Abstract
Regulation of patterning and morphogenesis during embryonic development depends on tissue-specific signaling by retinoic acid (RA), the active form of Vitamin A (retinol). The first enzymatic step in RA synthesis, the oxidation of retinol to retinal, is thought to be carried out by the ubiquitous or overlapping activities of redundant alcohol dehydrogenases. The second oxidation step, the conversion of retinal to RA, is performed by retinaldehyde dehydrogenases. Thus, the specific spatiotemporal distribution of retinoid synthesis is believed to be controlled exclusively at the level of the second oxidation reaction. In an N-ethyl-N-nitrosourea (ENU)-induced forward genetic screen we discovered a new midgestation lethal mouse mutant, called trex, which displays craniofacial, limb, and organ abnormalities. The trex phenotype is caused by a mutation in the short-chain dehydrogenase/reductase, RDH10. Using protein modeling, enzymatic assays, and mutant embryos, we determined that RDH10trex mutant protein lacks the ability to oxidize retinol to retinal, resulting in insufficient RA signaling. Thus, we show that the first oxidative step of Vitamin A metabolism, which is catalyzed in large part by the retinol dehydrogenase RDH10, is critical for the spatiotemporal synthesis of RA. Furthermore, these results identify a new nodal point in RA metabolism during embryogenesis.
Keywords: Retinoic acid, Vitamin A, orofacial cleft, limb
Retinoic acid (RA), the active derivative of Vitamin A, plays an important role in patterning and morphogenesis of a developing embryo. Insufficient or excessive RA levels cause abnormalities in craniofacial, skeletal, and limb development as well as malformations of multiple organs, including kidney, lung, heart, eyes, and the urogenital system (Clagett-Dame and DeLuca 2002). Therefore, it is essential to understand the mechanisms controlling Vitamin A metabolism and the developmental production of RA. Unlike most morphogens, which are peptides, RA is a small molecule. Thus, the specific levels of RA within various tissues of an embryo are regulated through a complex balance among synthesis by oxidative enzymes, storage and transport via retinoid-binding proteins, and by enzymatic degradation. The interplay among these aspects of RA metabolism and distribution are complex and not yet fully understood.
RA is synthesized from Vitamin A, also known as all-trans-retinol, via two consecutive enzymatic reactions (Duester et al. 2003). In the first reaction, all-trans-retinol is oxidized to all-trans-retinal. This reaction is thought to be catalyzed primarily by members of the alcohol dehydrogenase (ADH) family, specifically ADH1, ADH4, and ADH7 (formerly known as ADH3). It has been demonstrated that ADH1, ADH4, and ADH7 each play a role in RA metabolism, but only in postnatal growth and development (Deltour et al. 1999; Molotkov et al. 2002a). The ubiquitous expression pattern of ADH7 during embryogenesis (Molotkov et al. 2002b) has led to the notion that the enzymatic conversion of retinol to retinal occurs more or less uniformly and plays no role in the spatial or temporal regulation of RA synthesis during embryonic development (Duester et al. 2003). Another family of enzymes able to catalyze the oxidation of retinol to retinal is the short-chain dehydrogenase/reductase (SDR) family. Two members of the SDR family, RDH5 and RDH11, have been shown to play a role in the vision cycle in adult mice (Shang et al. 2002; Kasus-Jacobi et al. 2005; Kim et al. 2005). To date, no phenotype has been observed in embryos lacking ADH or SDR enzymes, implying that none are individually required for the prenatal synthesis of RA. The failure to identify any enzyme required for embryonic conversion of retinol to retinal has led to the conclusion that the oxidation of retinol to retinal within an embryo is catalyzed by the redundant and overlapping activities of ADH and SDR enzymes (Duester 2000).
The second step in the synthesis of RA from retinol is the oxidation of all-trans-retinal into RA. This reaction is catalyzed by the retinaldehyde dehydrogenases, RALDH1, RALDH2, and RALDH3 (ALDH1A1, ALDH1A2, and ALDH1A3), each of which is active in specific spatiotemporal domains during embryogenesis. RALDH2 is considered to be the predominant enzyme (Niederreither et al. 1999, 2002a; Dupe et al. 2003; Fan et al. 2003), as it is responsible for the production of nearly all embryonic RA and is essential for forelimb development, somitogenesis, and organogenesis of the heart, lungs, and kidneys (Niederreither et al. 1999, 2002b; Mic et al. 2002). RALDH1 and RALDH3 are primarily required for eye and nasal morphogenesis during embryonic development (Dupe et al. 2003; Fan et al. 2003; Matt et al. 2005). Whereas embryonic phenotypes have not been observed in mice with targeted deletions of Adh or Rdh genes, distinct developmental defects are observed in Raldh mutants, leading to the conclusion that the spatial and temporal regulation of embryonic RA metabolism occurs at the second oxidative step of RA synthesis (Duester et al. 2003).
In contrast to this view, we demonstrate in the present study that the first oxidative reaction in embryonic Vitamin A metabolism is absolutely essential for normal embryogenesis and is not catalyzed uniformly by ubiquitously expressed and redundant alcohol dehydrogenases. Instead, we show that RDH10, a retinol dehydrogenase that converts all-trans-retinol into all-trans-retinal, plays a critical role in embryonic RA synthesis. Loss of RDH10 function disrupts RA synthesis and leads to loss of RA signaling primarily in craniofacial tissues and trunk mesoderm. The reduced levels of RA cause orofacial, limb, and organ abnormalities and ultimately result in embryonic lethality. Our results therefore reveal that the retinol dehydrogenase RDH10 is critically required for Vitamin A metabolism and is essential for embryonic patterning, morphogenesis, and survival.
Results
Identification of a mouse mutant with defects in craniofacial and limb morphogenesis
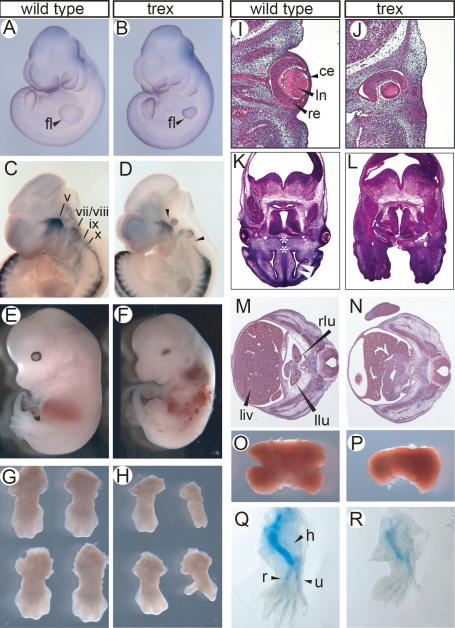
We undertook a search for novel genes regulating growth and pattern formation during mouse embryogenesis. To that end we performed a screen for recessive N-ethyl-N-nitrosourea (ENU)-induced mutations that produced morphological defects in limb and craniofacial structures at embryonic day 10.5 (E10.5). Here we describe one such mutant, named trex, which exhibits striking defects in growth and patterning of the forelimb, frontonasal process, pharyngeal arches, and various organs. The phenotypic hallmark of trex embryos at E10.5 is small, abnormally shaped forelimbs that resemble phocomelia (Fig. 1A,B). In contrast, the hindlimbs of trex embryos are relatively unaffected and morphologically indistinguishable from their wild-type littermates. Other defects observed in E10.5 trex embryos include variably misshapen optic vesicles, small otic vesicles that are often duplicated or displaced, absent caudal pharyngeal arches, and lung bud agenesis (data not shown). Formation of cranial ganglia, particularly the trigeminal, facial, vestibuloacoustic, and glossopharyngeal ganglia, are also abnormal in mutant embryos (Fig. 1C,D). Notably, the proximal region of each ganglion is specifically reduced in size or missing, consistent with defects in neural crest cell patterning (Fig. 1D, arrowheads).
Figure 1.
Developmental defects in trex mutant embryos from E10.5 to E13.5. Wild-type (A) and mutant (B) whole-mount embryos at E10.5; embryos hybridized with a Pax3 RNA probe to provide contrast. E10.5 wild-type (C) and mutant (D) embryos hybridized as whole mount with Sox10 RNA probe to visualize cranial ganglia. Arrowheads in D indicate reduction in proximal ganglia in mutant embryo. E12.5 wild-type (E) and mutant (F) embryos. Isolated forelimbs of wild-type (G) and mutant (H) embryos at E12.5. Transverse sections of wild-type (I) and mutant (J) embryos at eye level stained with haematoxylin and eosin; dorsal top. Transverse sections of wild-type (K) and mutant (L) embryos at E12.5 at the level of the frontonasal process, stained with haematoxylin and eosin; dorsal top. Asterisks indicate nasal septum; white arrowhead indicates nasal cavity. Transverse sections of wild-type (M) and mutant (N) embryos at the level of liver and lung bud stained with haematoxylin and eosin; dorsal right, ventral left. Dorsal view of isolated livers from wild-type (O) and mutant (P) embryos at E12.5; anterior top. Forelimbs of wild-type (Q) and mutant (R) embryos at E13.0 stained with Alcian Blue to reveal cartilage condensations. (fl) Forelimb bud; (v) trigeminal ganglion; (vii/viii) facial/vestibuloacoustic ganglia; (ix) glossopharyngeal ganglion; (x) vagal ganglion; (re) retina; (ln) lens; (ce) corneal ectoderm; (liv) liver; (rlu) right lung bud; (llu) left lung bud; (h) humerus cartilage; (r) radius cartilage; (u) ulna cartilage.
At E12.5–E13.0, morphological anomalies include persistently small, truncated forelimbs with autopods that are typically narrower and abnormally interdigitated (Fig. 1E–H). Eye development is also abnormal (Fig. 1I–J). In particular, the cornea and ventral half of the retina are absent and the lens is hypoplastic. Variable clefting of the frontonasal process is also observed, together with agenesis of the nasal septum and chambers (Fig. 1K,L). Organogenesis is also affected in trex embryos. The lungs are hypoplastic and fail to undergo growth and branching morphogenesis (Fig. 1M,N), and, similarly, the liver is typically smaller with fewer lobes (Fig. 1O,P). Gonad and metanephros rudiments are formed in mutant embryos, but are hypoplastic and underdeveloped relative to the corresponding organs of wild-type littermates at E13.0 (data not shown). Similarly, stomach and midgut are severely hypoplastic in mutant embryos and pancreas structures are not detected (data not shown). Trex mutant embryos die by approximately E13.0. Evidence of hemorrhage and pooled blood in mutant embryos (Fig. 1F) suggests that the cause of death is vascular defects. Embryos that survive to that stage have forelimbs with abnormal and reduced cartilage condensations of varying severity that often include agenesis of the radius (meromelia) (Fig. 1Q,R). This constellation of phenotypes is reminiscent of malformations that occur under conditions of Vitamin A deficiency in a variety of organisms.
Trex mutation maps to Rdh10
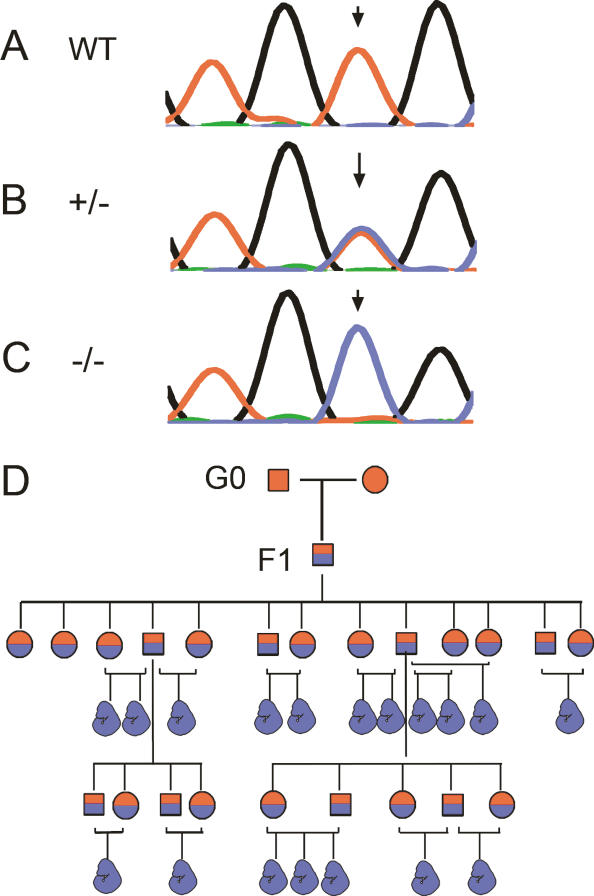
In order to identify the chromosomal locus of the trex mutation, we used a panel of microsatellite markers polymorphic between the mixed C57/BL6 and 129/Sv background of our mutagenized animals versus the FVB strain to which they were bred (Supplementary Table 1). With these markers we mapped the genomic interval associated with the trex phenotype to the proximal region of chromosome 1. The region was refined using selected single nucleotide polymorphisms. Subsequent sequencing of candidate genes revealed a mutation in the gene encoding retinol dehydrogenase 10 (Rdh10). The trex mutation occurs at position 251 of the Rdh10 gene with respect to the mouse RNA reference sequence NM 133,832. The wild-type allele is a T at nucleotide 251, whereas the trex mutant allele is a C at that position (Fig. 2A–C). The Rdh10 mutation segregates perfectly with the mutant phenotype. All embryos exhibiting the mutant phenotype are homozygous for the mutant allele (Fig. 2D). All carrier parent animals are heterozygous for the mutation except for the original mutagenized male, whose somatic tissue was wild-type, as expected. RDH10 is an enzyme of the SDR superfamily whose members catalyze the oxidation of retinol into retinal. Identification of the trex mutation as a defect in a gene involved in retinol metabolism suggests that deficiencies in Vitamin A oxidation and/or RA production are the principal cause of trex phenotypic anomalies and, further, that RDH10 may play an essential role in embryonic retinoid signaling.
Figure 2.
Point mutation in Rdh10 correlates with trex mutant phenotype. Sequence electrophereograms from Rdh10 exon 1 are shown. (A) Wild-type adult mouse. (B) Carrier adult mouse heterozygous for mutant allele. (C) trex mutant embryo homozygous for mutant allele. Arrows indicate the position of the mutated nucleotide. (D) Pedigree tree of first four generations following ENU mutagenesis. (Squares) Adult male; (circles) adult female; (embryo icon) phenotypically mutant embryos; (solid red) wild-type allele; (red and blue split) heterozygous for mutation; (solid blue) homozygous mutant allele.
Rdh10 is expressed in specific tissues of developing mouse embryos
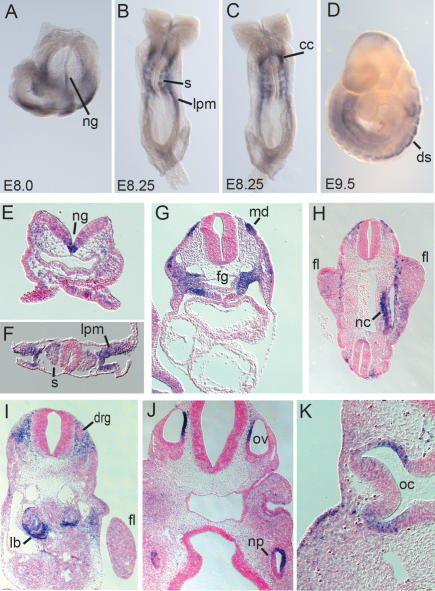
As a first step in understanding the mechanistic origins of the craniofacial, limb, and organ defects observed in trex embryos, we characterized the spatiotemporal expression of Rdh10 mRNA during normal embryogenesis. Rdh10 expression is first detected at E8.0, where it is present in the neural groove in the region of the prospective hindbrain and also in lateral mesoderm tissue (Fig. 3A). At E8.25, Rdh10 is expressed in the somites, ventral neural groove in the head region, cardiac crescent, lateral plate mesoderm, and developing body wall (Fig. 3B,C). Section in situ hybridization reveals that Rdh10 expression at E8.5 is concentrated in lateral plate mesoderm and the dorsal region of the somites as well as the floor plate of the neural tube and head mesenchyme (Fig. 3E,F). At E9.0, at the level of the heart, strong expression is observed in medial dermomyotomal lip of the somites and in dorsal pericardial mesoderm adjacent to the foregut (Fig. 3G). At E9.5, Rdh10 is expressed in dorsal paraxial (somite) mesoderm and lateral plate (body wall) mesoderm (Fig. 3D), and is also present in nephrogenic cord tissues and in proximal forelimb bud mesenchyme adjacent to somites 8–12 (Fig. 3H).
Figure 3.
Rdh10 mRNA expression in wild-type embryos at midgestation. (A–D) Whole-mount RNA ISH with Rdh10. (A) Ventral view at E8.0 showing expression in neural groove and lateral tissues. Dorsal view (B) and ventral view (C) at E8.5 showing expression in neural groove, somites, lateral mesoderm, and cardiac crescent. (D) Lateral view at E9.5 showing expression in dorsal somites and lateral mesoderm. (E–K) Rdh10 ISH on transversely sectioned embryos. (E) Head-level section through E8.5 somite shows expression in ventral groove and head mesenchyme. (F) Trunk section anterior to the heart of E8.5 shows strong expression in dorsal somites and lateral plate mesoderm. (G) Heart-level section of E9.0 embryos shows strong expression in the dorsal dermomyotome lip and the mesenchyme surrounding the foregut. (H) Section below heart level of E9.5 embryo shows expression in the trunk mesoderm at the base of the forelimb bud and nephrogenic cord region. (I) Trunk-level section of E10.5 embryo shows expression in lung buds, the mesenchyme surrounding the dorsal root ganglia, and the mesoderm adjacent to the lung bud. (J) Head-level section of E10.5 embryo showing expression in medial otic vesicles and lateral nasal pits. (K) Head-level section of E10.5 embryo shows expression in medial otic vesicles. (ng) Neural groove; (s) somite; (lpm) lateral plate mesoderm; (cc) cardiac crescent; (ds) dorsal somite; (md) medial dermomyotome lip; (fg) foregut; (fl) forelimb; (nc) nephrogenic cord; (drg) dorsal root ganglion; (lb) lung bud; (np) nasal pit; (oc) optic cup.
At E10.5, Rdh10 is expressed in lung buds and in mesodermal tissue surrounding the dorsal root ganglia and adjacent to the forelimb bud (Fig. 3I). Strong expression is also observed in the developing mesonephros (data not shown). Diffuse expression is observed in the forelimb at this stage, with higher signal in the proximal portion of the limb than the middle or distal limb (data not shown). Rdh10 is also expressed in the medial otic vesicle, in lateral nasal pits, and in the outer layer of the optic cup (Fig. 3J,K). Together, these data indicate that Rdh10 mRNA is expressed during embryogenesis in the precursors of tissues that are affected in Rdh10trex/trex mutant embryos, including the frontonasal processes, limbs, eyes, ears, and lungs. Thus, Rdh10, the only known gene encoding a retinol-to-retinal oxidizing enzyme shown to be essential for embryogenesis, is expressed in a unique pattern consistent with its mutant phenotype.
Amino acid replacement in RDH10 may destabilize the protein structural core
The trex point mutation is located in the coding sequence of exon 1 of the Rdh10 gene, and the mutant allele causes a cysteine-to-arginine substitution in the RDH10 protein. In order to evaluate the effect of such a substitution on protein function, we constructed a model of the mouse RDH10 protein (Fig. 4A). The model suggests that the mutated amino acid base is located in the first β-strand that occupies a central position in the β-sheet of the Rossman fold. In the mutant protein, the cysteine side chain is replaced by a larger arginine side chain, which has a potential to clash with side chains of several other residues, most notably Val 117 in strand 4 and phenylalanine 32 in the first α-helix. Thus, modeling suggests that the Rdh10trex mutation would likely cause destabilization of the mutant RDH10 protein.
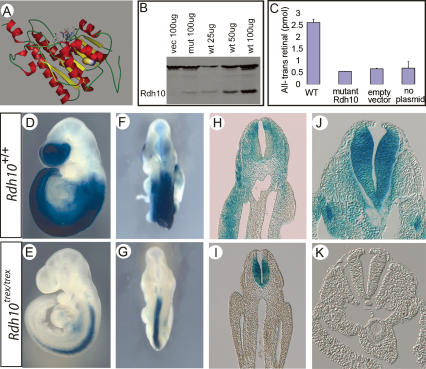
Figure 4.
Reduced stability and function of mutant RDH10 protein. (A) Model of mouse RDH10 protein suggests that mutated amino acid is in the core of the protein. Filled-ball side chain indicates wild-type cysteine, which is altered to arginine in mutant protein, and sticks denote NADP cofactor. The picture was produced using PyMOL package (DeLano 2002). (B,C) RDH10 protein level and catalytic activity were assayed from Cos-7 cells bearing wild-type or Rdh10trex mutant-expressing plasmids. The same samples were used for Western blotting and enzyme assay. The level of RNA produced by wild-type and mutant plasmids in three independent assays was normalized to empty vector. The relative quantitation value for wild-type plasmid was Log(10) 5.80 (standard error, 0.137), and for mutant plasmid was Log(10) 5.90 (standard error, 0.41). (B) The steady-state level of RDH10 protein produced by mutant plasmid is much lower than the level of protein produced by wild type as assessed by Western blot analysis. (C) Catalytic activity of conversion of all-trans-retinol into all-trans-retinal is not detectable by protein produced by mutant plasmid. (D–K) Retinoic acid signaling assayed using RARE–lacZ transgenic mice at E9.5 reveals loss of retinoic acid signaling from trunk mesoderm of mutant embryos. Lateral (D) and dorsal (F) view of wild-type embryo showing strong retinoic acid signaling in the frontonasal process and throughout the trunk mesoderm and neural tube. Lateral (E) and dorsal (G) view of Rdh10trex/trex mutant embryo show retinoic acid signaling is significantly reduced, particularly in trunk and possibly forebrain. (H,J) Transverse section through wild-type embryo at the level of the forelimb (H) and at the level of an epithelialized somite (J) shows retinoic acid signaling in neural ectoderm and trunk mesoderm. (I,K) Transverse section through mutant embryo at the level of the forelimb (I) shows retinoic acid signaling only in neural ectoderm, and at the level of an epithelialized somite (K), retinoic acid signaling is not detectable in any tissue.
To determine if the mutant RDH10 protein is indeed less stable than wild-type, we tested the steady-state protein level of the wild-type and mutant proteins in cultured cells. COS-7 cells were transfected with expression plasmids containing either a wild-type or trex mutant version of the Rdh10 gene. Both the wild-type and trex mutant versions of the plasmids were transfected at equal efficiency and transcribed equally well, as assayed by quantitative real-time PCR (data not shown). Cells bearing mutant plasmid, however, contained only ∼25% of the steady-state level of membrane-fraction RDH10 protein relative to cells bearing wild-type plasmid, as assayed by Western blot analysis (Fig. 4B). These data suggest that the mutant RDH10 protein is markedly less stable than the wild-type protein. Although the protein model suggests that the reduced level of RDH10trex mutant protein is likely due to reduced stability, it is also possible that the reduction in protein level is due to another mechanism such as decreased translation.
RDH10 is a retinol dehydrogenase known to be expressed in retinal pigment epithelium cells of the adult eye and Muller cells (Wu et al. 2002, 2004). Functional enzyme analysis indicates that the protein is localized to the microsomal fraction of cells and that it acts to specifically oxidize all-trans-retinol into all-trans-retinal in a NADP-dependent manner. To determine the amount of activity of mutant RDH10, proteins from cells bearing plasmids producing wild-type or mutant RDH10 protein were assayed for activity by HPLC. In these assays, membrane proteins from COS-7 cells expressing wild-type RDH10 produced an average of 2.6 pmol of all-trans-retinal from all-trans-retinol substrate (Fig. 4C), whereas no activity was detected in the fraction representing an equal amount of total membrane protein from cells expressing the mutant form of RDH10. The lack of RDH10 activity in the membrane extracts from the Rdh10trex-transfected cells is likely due to a combination of reduced protein stability and reduced enzymatic activity.
We next assessed the in vivo activity of the mutant protein by examining Rdh10trex/trex embryos carrying a RA response element–lacZ reporter transgene (RARE–lacZ). With this reporter transgene, RA signaling can be detected by the presence of β-galactosidase activity (Rossant et al. 1991), although expression of the transgene in the forebrain is subject to inactivation and is therefore a somewhat unreliable indicator of RA signaling in that tissue (Luo et al. 2004). In wild-type embryos at E9.5, RA signaling is detected in the frontonasal and forebrain region as well as in the trunk, where it is present in mesoderm and neural ectoderm tissues (Fig. 4D,F,H,J). Strikingly, mutant embryos lacked most or all RA signaling in the trunk mesoderm (Fig. 4E,G,I,K). Although the RARE–lacZ transgene is an imperfect reporter in forebrain tissues, RA signaling is substantially reduced in these head regions in mutant embryos (Fig. 4E). RA signaling is also markedly reduced in the neural tube of mutant embryos, particularly in the caudal region (Fig. 4E,K). The precise degree of deficit in RA signaling, however, varies slightly between mutant embryos and from side to side within an embryo, corresponding with variations in the morphological phenotype of Rdh10trex/trex mutant embryos. These in vivo observations correlate perfectly with our in vitro data demonstrating that RDH10 is a key regulator of RA production. Importantly, these data indicate that the craniofacial, limb, and organ defects observed in Rdh10trex/trex mutant embryos arise from the depletion of retinoid signaling, particularly in trunk mesoderm and frontonasal tissues including possibly the forebrain.
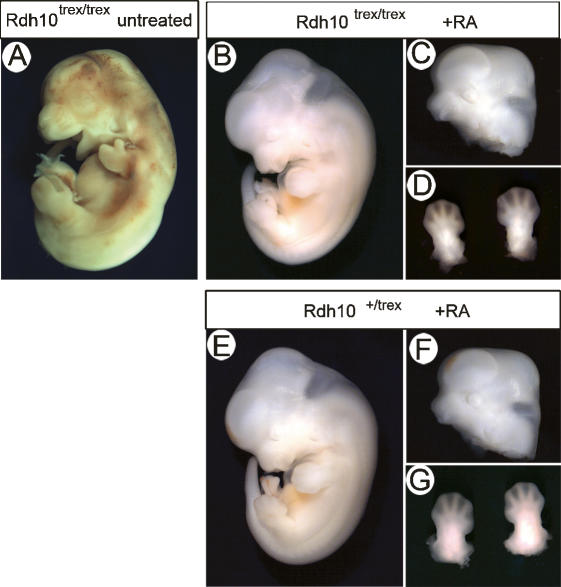
RA in the maternal diet rescues the Rdh10trex/trex mutant phenotype
Our results point toward RDH10 being a critical determinant of RA production. To conclusively demonstrate that the craniofacial, limb, and organ defects observed in Rdh10trex/trex embryos are caused by a deficiency in RA production, we studied the effect of adding supplemental RA to the maternal diet on the development of mutant embryos.
In the absence of RA supplementation, very few Rdh10trex/trex embryos survive until E13.0, and those that do have striking craniofacial and forelimb defects (Fig. 5A). In contrast, when the diet of pregnant females was supplemented with RA from E7.5 to E10.5 and pregnancy was allowed to proceed to E13.0, Rdh10trex/trex embryos survived at the normal Mendelian frequency of 25%. Moreover, with RA supplementation the orofacial clefting and limb outgrowth anomalies were rescued such that Rdh10trex/trex embryos appeared morphologically indistinguishable from their wild-type littermates (Fig. 5B–G). RA-supplemented Rdh10trex/trex embryos displayed normal craniofacial morphology and fully elongated forelimbs with proper interdigitation. Thus, supplemental RA can completely rescue the overt limb and craniofacial abnormalities of trex mutant embryos, conclusively demonstrating that mutation of Rdh10 and resulting deficiency of retinoid signaling are responsible for the trex phenotype. Although we have yet to analyze development of these rescued Rdh10trex/trex embryos at birth, our expectation is that the diminishment of cardiovascular anomalies will be compatible with postnatal viability.
Figure 5.
Mutant phenotype rescued by supplementing maternal diet with retinoic acid. (A) Rdh10trex/trex mutant embryo without retinoic acid supplementation is dead and resorbing at E13.0. (B) Rdh10trex/trex mutant embryo at E13.0 rescued with maternal retinoic acid supplementation looks indistinguishable from wild-type littermate (shown in E). Craniofacial morphology (C) and forelimb development (D) of Rdh10trex/trex mutant embryo rescued by maternal retinoic acid supplement are indistinguishable from wild-type littermate (shown in F,G).
Rdh10 is required for normal patterning of the forelimb bud
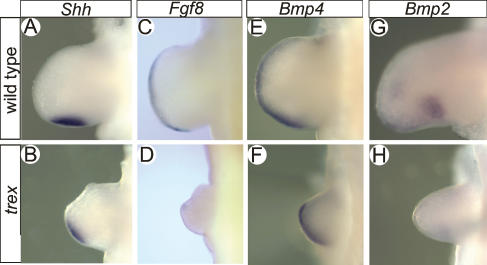
To gain further insight into the mechanism by which the trex mutation affects forelimb patterning, we examined the expression of several genes encoding signaling molecules known to be important for limb outgrowth and patterning. SHH signaling is critical for patterning the anterior–posterior axis of the limb. In wild-type embryos, Shh is initially expressed in a narrow medio–posterior domain of the limb bud that demarcates the zone of polarizing activity (ZPA) (Riddle et al. 1993). In E10.5 mutant trex forelimbs, we observed that the domain of Shh expression was reduced in size and shifted disto–anteriorly relative to controls (Fig. 6A,B). The mislocalized domain of Shh expression in mutant embryos indicates that loss of RDH10 affects both anterior–posterior and proximo–distal patterning in the forelimb by influencing positioning of the ZPA.
Figure 6.
Abnormal signaling in forelimbs of Rdh10trex/trex mutant embryos. Whole-mount RNA ISH of wild-type (A,C,E,G) and Rdh10trex/trex mutant (B,D,F,H) embryos at E10.5 hybridized with Shh (A,B), Fgf8 (C,D), Bmp4 (E,F), or Bmp2 (G,H) probes.
FGF8 signaling is essential for proximo–distal limb outgrowth, and Fgf8 is normally expressed in the apical ectodermal ridge (AER) (Martin 1998). In Rdh10trex/trex forelimbs, the pattern of Fgf8 expression is dramatically reduced relative to wild-type controls, which is indicative of a failure to properly maintain the AER (Fig. 6C,D). Interestingly, the degree of severity of the limb truncation phenotype correlates with the extent of reduction in Fgf8 expression levels. Therefore, although not required for the initiation of limb bud formation, RDH10 is essential for proximo–distal outgrowth of the limb through its key role in maintaining FGF8 activity and AER integrity.
BMP signaling is another key pathway that regulates the formation and maturation of the AER (Tickle 2003). Bmp4 is expressed in the AER and underlying distal mesenchyme in wild-type embryos at E10.5. In Rdh10trex/trex embryos, Bmp4 expression was only modestly altered from the wild-type pattern, being perhaps slightly elevated in the posterior and distal edge of the limb relative to wild-type forelimbs (Fig. 6E,F). Bmp2 stimulates chondrocyte proliferation and differentiation (Minina et al. 2001) and is implicated in regulating digit growth and size (Sears et al. 2006). Bmp2 is normally expressed in the mesenchyme underlying the posterior AER of E10.5 limbs in a domain adjacent to the ZPA. Interestingly, Bmp2 expression is down-regulated in Rdh10trex/trex forelimb mesenchyme compared with wild-type (Fig. 6G,H), a pattern that correlates well with the digit anomalies and aberrant cartilage differentiation observed at E13.0. Thus, our data suggest that RDH10 is required for proper regulation of components of the BMP signaling pathway and that the abnormal growth and patterning of Rdh10trex/trex mutant limbs is due in part to a reduction of Bmp2 but not Bmp4 levels. Collectively, these gene expression analyses, in combination with our morphological observations, suggest that loss of RDH10 does not affect the initial induction of forelimb development, but rather impacts on proximo–distal growth, anterior–posterior patterning, and subsequent differentiation of the forelimb.
Mesodermal RA is not required for bilateral symmetry during somitogenesis
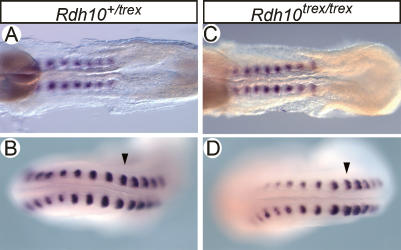
The surprising observation that RA synthesis is specifically depleted in the paraxial mesoderm tissues of Rdh10trex/trex embryos was particularly intriguing since a number of recent studies have suggested that RA in mesoderm restricts FGF8 activity and controls bilateral symmetry during the reiterated formation of somites (Kawakami et al. 2005; Vermot and Pourquie 2005). We therefore examined Rdh10trex/trex embryos for evidence of a disruption to the symmetry of the segmentation process by in situ hybridization with Uncx4.1, a gene that marks the caudal half of each newly formed somite (Mansouri et al. 2000). Interestingly, the pattern of Unxc4.1 expression revealed uniformly symmetrical somites in Rdh10trex/trex embryos at the eight- to 15-somite stage (n = 5) (Fig. 7). Similarly, no evidence of asymmetric somitogenesis is observed in mutant embryos with >15 somites and, in accord with this, no asymmetry is found in vertebrae at E13.0 (data not shown). Thus, although the Rdh10trex/trex embryos are specifically deficient in mesodermal retinoid signaling, they do not exhibit any early or late defects in the bilateral symmetry of somite formation and differentiation. Collectively, this argues that RA regulating bilateral symmetry during somitogenesis may be located in nonmesodermal tissues.
Figure 7.
Rdh10trex/trex mutant embryos form somites symmetrically. Uncx4.1 RNA ISH on 15-somite embryos. (A) Heterozygous wild-type eight-somite embryo. (B) Heterozygous wild-type 15-somite embryo. (C) Mutant Rdh10trex/trex eight-somite embryo. (D) Mutant Rdh10trex/trex 15-somite embryo.
Discussion
The retinol dehydrogenase RDH10 is required for embryonic RA synthesis
The phenotype of trex mutant mouse embryos is characterized by defects in limb outgrowth and differentiation, hypoplasia of the lungs and liver, craniofacial anomalies such as orofacial clefting, and ventral agenesis of the eye. The mutation results in lethality by E13.0, presumably as a result of vascular defects. The mechanistic basis for these developmental abnormalities stems from a deficiency in retinoid signaling, specifically in craniofacial and trunk mesoderm tissues. We determined that the trex mutation was a lesion in the gene encoding the retinol dehydrogenase RDH10 and showed that Rdh10 mRNA is expressed in the frontonasal, eye, ear, lung, lateral plate, and forelimb of midgestation embryos during developmental stages relevant for tissues affected by the mutation. Importantly, the developmental anomalies observed in Rdh10trex/trex embryos could be rescued through RA supplementation of the maternal diet. Thus, our data demonstrate a novel, essential role for RDH10 in craniofacial, limb, and organ development. Furthermore, our analyses reveal for the first time that the activity of a retinol dehydrogenase is required during embryogenesis to mediate the first step of Vitamin A metabolism.
The synthesis of RA from Vitamin A occurs in a two-step oxidative process and, over the past several years, attention has centered on the second oxidation reaction, in which RALDH enzymes convert retinal into RA. The focus on the second reaction was due largely to the observation that Raldh2 mutant embryos are nearly completely devoid of RA production and exhibit spectacular heart, limb, and axial developmental anomalies (Niederreither et al. 1999). Such phenotypes contrast with the absence of any identifiable developmental defect associated with previously examined mutations in Adh and Rdh genes. Collectively, these observations led to the conclusion that the first step in RA synthesis was mediated by the overlapping activity of functionally redundant ADH and possibly RDH enzymes, and, as a result, developmental regulation of RA synthesis was thought to be controlled solely at the level of the second oxidation by RALDH enzymes. Our analyses of trex mutant embryos and the identification and characterization of the essential embryonic function of the retinol dehydrogenase RDH10 challenge the notion that oxidation of retinol to retinal is mediated primarily by redundant alcohol dehydrogenases and occurs uniformly throughout the embryo.
Comparison of Raldh2−/− and Rdh10trex/trex phenotypes
The inability of transfected cells producing mutant RDH10 to oxidize all-trans-retinol into all-trans-retinal suggests that the reduction in RA signaling in mutant embryos is caused by reduced levels of all-trans-retinal. Thus, the defect in Rdh10trex/trex embryos is insufficient levels of substrate for RALDH2. The phenotype of Raldh2−/− mutants is clearly more severe than that of Rdh10trex/trex. Whereas Raldh2−/− embryos die around E9.5, the Rdh10trex/trex embryos can survive as late as E13.0. The more severe phenotype observed in Raldh2−/− embryos is consistent with the fact that Raldh2−/− embryos suffer a more dramatic loss of RA signaling than Rdh10trex/trex embryos (cf. Fig. 4 and Niederreither et al. 1999). The milder phenotype of Rdh10trex/trex embryos could be due to residual partial activity of the RDH10trex mutant enzyme. Although the mutant enzyme does not have detectable activity when expressed in COS-7 cells, it is possible that some residual function may be retained in vivo. Alternatively, the low levels of retinal RALDH2–substrate present in Rdh10trex/trex embryos could be produced by other RDH enzymes and/or by ADH enzymes. However, if there are redundant, isofunctional enzymes, they are clearly not able to completely compensate for the loss of RDH10-mediated all-trans-retinal production within the many affected tissues of Rdh10trex/trex mutant embryos.
Both Rdh10trex/trex and Raldh2−/− mutant embryos exhibit forelimb defects, the former having small, truncated forelimbs with absent and abnormally patterned cartilage elements, while the latter lack forelimb outgrowth entirely. Interestingly, Raldh2−/− embryos rescued with low doses of RA display small and abnormally patterned forelimbs similar to those of Rdh10trex/trex embryos. In both Raldh2−/−-rescued (Niederreither et al. 2002b; Mic and Duester 2003; Mic et al. 2004) and Rdh10trex/trex embryos, the ZPA domain, as defined by Shh expression, is ectopically shifted disto–anteriorly relative to its normal position. Similarly, both Raldh2−/−-rescued and Rdh10trex/trex embryos fail to maintain the AER, as marked by Fgf8 expression. In contrast, differences are observed between Raldh2−/−-rescued and Rdh10trex/trex embryo forelimbs with respect to BMP signaling. Rdh10trex/trex mutant embryos display sharply reduced Bmp2 expression in the forelimb, whereas Raldh2−/−-rescued embryos express Bmp2 and Bmp4 at normal levels but in inappropriate regions of the limb. The distinct effects on BMP activity may reflect differences in residual RA signaling in Rdh10trex/trex mutants compared with partially rescued Raldh2−/− mutants. Nonetheless, the comparable limb phenotype in Rdh10trex/trex and Raldh2−/−-rescued embryos is mechanistically due to the absence of trunk mesodermal RA. These results demonstrate that reduced RA levels in Rdh10trex/trex and partially rescued Raldh2−/− mutants lead to defects in proximo–distal outgrowth and anterior–posterior patterning of forelimbs.
Spatial and temporal regulation of RA synthesis
It is noteworthy that the expression patterns of Rdh10 and Raldh2 are similar but not identical (cf. Fig. 2 and Niederreither et al. 1997). For example, both genes are expressed in mesoderm during gastrulation. However, Raldh2 expression is detected as early as E7.0–E7.5 in the posterior half of the embryo, whereas Rdh10 is first detected a bit later, at approximately E8.0, in a domain that is shifted anteriorly from that of Raldh2. During early somitogenesis, both genes are expressed in somites and trunk mesoderm; however, Rdh10 is expressed in the neural groove of the head region at that stage, whereas the neurectodermal expression of Raldh2 is restricted to the optic vesicle. These differences in expression pattern suggest that first and second reactions in synthesis of RA may be carried out in different tissues. Thus, like RA, the all-trans-retinal produced by RDH10 may diffuse or be transported between different tissues of the embryo.
The site of RA synthesis may play a role in temporal and spatial control of RA signaling, but transport, diffusion, cellular uptake, and degradation clearly also contribute significantly to the regulation of timing and tissue specificity of RA signaling. The importance of these post-synthetic metabolic regulatory mechanisms is illustrated by the fact that embryos with RA synthesis defects can be rescued by supplementation with exogenous RA, which produces uniform levels of RA within an embryo (Niederreither et al. 2000; Mic et al. 2002). Moreover, post-synthetic regulation is further implicated by the fact that rescue of RA-depleted embryos is effective over a range of RA concentrations and in spite of fluctuation in RA levels during supplementation. The robustness of embryonic patterning to spatial and temporal variation in supplemental RA concentrations is likely due primarily to the RA degrading activity of the Cyp26 enzymes, as demonstrated recently for zebrafish hindbrain patterning (Hernandez et al. 2007).
It is noteworthy that the distribution of RA signaling in the Rdh10trex/trex mutant embryos is very similar to the pattern observed in Raldh2−/− embryos rescued by a pulse of RA supplementation (cf. Fig. 4E,G and Mic et al. 2002). In both cases, RA signaling in the trunk mesoderm is severely reduced, and the remaining activity is predominantly restricted to the ventral neural tube and, to a lesser extent, to organs and adjacent mesoderm. The similarity of the RA distribution in the two types of embryos suggests that post-synthetic mechanisms of RA regulation act to preferentially restrict RA signaling to the ventral neural tube and organs when RA concentrations are suboptimal.
The spatial distribution of RA signaling that remains in Rdh10trex/trex embryos, which is similar to the pattern in RA-rescued Raldh2−/− embryos, is interesting in light of the fact that Rdh10trex/trex embryos exhibit no somite symmetry defects. In the absence of RA rescue, Raldh2−/− embryos exhibit asymmetry in somite formation, an observation that fostered the idea that RA is essential for ensuring bilateral symmetry during somitogenesis. It is important to point out that the asymmetry of somite formation observed in Raldh2−/− mutant embryos occurs only during a limited time window (eight to 15 somites), and, to date, this early stage asymmetry has not been correlated with any subsequent vertebral defects. The relevant source of the retinoid signal for regulating symmetry of somitogenesis has been widely assumed to be the RA present in the trunk mesoderm, which is absent in Raldh2−/− mutants. Rdh10trex/trex embryos also lack mesodermal retinoid signaling but, in stark contrast to Raldh2−/− mutants, Rdh10trex/trex mutants display no evidence of anomalies in somite symmetry. Early somitogenesis and later vertebral differentiation appear to be symmetrical in Rdh10trex/trex embryos. These data argue compellingly that mesodermal retinoid signaling is not required for symmetrical somite segmentation. In support of this idea, a recent study has proposed that symmetry of somite formation may be regulated by retinoid signaling derived from the neurectoderm (Sirbu and Duester 2006).
In summary, Vitamin A is essential for craniofacial, limb, and organ development during embryogenesis. Insufficient RA signaling, via addition of inhibitors or by nutritional Vitamin A deprivation, results in a remarkably similar spectrum of defects in pig, chick, quail, rat, and mouse (Helms et al. 1996; Morriss-Kay and Sokolova 1996; Stratford et al. 1996; Power et al. 1999). Vitamin A deficiency is also a likely cause of embryopathies in humans and may account for some of the congenital malformations that occur with increased incidence in the developing world, where dietary sources of Vitamin A are inadequate. Conversely, exposure of pregnant mothers to excessive amounts of Vitamin A or RA via, for example, the acne drug Accutane, can also cause embryonic abnormalities in a developing embryo. Thus, understanding the regulation of Vitamin A metabolism is an important issue in human health.
The demonstration that RDH10 is a critical determinant of RA synthesis identifies the oxidation of retinol to retinal as a new nodal point in Vitamin A metabolism during embryogenesis. Consequently, Rdh10 is a promising candidate for clinical genetic screens aimed at identifying the primary genes or modifiers of syndromic and nonsyndromic human limb and craniofacial anomalies such as phocomelia and orofacial clefting.
Materials and methods
Mouse ENU mutagenesis screen for mutations affecting midgestation craniofacial and limb morphogenesis
Our screen for recessive mutations was modeled after successful screens performed in other laboratories (Garcia-Garcia et al. 2005). In this case, mutations were induced in mice of a mixed strain background containing a combination of C57/BL6 and 129S1/Sv genomes. To induce mutations, male mice 8–12 wk of age were injected intraperitoneally once per week for 3 wk with a 100 mg/kg dose of ENU as previously described (Justice 2000). These mutagenized males, designated generation 0 (G0) animals, were subsequently bred to wild-type FVB/NJ female mice to produce animals heterozygous for the induced mutations. Individual males of this first intercross generation were considered founder animals of mutant lines. Founder males were crossed with FVB/NJ females to produce generation 2 (G2). G2 females were backcrossed to their fathers, and their litters were examined at E10.5 for morphological defects to identify lines that yielded reproducible limb or craniofacial abnormalities. From 50 founders, 11 lines were generated that exhibited consistent and reproducible abnormal craniofacial and/or limb morphology. Individual mutant lines were maintained by backcrossing heterozygous carrier animals to FVB/NJ.
Mapping mutations
Since mutant lines were maintained by backcrossing to FVB/NJ, the chromosomal position of each mutation could be mapped by identification of the region of C57/BL6x129/Sv genome that cosegregated with the mutant phenotype. To that end we utilized a panel of microsatellite markers that were polymorphic between the mixed C57/BL6x129/Sv background versus the FVB strain to which they were bred (Supplementary Table 1). For each microsatellite marker, an FVB/NJ allele could be unequivocally distinguished from a C57/BL6 allele or a 129/Sv allele. Microsatellites chosen for this mapping panel did not necessarily distinguish C57/BL6 and 129Sv alleles from each other. Each chromosome of the mouse genome, with the exception of the Y chromosome, was represented by several microsatellite markers. PCR reactions were multiplexed such that 140 microsatellite markers could be analyzed in 48 individual reactions. Fluorescently labeled primers for PCR amplification of microsatellite markers were purchased as a set from ABI (Mouse Mapping Primers version 1.0). Of the 314 markers available, 140 were used for mapping polymorphisms between the mixed C57BL6x129Sv mutagenized strain background and the FVB backcross breeding strain. PCR products were analyzed with a 3730 DNA Analyzer and GeneMapper software. Yolk sac DNA samples from phenotypically mutant embryos and ear biopsy DNA samples from carrier parent animals were screened with the microsatellite panel to determine the region of the C56BL/6x129Sv genome that was genetically linked to the mutant phenotype.
For the trex mutation, the genomic interval associated with the mutant phenotype was initially mapped to the proximal region of chromosome 1 using the microsatellite panel. The region was then further refined using selected single-nucleotide polymorphisms. Once the genomic interval was limited to a span containing a manageable number of genes, transcripts from candidate genes were amplified and sequenced in order to identify alterations from wild type.
Cartilage staining
Cartilage staining of E13.5 embryos was performed as follows: Embryos were fixed overnight in 95% ethanol at 4°C and then stained for 48 h in 0.4% Alcian Blue/70% ethanol. Soft tissues were then dissolved in 1% KOH for 20 min, followed by 0.25% KOH for 30 min. Tissues were cleared for 1 h to overnight through a 20%, 30%, and 50% glycerol series, each containing 0.25% KOH.
Histological sections and whole-mount and section RNA in situ hybridization
For histological sectioning, embryos were fixed overnight in 4% PFA, equilibrated in ethanol, and embedded in Paraplast wax. The embryos were serially sectioned at 5–7 μm in transverse plane and stained with hematoxylin and eosin. Whole-mount and section in situ hybridization were performed as previously described (Nagy et al. 2003) using digoxigenin-labeled probes and detecting hybridization with BM-Purple or NBT-BCIP. Section mRNA in situ hybridization was also performed using an automated mRNA in situ hybridization protocol for a Ventana Discovery instrument (Nitta et al. 2003), with the exceptions that tissues were fixed in 4% paraformaldehyde and signal revelation was performed manually.
Model of RDH10
The model of mouse RDH10 was created using SWISS-MODEL (Schwede et al. 2003). Known proteins on which the model was based were human 17-β-hydroxysteroid dehydrogenase type XI (PDB ID 1YB1) and β-ketoacyl-[ACP] reductase from Escherichia coli in complex with NADP+ (PDB ID 1Q7B).
Retinol dehydrogenase enzyme assay
Retinol dehydrogenase activity assays were done under dim red light. For each assay, 125 μg of total membrane proteins from COS-7 cells expressing RDH10 in 0.1 M sodium phosphate buffer (pH 7.4) were added into 200 μL of the activity assay buffer containing 0.1 M sodium phosphate (pH 7.4), 0.5% BSA, and 1 mM NADP. The reaction was started by the addition of 2 μCi of all-trans-[11,12-3H]-retinol (specific activity 47 Ci/mM; Perkin Elmer) in 2 μL of dimethyl formamide. After 20 min the reaction was stopped by the addition 300 μL of ice-cold methanol. Retinoids were extracted and separated by normal phase HPLC as described previously (Wu et al. 2004). Elution peaks were identified by spiking with corresponding retinoid standards. The activity was calculated from the area of all-trans-[3H]-retinal peak using synthetic all-trans-[3H]-retinal as a standard for calibration.
RA reporter transgenic mouse
The RARE–lacZ transgenic mice (Rossant et al. 1991) were a generous gift of Dr. Janet Rossant (Hospital for Sick Children, Toronto, Ontario, Canada) via the laboratory of Dr. Ursula Drager (Shriver Center, University of Massachusetts Medical School, Waltham, MA). Staining for β-galactosidase activity was performed using reagents from Specialty Media.
RA rescue
For the RA rescue experiments, RA was administered via supplementation of the maternal diet. RA was mixed with Transgel food/water gel packs (Charles River) in mylar packets to protect from light. In order to minimize stress to pregnant females during the time of supplementation, Transgel was substituted for maternal diet beginning at E0.5 and was maintained until the end of supplementation at E11.5. From E7.5 to E10.5, all-trans-RA (Sigma) was mixed into the Transgel. Pregnant females were provided with an excess of Transgel that they could consume ad libidinum. The dose of RA was based the observation that pregnant females consumed or distributed ∼20 g of Transgel per day. At E7.5, RA was supplemented at a dose of 550 μg of RA/20 μg of Transgel per day. From E8.5 through E10.5, RA was supplemented at a dose of 1.25 mg of RA/20 μg of Transgel per day. RA Transgel packs were replaced fresh once per day.
Acknowledgments
We are indebted to Andy Salinger and Monica Justice for their assistance in initiating our mutagenesis screen. We greatly appreciate the technical expertise of Teresa Terrel, Rodney McCay, Michael Morgan, and Lacey Ellington. Research in the P.A.T. Laboratory is supported by National Institute of Dental and Craniofacial Research Grant R01 DE 016082-01, the Hudson Foundation, and the Stowers Institute for Medical Research.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1533407
References
- Clagett-Dame M., DeLuca H.F., DeLuca H.F. The role of vitamin A in mammalian reproduction and embryonic development. Annu. Rev. Nutr. 2002;22:347–381. doi: 10.1146/annurev.nutr.22.010402.102745E. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. The PyMOL molecular graphics system. DeLano Scientific LLC; Palo Alto, CA: 2002. [Google Scholar]
- Deltour L., Foglio M.H., Duester G., Foglio M.H., Duester G., Duester G. Metabolic deficiencies in alcohol dehydrogenase Adh1, Adh3, and Adh4 null mutant mice. Overlapping roles of Adh1 and Adh4 in ethanol clearance and metabolism of retinol to retinoic acid. J. Biol. Chem. 1999;274:16796–16801. doi: 10.1074/jbc.274.24.16796. [DOI] [PubMed] [Google Scholar]
- Duester G. Families of retinoid dehydrogenases regulating vitamin A function: Production of visual pigment and retinoic acid. Eur. J. Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- Duester G., Mic F.A., Molotkov A., Mic F.A., Molotkov A., Molotkov A. Cytosolic retinoid dehydrogenases govern ubiquitous metabolism of retinol to retinaldehyde followed by tissue-specific metabolism to retinoic acid. Chem. Biol. Interact. 2003;143–144:201–210. doi: 10.1016/s0009-2797(02)00204-1. [DOI] [PubMed] [Google Scholar]
- Dupe V., Matt N., Garnier J.M., Chambon P., Mark M., Ghyselinck N.B., Matt N., Garnier J.M., Chambon P., Mark M., Ghyselinck N.B., Garnier J.M., Chambon P., Mark M., Ghyselinck N.B., Chambon P., Mark M., Ghyselinck N.B., Mark M., Ghyselinck N.B., Ghyselinck N.B. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. 2003;100:14036–14041. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Molotkov A., Manabe S., Donmoyer C.M., Deltour L., Foglio M.H., Cuenca A.E., Blaner W.S., Lipton S.A., Duester G., Molotkov A., Manabe S., Donmoyer C.M., Deltour L., Foglio M.H., Cuenca A.E., Blaner W.S., Lipton S.A., Duester G., Manabe S., Donmoyer C.M., Deltour L., Foglio M.H., Cuenca A.E., Blaner W.S., Lipton S.A., Duester G., Donmoyer C.M., Deltour L., Foglio M.H., Cuenca A.E., Blaner W.S., Lipton S.A., Duester G., Deltour L., Foglio M.H., Cuenca A.E., Blaner W.S., Lipton S.A., Duester G., Foglio M.H., Cuenca A.E., Blaner W.S., Lipton S.A., Duester G., Cuenca A.E., Blaner W.S., Lipton S.A., Duester G., Blaner W.S., Lipton S.A., Duester G., Lipton S.A., Duester G., Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell. Biol. 2003;23:4637–4648. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia M.J., Eggenschwiler J.T., Caspary T., Alcorn H.L., Wyler M.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Eggenschwiler J.T., Caspary T., Alcorn H.L., Wyler M.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Caspary T., Alcorn H.L., Wyler M.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Alcorn H.L., Wyler M.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Wyler M.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Huangfu D., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Rakeman A.S., Lee J.D., Feinberg E.H., Timmer J.R., Lee J.D., Feinberg E.H., Timmer J.R., Feinberg E.H., Timmer J.R., Timmer J.R., et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc. Natl. Acad. Sci. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms J.A., Kim C.H., Eichele G., Thaller C., Kim C.H., Eichele G., Thaller C., Eichele G., Thaller C., Thaller C. Retinoic acid signaling is required during early chick limb development. Development. 1996;122:1385–1394. doi: 10.1242/dev.122.5.1385. [DOI] [PubMed] [Google Scholar]
- Hernandez R.E., Putzke A.P., Myers J.P., Margaretha L., Moens C.B., Putzke A.P., Myers J.P., Margaretha L., Moens C.B., Myers J.P., Margaretha L., Moens C.B., Margaretha L., Moens C.B., Moens C.B. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice M.J. Mutagenesis of the mouse germline. In: Jackson I.J., Abbot C.M., Abbot C.M., editors. Mouse genetics and transgenics: A practical approach. Oxford University Press; Oxford: 2000. pp. 185–245. [Google Scholar]
- Kasus-Jacobi A., Ou J., Birch D.G., Locke K.G., Shelton J.M., Richardson J.A., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Ou J., Birch D.G., Locke K.G., Shelton J.M., Richardson J.A., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Birch D.G., Locke K.G., Shelton J.M., Richardson J.A., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Locke K.G., Shelton J.M., Richardson J.A., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Shelton J.M., Richardson J.A., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Richardson J.A., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Valenzuela D.M., Yancopoulos G.D., Edwards A.O., Yancopoulos G.D., Edwards A.O., Edwards A.O. Functional characterization of mouse RDH11 as a retinol dehydrogenase involved in dark adaptation in vivo. J. Biol. Chem. 2005;280:20413–20420. doi: 10.1074/jbc.M413789200. [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Raya A., Raya R.M., Rodriguez-Esteban C., Belmonte J.C., Raya A., Raya R.M., Rodriguez-Esteban C., Belmonte J.C., Raya R.M., Rodriguez-Esteban C., Belmonte J.C., Rodriguez-Esteban C., Belmonte J.C., Belmonte J.C. Retinoic acid signalling links left–right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- Kim T.S., Maeda A., Maeda T., Heinlein C., Kedishvili N., Palczewski K., Nelson P.S., Maeda A., Maeda T., Heinlein C., Kedishvili N., Palczewski K., Nelson P.S., Maeda T., Heinlein C., Kedishvili N., Palczewski K., Nelson P.S., Heinlein C., Kedishvili N., Palczewski K., Nelson P.S., Kedishvili N., Palczewski K., Nelson P.S., Palczewski K., Nelson P.S., Nelson P.S. Delayed dark adaptation in 11-cis-retinol dehydrogenase-deficient mice: A role of RDH11 in visual processes in vivo. J. Biol. Chem. 2005;280:8694–8704. doi: 10.1074/jbc.M413172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T., Wagner E., Grun F., Drager U.C., Wagner E., Grun F., Drager U.C., Grun F., Drager U.C., Drager U.C. Retinoic acid signaling in the brain marks formation of optic projections, maturation of the dorsal telencephalon, and function of limbic sites. J. Comp. Neurol. 2004;470:297–316. doi: 10.1002/cne.20013. [DOI] [PubMed] [Google Scholar]
- Mansouri A., Voss A.K., Thomas T., Yokota Y., Gruss P., Voss A.K., Thomas T., Yokota Y., Gruss P., Thomas T., Yokota Y., Gruss P., Yokota Y., Gruss P., Gruss P. Uncx4.1 is required for the formation of the pedicles and proximal ribs and acts upstream of Pax9. Development. 2000;127:2251–2258. doi: 10.1242/dev.127.11.2251. [DOI] [PubMed] [Google Scholar]
- Martin G.R. The roles of FGFs in the early development of vertebrate limbs. Genes & Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- Matt N., Dupe V., Garnier J.M., Dennefeld C., Chambon P., Mark M., Ghyselinck N.B., Dupe V., Garnier J.M., Dennefeld C., Chambon P., Mark M., Ghyselinck N.B., Garnier J.M., Dennefeld C., Chambon P., Mark M., Ghyselinck N.B., Dennefeld C., Chambon P., Mark M., Ghyselinck N.B., Chambon P., Mark M., Ghyselinck N.B., Mark M., Ghyselinck N.B., Ghyselinck N.B. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development. 2005;132:4789–4800. doi: 10.1242/dev.02031. [DOI] [PubMed] [Google Scholar]
- Mic F.A., Duester G., Duester G. Patterning of forelimb bud myogenic precursor cells requires retinoic acid signaling initiated by Raldh2. Dev. Biol. 2003;264:191–201. doi: 10.1016/s0012-1606(03)00403-2. [DOI] [PubMed] [Google Scholar]
- Mic F.A., Haselbeck R.J., Cuenca A.E., Duester G., Haselbeck R.J., Cuenca A.E., Duester G., Cuenca A.E., Duester G., Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129:2271–2282. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mic F.A., Sirbu I.O., Duester G., Sirbu I.O., Duester G., Duester G. Retinoic acid synthesis controlled by Raldh2 is required early for limb bud initiation and then later as a proximodistal signal during apical ectodermal ridge formation. J. Biol. Chem. 2004;279:26698–26706. doi: 10.1074/jbc.M401920200. [DOI] [PubMed] [Google Scholar]
- Minina E., Wenzel H.M., Kreschel C., Karp S., Gaffield W., McMahon A.P., Vortkamp A., Wenzel H.M., Kreschel C., Karp S., Gaffield W., McMahon A.P., Vortkamp A., Kreschel C., Karp S., Gaffield W., McMahon A.P., Vortkamp A., Karp S., Gaffield W., McMahon A.P., Vortkamp A., Gaffield W., McMahon A.P., Vortkamp A., McMahon A.P., Vortkamp A., Vortkamp A. BMP and Ihh/PTHrP signaling interact to coordinate chondrocyte proliferation and differentiation. Development. 2001;128:4523–4534. doi: 10.1242/dev.128.22.4523. [DOI] [PubMed] [Google Scholar]
- Molotkov A., Deltour L., Foglio M.H., Cuenca A.E., Duester G., Deltour L., Foglio M.H., Cuenca A.E., Duester G., Foglio M.H., Cuenca A.E., Duester G., Cuenca A.E., Duester G., Duester G. Distinct retinoid metabolic functions for alcohol dehydrogenase genes Adh1 and Adh4 in protection against vitamin A toxicity or deficiency revealed in double null mutant mice. J. Biol. Chem. 2002a;277:13804–13811. doi: 10.1074/jbc.M112039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A., Fan X., Deltour L., Foglio M.H., Martras S., Farres J., Pares X., Duester G., Fan X., Deltour L., Foglio M.H., Martras S., Farres J., Pares X., Duester G., Deltour L., Foglio M.H., Martras S., Farres J., Pares X., Duester G., Foglio M.H., Martras S., Farres J., Pares X., Duester G., Martras S., Farres J., Pares X., Duester G., Farres J., Pares X., Duester G., Pares X., Duester G., Duester G. Stimulation of retinoic acid production and growth by ubiquitously expressed alcohol dehydrogenase Adh3. Proc. Natl. Acad. Sci. 2002b;99:5337–5342. doi: 10.1073/pnas.082093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriss-Kay G.M., Sokolova N., Sokolova N. Embryonic development and pattern formation. FASEB J. 1996;10:961–968. doi: 10.1096/fasebj.10.9.8801178. [DOI] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R.R., Gertsenstein M., Vintersten K., Behringer R.R., Vintersten K., Behringer R.R., Behringer R.R. Manipulating the mouse embryo. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 2003. [Google Scholar]
- Niederreither K., McCaffery P., Drager U.C., Chambon P., Dollé P., McCaffery P., Drager U.C., Chambon P., Dollé P., Drager U.C., Chambon P., Dollé P., Chambon P., Dollé P., Dollé P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- Niederreither K., Subbarayan V., Dollé P., Chambon P., Subbarayan V., Dollé P., Chambon P., Dollé P., Chambon P., Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Niederreither K., Vermot J., Schuhbaur B., Chambon P., Dollé P., Vermot J., Schuhbaur B., Chambon P., Dollé P., Schuhbaur B., Chambon P., Dollé P., Chambon P., Dollé P., Dollé P. Retinoic acid synthesis and hindbrain patterning in the mouse embryo. Development. 2000;127:75–85. doi: 10.1242/dev.127.1.75. [DOI] [PubMed] [Google Scholar]
- Niederreither K., Vermot J., Fraulob V., Chambon P., Dolle P., Vermot J., Fraulob V., Chambon P., Dolle P., Fraulob V., Chambon P., Dolle P., Chambon P., Dolle P., Dolle P. Retinaldehyde dehydrogenase 2 (RALDH2)-independent patterns of retinoic acid synthesis in the mouse embryo. Proc. Natl. Acad. Sci. 2002a;99:16111–16116. doi: 10.1073/pnas.252626599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K., Vermot J., Schuhbaur B., Chambon P., Dolle P., Vermot J., Schuhbaur B., Chambon P., Dolle P., Schuhbaur B., Chambon P., Dolle P., Chambon P., Dolle P., Dolle P. Embryonic retinoic acid synthesis is required for forelimb growth and anteroposterior patterning in the mouse. Development. 2002b;129:3563–3574. doi: 10.1242/dev.129.15.3563. [DOI] [PubMed] [Google Scholar]
- Nitta H., Kishimoto J., Grogan T.M., Kishimoto J., Grogan T.M., Grogan T.M. Application of automated mRNA in situ hybridization for formalin-fixed, paraffin-embedded mouse skin sections: Effects of heat and enzyme pretreatment on mRNA signal detection. Appl. Immunohistochem. Mol. Morphol. 2003;11:183–187. doi: 10.1097/00129039-200306000-00017. [DOI] [PubMed] [Google Scholar]
- Power S.C., Lancman J., Smith S.M., Lancman J., Smith S.M., Smith S.M. Retinoic acid is essential for Shh/Hoxd signaling during rat limb outgrowth but not for limb initiation. Dev. Dyn. 1999;216:469–480. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<469::AID-DVDY15>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Riddle R.D., Johnson R.L., Laufer E., Tabin C., Johnson R.L., Laufer E., Tabin C., Laufer E., Tabin C., Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Rossant J., Zirngibl R., Cado D., Shago M., Goguere V., Zirngibl R., Cado D., Shago M., Goguere V., Cado D., Shago M., Goguere V., Shago M., Goguere V., Goguere V. Expression of retinoic acid response element–hsp lacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes & Dev. 1991;5:1333–1344. doi: 10.1101/gad.5.8.1333. [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C., Kopp J., Guex N., Peitsch M.C., Guex N., Peitsch M.C., Peitsch M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears K.E., Behringer R.R., Rasweiler IV J.J., Niswander L.A., Behringer R.R., Rasweiler IV J.J., Niswander L.A., Rasweiler IV J.J., Niswander L.A., Niswander L.A. Development of bat flight: Morphologic and molecular evolution of bat wing digits. Proc. Natl. Acad. Sci. 2006;103:6581–6586. doi: 10.1073/pnas.0509716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E., Lai K., Packer A.I., Paik J., Blaner W.S., de Morais Vieira M., Gouras P., Wolgemuth D.J., Lai K., Packer A.I., Paik J., Blaner W.S., de Morais Vieira M., Gouras P., Wolgemuth D.J., Packer A.I., Paik J., Blaner W.S., de Morais Vieira M., Gouras P., Wolgemuth D.J., Paik J., Blaner W.S., de Morais Vieira M., Gouras P., Wolgemuth D.J., Blaner W.S., de Morais Vieira M., Gouras P., Wolgemuth D.J., de Morais Vieira M., Gouras P., Wolgemuth D.J., Gouras P., Wolgemuth D.J., Wolgemuth D.J. Targeted disruption of the mouse cis-retinol dehydrogenase gene: Visual and nonvisual functions. J. Lipid Res. 2002;43:590–597. [PubMed] [Google Scholar]
- Sirbu I.O., Duester G., Duester G. Retinoic-acid signalling in node ectoderm and posterior neural plate directs left–right patterning of somitic mesoderm. Nat. Cell Biol. 2006;8:271–277. doi: 10.1038/ncb1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford T., Horton C., Maden M., Horton C., Maden M., Maden M. Retinoic acid is required for the initiation of outgrowth in the chick limb bud. Curr. Biol. 1996;6:1124–1133. doi: 10.1016/s0960-9822(02)70679-9. [DOI] [PubMed] [Google Scholar]
- Tickle C. Patterning systems—From one end of the limb to the other. Dev. Cell. 2003;4:449–458. doi: 10.1016/s1534-5807(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Vermot J., Pourquie O., Pourquie O. Retinoic acid coordinates somitogenesis and left–right patterning in vertebrate embryos. Nature. 2005;435:215–220. doi: 10.1038/nature03488. [DOI] [PubMed] [Google Scholar]
- Wu B.X., Chen Y., Fan J., Rohrer B., Crouch R.K., Ma J.X., Chen Y., Fan J., Rohrer B., Crouch R.K., Ma J.X., Fan J., Rohrer B., Crouch R.K., Ma J.X., Rohrer B., Crouch R.K., Ma J.X., Crouch R.K., Ma J.X., Ma J.X. Cloning and characterization of a novel all-trans retinol short-chain dehydrogenase/reductase from the RPE. Invest. Ophthalmol. Vis. Sci. 2002;43:3365–3372. [PubMed] [Google Scholar]
- Wu B.X., Moiseyev G., Chen Y., Rohrer B., Crouch R.K., Ma J.X., Moiseyev G., Chen Y., Rohrer B., Crouch R.K., Ma J.X., Chen Y., Rohrer B., Crouch R.K., Ma J.X., Rohrer B., Crouch R.K., Ma J.X., Crouch R.K., Ma J.X., Ma J.X. Identification of RDH10, an all-trans retinol dehydrogenase, in retinal Muller cells. Invest. Ophthalmol. Vis. Sci. 2004;45:3857–3862. doi: 10.1167/iovs.03-1302. [DOI] [PubMed] [Google Scholar]