identification of two novel NMD factors that are highly conserved during evolution. Interestingly, these two factors have a role in NMD not only in C. elegans but also in human cultured cells. Both smgl-1 (NAG) and smgl-2 (DHX34) are genes required for viability in C. elegans and, since NMD is not an essential process in this organism, this suggests that they may be involved in additional cellular functions. Interestingly, many cellular functions other than NMD have been described for several smg genes. For instance, three smg genes, smg-2, smg-5, and smg-6, were shown to be required for persistence of RNAi in C. elegans, establishing a connection between mRNA surveillance and RNAi (Domeier et al. 2000). In the case of SMG-2/UPF1, this essential NMD factor also has been shown to be required for genome stability, since its depletion induces cell cycle arrest early in S phase (Azzalin and Lingner 2006a; for review, see Azzalin and Lingner 2006b). Moreover, SMG-2/UPF1 is also involved in alternative mRNA degradation pathways, which are NMD-independent, by being recruited to 3′UTRs by the RNA-binding protein Staufen 1 (Kim et al. 2005) or to the 3′ end of histone mRNAs via the stem–loop-binding protein (Kaygun and Marzluff 2005). The hSMG1 gene, which functions in UPF1/SMG-2 phosphorylation, has been shown to have a role in genotoxic stress (Brumbaugh et al. 2004). In the case of DHX34, it is possible to hypothesize that the RNA helicase activity of this protein is involved both in NMD and in other aspects of RNA metabolism that are essential for the cell.
An important aspect of this study is that we identified two novel highly conserved factors that are required for the NMD in an organism where this pathway is defined independently of splicing (C. elegans) and in humans, where exon boundaries play an important, if not essential, role for this surveillance mechanism. Future work will elucidate whether these factors are recruited to the NMD complex in different manners in both organisms.
Materials and methods
Reporters
A series of NMD reporters based on the lacZ∷GFP reporter vector pDP96.04 (Fire et al. 1990; Roberts et al. 2003), which is driven by the ubiquitous sec-23 promoter, were created either in a wild-type version or harboring a PTC in different positions of the LacZ gene. Nonsense mutations were created by site-directed mutagenesis of the wild-type reporter. In the PTCx construct, a C-to-T change was introduced at position 2193, creating a TAA stop codon. In the PTCa reporter, a C-to-T change at position 5172 resulted in a TAA stop codon, whereas in the PTCb reporter, a G-to-A change was introduced at position 5338, creating a TAG stop codon. The PTCxΔIN and PTCaΔIN reporters carry the same mutation as PTCx and PTCa, respectively; however, the LacZ part of the reporter was replaced by the intronless LacZ gene from vector pPD95.93 (Fire et al. 1990).
Transgenic strains
Transgenic strains were created by microinjections. Young adults from a DR96 strain carrying a mutant allele of unc-76 gene were injected into their gonads with 10–20 ng/μL of the appropriate reporter, together with 20 ng/μL of the unc-76 rescue plasmid and 100 ng/μL of carrier DNA. The F1 progeny carrying the transgene were identified by rescued movement and transferred onto fresh plates, 10–20 individuals per plate. From each plate one F2 animal with wild-type movement was selected to establish a line, and for each reporter three to five lines were generated. For use in the genome-wide RNAi screen, the PTCx reporter was integrated into the genome. A transgenic line with lower transmission carrying the PTCx reporter was γ-irradiated, and a worm with 100% transmission of the reporter was backcrossed four times with the Bristol N2 strain to establish the PTCxi strain.
dsRNA preparation and microinjection
Templates for RNA synthesis were generated by PCR from C. elegans genomic DNA using gene-specific primers with T3 and T7 promoter sequences added onto forward (F) and reverse (R) primers, respectively. PCR products were gel-purified and used as templates for in vitro RNA synthesis with T3 and T7 RNA polymerase (Boehringer Mannheim) following instructions from the manufacturers. RNA was dissolved in sterile water. DsRNA was assembled by mixing equal amounts of sense and antisense RNA followed by incubation for 10 min at 68°C and then for 30 min at 37°C. For each gene, 10–15 young adult hermaphrodites were injected with dsRNA into the gonad. Injected worms were left to recover and egg lay for 16 h. Then, injected animals were transferred onto individual plates and the phenotype was observed in the F1 progeny. The affected progeny were examined using DIC or fluorescence microscopy.
Primers used were the following: T3 sequence, ATTAACCC TCACTAAAGGGAAG; T7 sequence, TAATACGACTCACTA TAGG; eIF4AIIIF, GATGATATGGCAACAGTGG; eIF4AIIIR, CTTGTGGAACATCTAATCC; smg-2F, AAATCAGTGATGG AGTCG; smg-2R, ATCAGCCGCCGATGAGCACG; F20G4.1F, GAAGTTGTTTGGATGCGGC; F20G4.1R, TCACTGCATCA CGTAATCG; Y37E11AM.1F, CGCACTTTCCCATTCATCG; Y37E11AM.1R, GATCAATCCTCCTCTGCCG. Primers used for Y14 and the Ref1, Ref2, and Ref3 genes are as described in Longman et al. (2003). Primers used for SR proteins and hnRNPA1 genes are as described in Longman et al. (2000).
RT–PCR
RT–PCR was performed using SuperScriptIII One-Step RT–PCR Kit (Invitrogen) following the manufacturer’s instructions. To test the efficiency of RNAi treatment by RT–PCR, 100-μL reactions were prepared for wild-type and RNAi-treated samples. These reactions were split into identical fractions and RT–PCR analysis was performed to compare RNA levels corresponding to either a control gene or the gene(s) targeted in the RNAi experiment. After 23–28 cycles of amplification, RT–PCR products were loaded on ethidium bromide-stained agarose gel. Primers used for RT–PCR analysis were the same as the ones used for preparation of dsRNA fragments without the T3 and T7 sequences.
Genome-wide RNAi screen
The RNAi library used has been previously described (Fraser et al. 2000; Kamath et al. 2003). It was created in the laboratory of Julie Ahringer (The Gurdon Institute, Cambridge, UK) and is distributed via MRC GeneService (http://www.geneservice.co.uk/home). RNAi feeding was performed as previously described (van Haaften et al. 2004), with minor modifications. Briefly, RNAi was performed in liquid format by feeding synchronized population of PTCxi L1 larvae with bacterial clones expressing dsRNA corresponding to individual genes in 96-well plates. Plates were then scored for the appearance of GFP expression, indicating that the depleted protein is required for NMD in C. elegans.
RNAi in HeLa cells
HeLa cells were grown in DMEM (Invitrogen) supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin. siRNA-mediated depletion was performed in six-well plate format using DharmaFECT1 for all siRNA transfections, following the manufacturer’s instructions. HeLa cells that were 30%–40% confluent were transfected with the following siRNAs (100 nM each): Dharmacon siGenome SMARTpool M-020986-00-0005 (NAG-I), ON-TARGETplus SMARTpool L-020986-00-0005 (NAG-II), siGenome SMARTpool M-032233-00-0005 (DHX34-I), ON-TARGETplus SMARTpool L-032233-01-005 (DHX34-II). For depletion of hUpf1, a combination of two previously described RNA oligos was used: hUPF1-I, GAGAATCGCCTACTTCACT; and hUPF1-II RNA, GATGCAGTTCCGCTCCATT (Paillusson et al. 2005; Azzalin and Lingner 2006a). The efficiency of the knockdown was assessed by real-time RT–PCR.
Transient transfection assays
Forty-eight hours after siRNA depletion, HeLa cells were transfected using Lipofectamine 2000 following the manufacturer’s instructions. For each experiment, 0.2 μg of each of the β-globin reporters (either β39 or βwt) were cotransfected with 0.2 μg of an EGFP vector (Clontech) as a transfection control. β-Globin reporters β39 and βwt were described previously in Lykke-Andersen et al. (2000). Twenty-four hours after transfection, growth medium was removed and cells were washed with PBS. Total RNA was prepared by resuspending cells in 0.5 mL of TRI Reagent (Sigma) according to the manufacturer’s instructions. Isolated RNA was resuspended in 150 μL of water, and 1 μL was used for each RT–PCR reaction.
Western blotting
Cells from six-well plates were resuspended in 500 μL of Laemmli buffer. Then, 15 and 30 μL of each sample were subjected to SDS/polyacrylamide gel electrophoresis followed by Western blotting using mouse anti-hUpf1 antibody (1:2000) (Lykke-Andersen et al. 2000) or rabbit anti-GAPDH antibody (1:2000). Proteins were visualized by ECL (Pierse).
Real-time RT–PCR
RT–PCR was performed using SuperScript III Platinum SYBR Green One-Step qRT–PCR Kit (Invitrogen) following the manufacturer’s instructions. Real-time RT–PCT was run on an MJ Research Chromo4 Detector. Generally, 1 μL of sample RNA was used in a 20-μL reaction, and each sample was run in duplicates. To assess the level of siRNA-mediated depletion levels of UPF1, NAG and DHX34 mRNAs were normalized against GAPDH mRNA. The level of β-globin reporter mRNA was normalized against EGFP mRNA. For each independent experiment, samples were analyzed two to four times. Primers used were the following: βglobF2, CATGGTGCATCTGACTCCTG; βglobR2, TTAGGGTTGCCCATAACAGC; EGFPF, ACGTAA ACGGCCACAAGTTC; EGFPR, AAGTCGTGCTGCTTCAT GTG; GAPDHF, GAGTCAACGGATTTGGTCGT; GAPDHR1, TTGATTTTGGAGGGATCTCG; NAGF3, GAATGGTTGTT GAGCCGAAT; NAGR3, GCTCCATCCCAAGTCGAATA; DHX34F2, GAGCACCAGGTGGTGGTAGT; DHX34R2, CGA CCTGTGAGCCATACTGA; Upf1F3, CCCTCCAGAATGGT GTCACT; Upf1R3, CTTCAGCAACTTCGTGGTGA
Acknowledgments
We are grateful to Phillippe Gautier (Medical Research Council [MRC] HGU), Colin Semple (MRC HGU), and Chris Ponting (Oxford) for help with bioinformatic analysis. We thank Nick Hastie for helpful discussions. J.F.C. and D.L. were supported by the MRC. We thank Jens Lykke-Andersen for the generous gift of the β-globin reporters (β39 and βwt) and the anti-Upf1 antibody.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.417707
References
- Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A., Kervestin S., Mangus D.A., Ghosh S., Jacobson A., Mangus D.A., Ghosh S., Jacobson A., Ghosh S., Jacobson A., Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- Amrani N., Dong S., He F., Ganesan R., Ghosh S., Kervestin S., Li C., Mangus D.A., Spatrick P., Jacobson A., Dong S., He F., Ganesan R., Ghosh S., Kervestin S., Li C., Mangus D.A., Spatrick P., Jacobson A., He F., Ganesan R., Ghosh S., Kervestin S., Li C., Mangus D.A., Spatrick P., Jacobson A., Ganesan R., Ghosh S., Kervestin S., Li C., Mangus D.A., Spatrick P., Jacobson A., Ghosh S., Kervestin S., Li C., Mangus D.A., Spatrick P., Jacobson A., Kervestin S., Li C., Mangus D.A., Spatrick P., Jacobson A., Li C., Mangus D.A., Spatrick P., Jacobson A., Mangus D.A., Spatrick P., Jacobson A., Spatrick P., Jacobson A., Jacobson A. Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 2006;34:39–42. doi: 10.1042/BST20060039. [DOI] [PubMed] [Google Scholar]
- Anders K.R., Grimson A., Anderson P., Grimson A., Anderson P., Anderson P. SMG-5, required for C. elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin C.M., Lingner J., Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 2006a;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Azzalin C.M., Lingner J., Lingner J. The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle. 2006b;5:1496–1498. doi: 10.4161/cc.5.14.3093. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I., Izaurralde E., Izaurralde E. Quality control of gene expression: A stepwise assembly pathway for the surveillance complex that triggers nonsense-mediated mRNA decay. Genes & Dev. 2006;20:391–398. doi: 10.1101/gad.1407606. [DOI] [PubMed] [Google Scholar]
- Bejsovec A., Anderson P., Anderson P. Myosin heavy-chain mutations that disrupt Caenorhabditis elegans thick filament assembly. Genes & Dev. 1988;2:1307–1317. doi: 10.1101/gad.2.10.1307. [DOI] [PubMed] [Google Scholar]
- Brumbaugh K.M., Otterness D.M., Geisen C., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T., Otterness D.M., Geisen C., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T., Geisen C., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T., Brognard J., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T., Li X., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T., Lejeune F., Tibbetts R.S., Maquat L.E., Abraham R.T., Tibbetts R.S., Maquat L.E., Abraham R.T., Maquat L.E., Abraham R.T., Abraham R.T. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Buhler M., Steiner S., Mohn F., Paillusson A., Muhlemann O., Steiner S., Mohn F., Paillusson A., Muhlemann O., Mohn F., Paillusson A., Muhlemann O., Paillusson A., Muhlemann O., Muhlemann O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Cali B.M., Kuchma S.L., Latham J., Anderson P., Kuchma S.L., Latham J., Anderson P., Latham J., Anderson P., Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E., Izaurralde E., Izaurralde E. Nonsense-mediated mRNA decay: Molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Culbertson M.R., Leeds P.F., Leeds P.F. Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 2003;13:207–214. doi: 10.1016/s0959-437x(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Dibb N.J., Brown D.M., Karn J., Moerman D.G., Bolten S.L., Waterston R.H., Brown D.M., Karn J., Moerman D.G., Bolten S.L., Waterston R.H., Karn J., Moerman D.G., Bolten S.L., Waterston R.H., Moerman D.G., Bolten S.L., Waterston R.H., Bolten S.L., Waterston R.H., Waterston R.H. Sequence analysis of mutations that affect the synthesis, assembly and enzymatic activity of the unc-54 myosin heavy chain of Caenorhabditis elegans. J. Mol. Biol. 1985;183:543–551. doi: 10.1016/0022-2836(85)90170-6. [DOI] [PubMed] [Google Scholar]
- Domeier M.E., Morse D.P., Knight S.W., Portereiko M., Bass B.L., Mango S.E., Morse D.P., Knight S.W., Portereiko M., Bass B.L., Mango S.E., Knight S.W., Portereiko M., Bass B.L., Mango S.E., Portereiko M., Bass B.L., Mango S.E., Bass B.L., Mango S.E., Mango S.E. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science. 2000;289:1928–1931. doi: 10.1126/science.289.5486.1928. [DOI] [PubMed] [Google Scholar]
- Fire A., Harrison S.W., Dixon D., Harrison S.W., Dixon D., Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Fraser A.G., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Kamath R.S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J., Martinez-Campos M., Sohrmann M., Ahringer J., Sohrmann M., Ahringer J., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Frischmeyer P.A., Dietz H.C., Dietz H.C. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Fukuhara N., Ebert J., Unterholzner L., Lindner D., Izaurralde E., Conti E., Ebert J., Unterholzner L., Lindner D., Izaurralde E., Conti E., Unterholzner L., Lindner D., Izaurralde E., Conti E., Lindner D., Izaurralde E., Conti E., Izaurralde E., Conti E., Conti E. SMG7 is a 14–3–3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D., Unterholzner L., Ciccarelli F.D., Bork P., Izaurralde E., Unterholzner L., Ciccarelli F.D., Bork P., Izaurralde E., Ciccarelli F.D., Bork P., Izaurralde E., Bork P., Izaurralde E., Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C.I., Ruiz-Echevarria M.J., Vasudevan S., Henry M.F., Peltz S.W., Ruiz-Echevarria M.J., Vasudevan S., Henry M.F., Peltz S.W., Vasudevan S., Henry M.F., Peltz S.W., Henry M.F., Peltz S.W., Peltz S.W. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- Grimson A., O’Connor S., Newman C.L., Anderson P., O’Connor S., Newman C.L., Anderson P., Newman C.L., Anderson P., Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA decay in Caenorhabditis elegans. Mol. Cell. Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Dong Y., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Shindo M., Liu W., Odell I., Ruvkun G., Lee S.S., Liu W., Odell I., Ruvkun G., Lee S.S., Odell I., Ruvkun G., Lee S.S., Ruvkun G., Lee S.S., Lee S.S. A systematic RNAi screen for longevity genes in C. elegans. Genes & Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P., Papp A., Pulak R., Ambros V., Anderson P., Pulak R., Ambros V., Anderson P., Ambros V., Anderson P., Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J.A., Neu-Yilik G., Hentze M.W., Kulozik A.E., Neu-Yilik G., Hentze M.W., Kulozik A.E., Hentze M.W., Kulozik A.E., Kulozik A.E. Nonsense-mediated decay approaches the clinic. Nat. Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Le Bot N., Moreno S., Sohrmann M., Moreno S., Sohrmann M., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kashima I., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Yamashita A., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Izumi N., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Kataoka N., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Morishita R., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Hoshino S., Ohno M., Dreyfuss G., Ohno S., Ohno M., Dreyfuss G., Ohno S., Dreyfuss G., Ohno S., Ohno S. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun H., Marzluff W.F., Marzluff W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Furic L., Desgroseillers L., Maquat L.E., Furic L., Desgroseillers L., Maquat L.E., Desgroseillers L., Maquat L.E., Maquat L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Gatfield D., Izaurralde E., Moore M.J., Gatfield D., Izaurralde E., Moore M.J., Izaurralde E., Moore M.J., Moore M.J. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P., Peltz S.W., Jacobson A., Culbertson M.R., Peltz S.W., Jacobson A., Culbertson M.R., Jacobson A., Culbertson M.R., Culbertson M.R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes & Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P., Wood J.M., Lee B.S., Culbertson M.R., Wood J.M., Lee B.S., Culbertson M.R., Lee B.S., Culbertson M.R., Culbertson M.R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelivelt M.J., Culbertson M.R., Culbertson M.R. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: A family affair—Active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Johnstone I.L., Caceres J.F., Johnstone I.L., Caceres J.F., Caceres J.F. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., Johnstone I.L., Caceres J.F., Johnstone I.L., Caceres J.F., Caceres J.F. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA. 2003;9:881–891. doi: 10.1261/rna.5420503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.D., Steitz J.A., Shu M.D., Steitz J.A., Steitz J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu M.D., Steitz J.A., Shu M.D., Steitz J.A., Steitz J.A. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Mango S.E. Stop making nonSense: The C. elegans smg genes. Trends Genet. 2001;17:646–653. doi: 10.1016/s0168-9525(01)02479-9. [DOI] [PubMed] [Google Scholar]
- Maquat L.E. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Maquat L.E., Li X., Li X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–456. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi S.M., Frischmeyer P.A., Mendell J.T., Kelly A.G., Lawler A.M., Dietz H.C., Frischmeyer P.A., Mendell J.T., Kelly A.G., Lawler A.M., Dietz H.C., Mendell J.T., Kelly A.G., Lawler A.M., Dietz H.C., Kelly A.G., Lawler A.M., Dietz H.C., Lawler A.M., Dietz H.C., Dietz H.C. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C., Meyers J.L., Martinez-Murillo F., Dietz H.C., Martinez-Murillo F., Dietz H.C., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Nagy E., Maquat L.E., Maquat L.E. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Nollen E.A., Garcia S.M., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., Garcia S.M., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., Kim S., Chavez A., Morimoto R.I., Plasterk R.H., Chavez A., Morimoto R.I., Plasterk R.H., Morimoto R.I., Plasterk R.H., Plasterk R.H. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.F., Carr B., Anders K.R., Grimson A., Anderson P., Carr B., Anders K.R., Grimson A., Anderson P., Anders K.R., Grimson A., Anderson P., Grimson A., Anderson P., Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol. Cell. Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson A., Hirschi N., Vallan C., Azzalin C.M., Muhlemann O., Hirschi N., Vallan C., Azzalin C.M., Muhlemann O., Vallan C., Azzalin C.M., Muhlemann O., Azzalin C.M., Muhlemann O., Muhlemann O. A GFP-based reporter system to monitor nonsense-mediated mRNA decay. Nucleic Acids Res. 2005;33:e54. doi: 10.1093/nar/gni052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof J., van Haaften G., Thijssen K., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H., Tijsterman M., van Haaften G., Thijssen K., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H., Tijsterman M., Thijssen K., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H., Tijsterman M., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H., Tijsterman M., Fraser A.G., Ahringer J., Plasterk R.H., Tijsterman M., Ahringer J., Plasterk R.H., Tijsterman M., Plasterk R.H., Tijsterman M., Tijsterman M. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes & Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R., Anderson P., Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Letunic I., Raes J., Bork P., Izaurralde E., Letunic I., Raes J., Bork P., Izaurralde E., Raes J., Bork P., Izaurralde E., Bork P., Izaurralde E., Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Raes J., Izaurralde E., Raes J., Izaurralde E., Izaurralde E. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem. Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Roberts B., Clucas C., Johnstone I.L., Clucas C., Johnstone I.L., Johnstone I.L. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol. Biol. Cell. 2003;14:4414–4426. doi: 10.1091/mbc.E03-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S., Linder P., Linder P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- Scott D.K., Board J.R., Lu X., Pearson A.D., Kenyon R.M., Lunec J., Board J.R., Lu X., Pearson A.D., Kenyon R.M., Lunec J., Lu X., Pearson A.D., Kenyon R.M., Lunec J., Pearson A.D., Kenyon R.M., Lunec J., Kenyon R.M., Lunec J., Lunec J. The neuroblastoma amplified gene, NAG: Genomic structure and characterisation of the 7.3 kb transcript predominantly expressed in neuroblastoma. Gene. 2003;307:1–11. doi: 10.1016/s0378-1119(03)00459-1. [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J., Gaitatzes C., Saxena K., Neer E.J., Saxena K., Neer E.J., Neer E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Tange T.O., Nott A., Moore M.J., Nott A., Moore M.J., Moore M.J. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Izaurralde E., Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol. Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- van Haaften G., Vastenhouw N.L., Nollen E.A., Plasterk R.H., Tijsterman M., Vastenhouw N.L., Nollen E.A., Plasterk R.H., Tijsterman M., Nollen E.A., Plasterk R.H., Tijsterman M., Plasterk R.H., Tijsterman M., Tijsterman M. Gene interactions in the DNA damage-response pathway identified by genome-wide RNA-interference analysis of synthetic lethality. Proc. Natl. Acad. Sci. 2004;101:12992–12996. doi: 10.1073/pnas.0403131101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw N.L., Fischer S.E., Robert V.J., Thijssen K.L., Fraser A.G., Kamath R.S., Ahringer J., Plasterk R.H., Fischer S.E., Robert V.J., Thijssen K.L., Fraser A.G., Kamath R.S., Ahringer J., Plasterk R.H., Robert V.J., Thijssen K.L., Fraser A.G., Kamath R.S., Ahringer J., Plasterk R.H., Thijssen K.L., Fraser A.G., Kamath R.S., Ahringer J., Plasterk R.H., Fraser A.G., Kamath R.S., Ahringer J., Plasterk R.H., Kamath R.S., Ahringer J., Plasterk R.H., Ahringer J., Plasterk R.H., Plasterk R.H. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 2003;13:1311–1316. doi: 10.1016/s0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- Wang J., Gudikote J.P., Olivas O.R., Wilkinson M.F., Gudikote J.P., Olivas O.R., Wilkinson M.F., Olivas O.R., Wilkinson M.F., Wilkinson M.F. Boundary-independent polar nonsense-mediated decay. EMBO Rep. 2002;3:274–279. doi: 10.1093/embo-reports/kvf036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer K., Zhu X.X., Lamb B.J., Kuick R., Ambros P.F., Kovar H., Thoraval D., Motyka S., Alberts J.R., Hanash S.M., Zhu X.X., Lamb B.J., Kuick R., Ambros P.F., Kovar H., Thoraval D., Motyka S., Alberts J.R., Hanash S.M., Lamb B.J., Kuick R., Ambros P.F., Kovar H., Thoraval D., Motyka S., Alberts J.R., Hanash S.M., Kuick R., Ambros P.F., Kovar H., Thoraval D., Motyka S., Alberts J.R., Hanash S.M., Ambros P.F., Kovar H., Thoraval D., Motyka S., Alberts J.R., Hanash S.M., Kovar H., Thoraval D., Motyka S., Alberts J.R., Hanash S.M., Thoraval D., Motyka S., Alberts J.R., Hanash S.M., Motyka S., Alberts J.R., Hanash S.M., Alberts J.R., Hanash S.M., Hanash S.M. Co-amplification of a novel gene, NAG, with the N-myc gene in neuroblastoma. Oncogene. 1999;18:233–238. doi: 10.1038/sj.onc.1202287. [DOI] [PubMed] [Google Scholar]
- Yamashita A., Ohnishi T., Kashima I., Taya Y., Ohno S., Ohnishi T., Kashima I., Taya Y., Ohno S., Kashima I., Taya Y., Ohno S., Taya Y., Ohno S., Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes & Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Krainer A.R., Krainer A.R. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]