Abstract
Early in life, there is a delicate and critical balance aimed to maintain low hormone responses derived from the stress responsive hypothalamic–pituitary–adrenal axis (HPA). However, in the infant rat hypothalamic corticotrophin-releasing hormone (CRH) stress responses to environmental events are clearly seen even though other elements of the HPA axis may have limited responses. In view of the role of CRH in mediating behavior associated with stress and anxiety, we considered the ontogeny and the effects of prolonged maternal deprivation (DEP) in brain areas that express CRH-related molecules outside the hypothalamus. We hypothesized that DEP would alter the ontogeny of CRH, CRH binding protein and CRH receptor 1 in prefrontal cortex, amygdala, septum and hippocampus, areas that are part of the CRH extra hypothalamic system, and that a differential modulation would be observed in response to restraint. We compared non-deprived animals to animals subjected to 24 h of DEP at 6, 12 and 18 days of life. We found (1) developmental patterns, which were idiosyncratic to the anatomical area examined, and (2) a temporal response of mRNA levels which was also site specific. The genomic changes are not always related to maternal deprivation status, in fact DEP enhanced, suppressed or had no consequence on the underlying ontogenic progression and restraint response of these CRH-related molecules. We conclude that the extra hypothalamic CRH system is a dynamic system responding to developmental and environmental demands challenging the basic assumption of stress hypo responsiveness in the infant rat. This modulation may have important repercussions on morphological organization and events leading to neuroprotection.
Keywords: Gene regulation, Stress response, Peptide, Receptor
1. Introduction
Ontogenic studies have documented a period within the first two weeks of life during which stressors elicit a rather minimal adrenal response in the infant rat. This period has been called the stress-hyporesponsive period (SHRP) (for a review, see Walker et al., 2001). However, recent research has shown the paraventricular nucleus of the hypothalamus (PVN), that guide the infant stress response, is not unresponsive to stress but rather hyper responsive when studied using short time frames (Dent et al., 2000a,b). Using the animal paradigm of prolonged maternal deprivation at several ages during the SHRP, Dent and co-workers showed that maternally deprived animals that are administered a mild stimulus of a saline injection have an endocrine response and increased corticotrophin-releasing hormone (CRH) biosynthesis in the PVN. Surprisingly, although non-deprived animals have minimal adrenal response to this stressor, a rapid induction of CRH mRNA transcription was observed in the PVN. In addition, the small elevation in glucocorticoid level was capable of suppressing the early induction in all ages studied (Dent et al., 2000b). These data suggest that during ontogeny central brain elements are clearly responsive to environmental events even though adrenal responses may be limited and that small but significant elevations in glucocorticoids can elicit negative regulation of these responses.
Although glucocorticoids inhibit hypothalamic PVN CRH neurons, their action elicits CRH synthesis and release in other brain areas in the adult animal. Elevation in glucocorticoids increase CRH release from the amygdala and CRH terminals projecting from this structure that can lead to the modulation of cardiovascular tone in response to a stressor (Makino et al., 1999). CRH-related gene expression changes in the amygdala and prefrontal cortical areas have also been associated with mechanisms that underlie enhancement or facilitation of the hypothalamic–pituitary–adrenal axis (HPA) response (Bhatnagar and Dallman, 1998; Herman and Cullinan, 1997; Herman et al., 1996). Evidence that CRH acts on other brain areas is the fact that when CRH is administered centrally to adult rats a wide spectrum of autonomic, electrophysiologic and behavioral effects are seen that appear to integrate perception and expression of fear or anxiety (Charney and Deutch, 1996; Gray and Bingaman, 1996). These effects are mediated through the interaction of CRH with CRH receptor 1 (CRHr1) and to a very limited extent, CRH receptor 2 (CRHr2), receptors that are present within cortical and subcortical layers, limbic structures and brainstem nuclei (Chalmers et al., 1996; Lewis et al., 2001). Yet, another CRH-related molecule that also participates in CRH neuromodulator actions in brain is the CRH binding protein (CRHbp). CRH binding protein has a high affinity for CRH that limits CRH receptor occupation (Seasholtz et al., 2001). Its expression has been found in a variety of brain regions, most notably the neocortex and hippocampus (Chalmers et al., 1996; Seasholtz et al., 2001). Thus, hypothalamic areas outside the PVN that contain CRH, CRH binding protein and/or CRH receptor 1 appear to play a pivotal role in endocrine and behavioral responses to stress.
The site-specific glucocorticoid modulation of the extra hypothalamic CRH system in the adult animal led us to consider the effects of stress in brain areas that express CRH-related molecules outside the PVN in the developing animal. However, the density and location of CRH receptors is altered through development. Some brain regions exhibit a decline in CRHr1 and r2 expression as the animal matures, whereas other regions show increased gene expression during specific stages of development (Avishai-Eliner et al., 1996; Eghbal-Ahmadi et al., 1998). To our knowledge, the expression of CRH binding protein early in development has not been described (Eckart et al., 2002) and the modulation of these CRH-related molecules by stress has been relatively unexplored during development. Therefore, we hypothesized that maternal deprivation would alter the ontogenic progression of CRH, CRHbp and CRHr1 and that a differential modulation would be observed in deprived vs. non-deprived animals dependent on the glucocorticoid rise observed when exposed to the relative moderate stressor of restraint. We chose ages representative of the varying adrenocortical response observed during the SHRP of which we have already reported the PVN CRH neuron's response after restraint (Dent et al., 2000a). Our goals were, therefore, three-fold: (1) to investigate CRH expression in brain regions outside the hypothalamus in 6-, 12- and 18-day-old animals; (2) to assess CRH modulation in the presence of a robust vs. a limited glucocorticoid response to an aversive stimulus typical of the responses observed in maternally deprived and non-deprived pups; and (3) to ascertain the regulation of the other components of the CRH system that can influence CRH physiologic action, i.e., binding protein and CRHr1.
2. Results
We examined cortex (CTX), amygdala (AMYG), septum (SEP) and hippocampus (HC) for the effect of DEP and modulation following restraint stress. We also examined the ontogenic progression of mRNA expression comparing the time 0 NDEP group across all the ages studied. An initial multi-factorial analysis of variance revealed no significant sex effect for all of the molecules within each anatomical region. This is consistent with other studies using this maternal deprivation paradigm early in life (Vazquez et al., 1993, 1996). The data were therefore collapsed across the sex variable for all molecules within each respective region tested. We present subsequent analyses for each anatomical site studied below. Fig. 1 depicts microphotographs that represent the in situ images from which the mRNA transcript expression were analyzed. Table 1 is a summary of all the results.
Fig. 1.
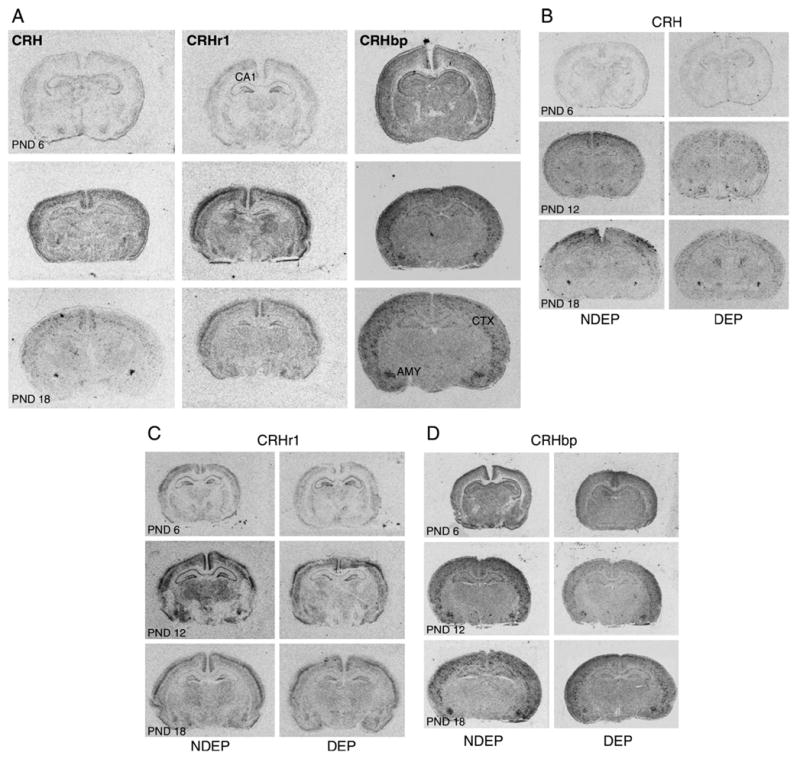
A representative microphotograph of mRNA localization via in-situ hybridization of CRH, CRHr1 and CRHbp on 6-, 12- and 18-day-old animals. Panel A depicts the age progression for each of these molecules in NDEP control animals. Panels B, C and D shows CRH, CRHr1 and CRHbp expression in NDEP and DEP animals across all ages studied. PND=postnatal, CTX=cortex, AMYG=amygdala, hippocampal pyramidal cell regions: cytoarchitectural area 1 (CA1), CA2, CA3, hippocampal granular cell region: dentate gyrus (DG).
Table 1.
Summary of the ontogenic progression and effects of a single 24-h maternal deprivation and response to restraint in the 6-, 12- and 18-day-old animal
| Cortex
|
Amygdala
|
Septum
|
Hippocampus
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecules | CRH | CRHbp | CRHr1 | CRH | CRHbp | CRHr1 | CRH | CRHbp | CRHr1 | CRH | CRHbp | CRHr1 |
| Ontogeny (0 time point, NDEP) | ||||||||||||
| Postnatal day 6 | Low | ↑ | Low | Low | Low | ↑ | Low | Low | Low- | Low CA1, CA3, DG | Low | Low CA1
Low CA3–4 |
| Postnatal day 12 | ↑ ↑ | ↑ ↑ | ↑ ↑ | ↑ | ↑ | No change | Low | No change | -Low- | No change | Low | ↓ CA1
↑ ↑ DG |
| Postnatal day 18 | ↑ ↑ | ↑ ↑ | ↓ ↓ | ↑ ↑ | ↑ ↑ | No change | ↓ | No change | ↓ ↓ | No change | ↓ CA1 | ↓ ↓ CA1
↓ DG |
| Maternal deprivation (0 time point, DEP) | ||||||||||||
| Postnatal day 6 | NE | NE | NE | NE | NE | NE | NE | NE | NE | ↑ CA1 | NE | NE |
| Postnatal day 12 | ↓ | NE | ↓ | NE | NE | NE | NE | NE | NE | ↓CA1 | NE | NE |
| Postnatal day 18 | ↓ | NE | NE | NE | ↓ | NE | NE | NE | NE | NE | NE | ↓CA3 |
| Restraint response | ||||||||||||
| Postnatal day 6 | NE | NE | NE | ↓DEP | NE | ↓NDEP and DEP | ↑ | ↑ | ↑NDEP | ↓CA1 DEP | NE | ↑CA1DEP, DG |
| Postnatal day 12 | ↓NDEP | ↑NDEP and DEP | ↓NDEP and DEP | ↓NDEP and DEP | NE | ↓NDEP | ↑ | ↑ | ↑NDEP | ↑DG | NE | ↓CA1 DEP, CA3 NDEP and DEP, DG |
| Postnatal day 18 | ↑DEP | NE | NE | ↓DEP | ↓NDEP and DEP | ↑NDEP | ↑ | ↑ | NE | NE | NE | ↑CA3 DEP |
NE=no effect.
2.1. Cortex
2.1.1. CRH mRNA
Cortical CRH mRNA data are presented in Fig. 2, Panel A. A three-way ANOVA revealed a main effect of age (F(2, 242)= 32.1, p <0.0001) and time after restraint (F(4, 242)=2.73, p=0.03). In addition, the following interactions were observed: an age by DEP treatment (F(2, 242)=3.40, p=0.03); an age by time after restraint stress (F(8, 242)=3.29, p=0.001); a DEP treatment by time after restraint (F(4, 242)=3.29, p=0.01); and an age by treatment by time interaction (F(8, 242)=2.9, p=0.005). Consequently, the ANOVA was repeated splitting by age and treatment as independent variables. This revealed a time effect for the 12- and 18-day-old animal (time: NDEP PND 12: F(4, 43)=9.15, p<0.0001, and DEP PND 18: F(4, 43)=3.31, p=0.02; see Panel A, Fig. 2). Specifically, the NDEP 12-day-old animal had significant down-regulation of cortical CRH mRNA at 15, 30 and 240 min. Similarly, the 18-day-old DEP animal showed a down-regulation at 30 min post-restraint.
Fig. 2.
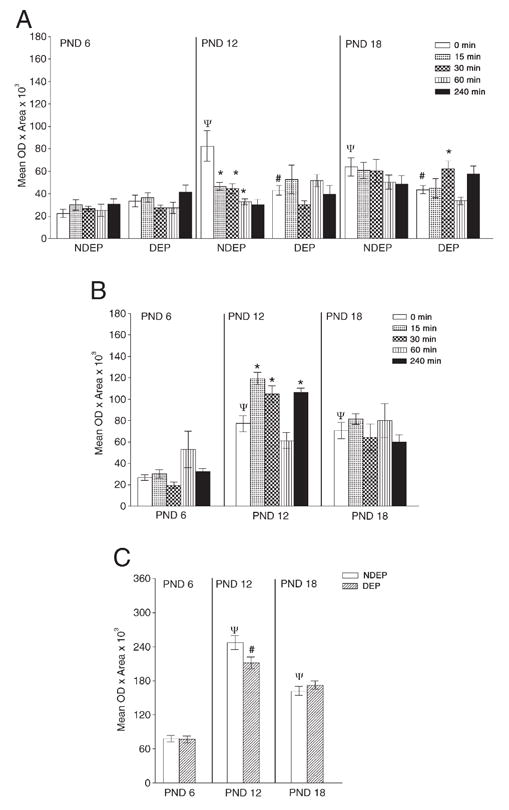
Densitometric analyses of CRH-related molecules in cortex. Cortical CRH mRNA (Panel A), CRH binding protein (CRHbp; Panel B), CRH receptor 1 (CRHr1; Panel C) are shown. NDEP and DEP rats were subjected to a 30-min period of physical restraint. Tissue was obtained at rest (time 0) and 15, 30, 60 and 240 min post-restraint. Data for each age were analyzed using ANOVA considering brain region, age, sex, DEP treatment and time of sacrifice. When no differences were found with a particular variable, the data were collapsed across this variable. Consequently, Panel A depicts all variables (except sex), Panel B is collapsed across the DEP variable and Panel C depicts an age effect only (data collapsed across time). Data are presented as mean±SEM. Post hoc analysis is represented as *p<0.05, significantly different from corresponding basal level (time 0); #p<0.05, significantly different from NDEP time point, p<0.05, significantly different from PND 6 time point.
We observed an ontogenic progression of cortical CRH mRNA expression when comparing time 0 of the NDEP animals across all the ages studied. The PND 6 NDEP animals displayed significantly lower CRH mRNA levels when compared to animals of older ages [see time 0, Fig. 2, Panel A (age NDEP 0 min, F(2, 22)=8.63, p=0.002)]. The DEP treatment had an effect on this progression in the 12- and 18-day-old animal.
2.1.2. CRH binding protein—cortical
CRHbp data are depicted in Fig. 2, Panel B. In this structure, a three-way ANOVA revealed a main effect of age (F(2, 129)= 155.29, p<0.0001) and time (F(4, 129)=4.10, p=0.004) without a DEP treatment effect. In addition, an interaction of age with time after restraint was detected (F(8, 129)=3.80, p=0.0005). The ANOVA was then performed splitting for age as independent variable revealing the effect on the 12- and 18-day-old animals (age: PND 12—F(4, 53)=6.88, p=0.0002; PND 18—F(4, 48)=2.73, p=0.04). Considering time as an independent variable, 15, 30, 60 and 240 min showed significance (time: 15 min, F(2, 29)= 58.98, p<0.0001; 30 min, F(2, 30)=41.21, p<0.0001; 60 min, F(2, 21)=11.17, p=0.0005; 240 min, F(2, 30)=45.73, p<0.0001—see Fig. 2, Panel B for post hoc significant comparisons across time).
The analysis of the ontogenic progression of CRHbp gene expression revealed an “inverted U” profile (age: 0 min—F(2, 34)=23.15, p<0.0001). The PND 12 NDEP animals showed significantly greater mRNA levels at time 0 when compared to PND 6 and 18 at this same time point (see time 0, PND 12 and 18, in Panel B, Fig. 2).
2.1.3. CRH receptor 1
Cortical CRHr1 is shown in Fig. 2, Panel C. A three-way ANOVA revealed a main effect of age (F(2, 223)=156.48, p<0.0001), with an age by DEP treatment interaction (F(2, 223)=3.59, p=0.03). A split by age analysis revealed the DEP effect in the 12-day-old animals (treatment: PND 12, F(1, 92)=4.93, p=0.03).
The ontogenic progression of CRHr1 gene expression mirrored the CRHbp “inverted U” profile (age, NDEP, 0 min: F(2, 19)=22.22, p<0.0001). The PND 12 and 18 NDEP animals showed significantly greater mRNA levels at time 0 when compared to PND 6 at this same time point (compare time 0, PND 12 and 18 with PND 6 in Panel B, Fig. 2).
In summary, the ontogenic progression of the molecules in cortex suggests that the occupation of CRHr1 receptor in the 12-day-old animal is buffered by increases in CRHbp levels at this age. Maternal deprivation, which can be construed as a chronic stress, affects ontogenic progression of cortical CRH and CRHr1, down-regulating their expression (NDEP vs. DEP, time 0 time point). We also observed that the 12-day-old NDEP animal, the CRH and CRHbp gene expression are particularly sensitive to acute restraint stress because both of these molecules decrease during the time course of this stressful manipulation. In contrast, CRHr1 remains unchanged through the restraint time course.
2.2. Amygdala
2.2.1. CRH mRNA
Within the amygdala, a main effect of age (F(2, 236)=200.87, p<0.0001) and time after restraint (F(4, 236)=8.30, p<0.0001) were observed. Interactions of time with DEP treatment (F(4, 236)=2.88, p=0.02), age with DEP treatment (F(2, 236)=6.06, p=0.003) and time with age and treatment (F(8, 236)=4.53, p<0.0001) were detected. The ANOVA was repeated splitting by age and DEP treatment as independent variables. This revealed a time effect in the DEP animals (time: PND 6—F(4, 38)=5.64, p=0.0012; PND 12—F(4, 38)=3.39, p=0.02; PND 18—F(4. 38)=3.14, p=0.02) and the 12-day-old NDEP control (F(4, 42)= 12.96, p<0.0001). Overall, the animals at these particular ages and conditions had a delayed decrease in CRH gene expression after restraint. These results are depicted in Fig. 3, Panel A.
Fig. 3.
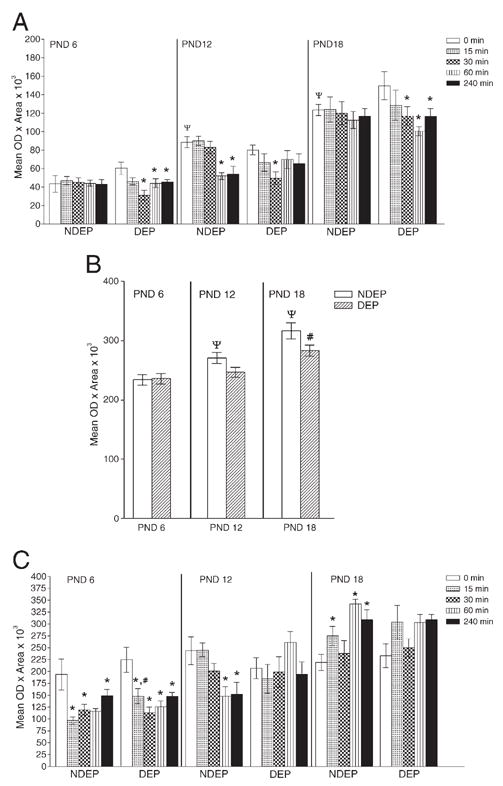
Densitometric analyses of CRH-related molecules in amygdala. Panel A depicts CRH mRNA with all variables tested, Panel B depicts CRHbp age and DEP treatment effect only (data collapsed across time), whereas Panel C shows CRHr1 depicting all variables. Data are presented as mean± SEM. Post hoc analysis is represented as *p<0.05, significantly different from corresponding basal level (time 0); #p<0.05, significantly different from NDEP time point, ψp<0.05, significantly different from PND 6 time point.
The ontogenic expression of CRH mRNA levels within the amygdala has a pattern of a gradual increase in basal CRH mRNA levels. This is evident comparing time 0 NDEP value across all ages (age: NDEP 0 min, F(2, 23)=31.58, p<0.0001).
2.2.2. CRH binding protein
A three-way ANOVA showed a main effect of age (F(2, 227)= 23.06, p<0.0001) and a deprivation treatment (F(1, 227)=4.99, p=0.03) without interactions. No stress-induced changes in the CRHbp mRNA expression resulted within the 2 h following physical restraint; consequently, the data were collapsed across the time after restraint variable. Fig. 3, Panel B shows these relations.
As with CRH mRNA, a stepwise ontogenic progression of CRHbp mRNA is observed with increasing age (age: NDEP F(2, 121)=14.91, p<0.0001; DEP F(2, 130)=8.12, p=0.0004). The maternal deprivation treatment altered this progression in the PND 18 animals only (see reduced mRNA expression in Panel B).
2.2.3. CRH receptor 1
Quantification data for CRHr1 mRNA expression in the amygdala are seen in Fig. 3, Panel C. A three-way ANOVA resulted in a main effect of age (F(2, 249)=98.98, p<0.0001), with an interaction of age by time following restraint (F=6.93, p<0.0001) and an age by time after restraint by DEP treatment interaction (F(8, 249)=3.202, p=0.002). The ANOVA was repeated splitting by age and treatment as independent variables. The NDEP control animals showed significant differences in response to the restraint (time: PND 6—F(4, 23)=5.17, p=0.002; PND 12—F(4, 40) = 4.75, p = 0.003, PND 18—F(4, 36) = 7.31, p =0.0002). Only DEP 6-day-old animals had a restraint response (time: PND 6—F(4, 42)=7.23, p=0.0002).
In summary, both CRH and CRH bp mRNA amygdala levels increase as the animal matures. The chronic stress of maternal deprivation down-regulates CRH bp mRNA levels in the 18-day-old animal only. In contrast, CRH and CRH r1 gene expression in NDEP and DEP animals is sensitive to acute restraint stress, but in an age and DEP state-specific manner (see Table 1, restraint response section).
2.3. Septum
2.3.1. CRH mRNA
Analysis of CRH mRNA expression in septum revealed a main effect of age that was opposite from cortex and amygdala (F(2, 251)=66.33, p<0.0001). A time effect was also detected (F(4. 251)= 8.66, p<0.0001) with an age by time interaction (F(8, 251)= 2.00, p=0.05). No DEP treatment effect or interactions were observed (F(1, 251)=0.439, p=0.51). The ANOVA was then performed considering age as independent variable revealing the effect on the 12- and 18-day-old animals (age: PND 12—F(4, 91)=7.47, p<0.0001; PND 18—F(4, 87)=3.94, p=0.005). Considering time as an independent variable, the times after restraint showed significance (time: 15 min, F(2, 29)=58.98, p<0.0001); 30 min, F(2, 30)=41.21, p<0.0001; 60 min, F(2, 21)=11.17, p=0.0005; 240 min, F(2, 30)=45.73, p<0.0001—Fig. 4, Panel A depicts these data collapsed across the DEP variable with post hoc significant comparisons across time.
Fig. 4.
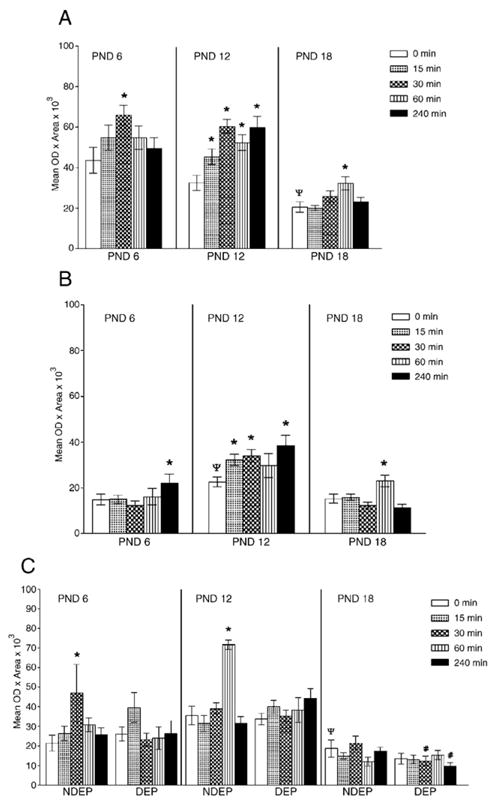
Densitometric analyses of CRH-related molecules in septum. CRH mRNA (Panel A) and CRH bp mRNA (Panel B) depict the response to acute restraint across ages (data collapsed across the DEP treatment variable). Panel C shows CRHr1 mRNA with all variables tested. Data are presented as mean±SEM. Post hoc analysis is represented as *p<0.05, significantly different from corresponding basal level (time 0); #p<0.05, significantly different from NDEP time point, ψp<0.05, significantly different from PND 6 time point.
Overall, we observed a significant decrease in CRH mRNA ontogenic expression with increasing age (age: 0 min—F(2, 52)= 6.94, p=0.002). The 18-day-old NDEP animal at time 0 had significantly lower levels when compared to 12- and 6-day-old pups (see time 0 NDEP, Panel A, Fig. 4).
2.3.2. CRH binding protein
A three-way ANOVA revealed a main effect of age (F(2, 137)= 56.09, p<0.0001) and time following restraint (F(4, 137)=3.03, p=0.02), with an age by time interaction (F(8, 137)=2.93, p=0.005). Maternal deprivation treatment was not significant. The ANOVA was again performed considering for age as an independent variable. Twelve- and 18-day-old animals again showed significant effect of age (age: PND 12—F(4, 55)=3.45, p=0.02; PND 18—F(4, 53)=6.26, p=0.0003). Again considering time as an independent variable times after restraint showed significance (time: 15 min, 30, 240 F(2, 29–32)=19.67–27.76, p<0.0001). Fig. 4, Panel B depicts these data collapsed across the DEP variable with post hoc significant comparisons across time.
We observed a significant increase in CRHbp mRNA ontogenic expression on the PND 12 animal only (time 0 min: F(2, 34)=4.17, p=0.02). Results are depicted in Fig. 4, Panel B (compare time 0 across ages).
2.3.3. CRH receptor 1
An age effect on CRHr1 mRNA expression was revealed with a three-way ANOVA (F(2, 218)=35.15, p<0.0001) with a significant time by DEP treatment interaction (F(4, 218)=2.47, p=0.04). The ANOVA was again performed considering for age and treatment as independent variables and the 12-day-old NDEP animals showed significant effect of time (time: F(4, 43)=2.45, p=0.05). Fig. 4, Panel C shows these results with post hoc comparisons.
Overall, low levels of CRH r1 are seen in septum. However, significantly higher levels are appreciated in the NDEP PND 12 animals at time 0 when compared to 6- and 12-day-old indicating an inverted U pattern of CRHr1 ontogenic progression.
In summary, the CRH and CRHr1 mRNA levels decrease as the animal matures. Chronic deprivation has a minimal effect in this structure. Whereas CRH and CRHr1 mRNA increases are observed in the 6- and 12-day-old NDEP animals in response to restraint stress, the DEP 18-day-old animal down-regulates CRHr1 in response to this acute stressor.
2.4. Hippocampus
2.4.1. CRH mRNA
A low CRH mRNA signal was detected in the hippocampus of all animals studied. A four-way ANOVA revealed a main effect of region (F(3, 924)=4.94, p=0.002); consequently, the ANOVA was performed for each different hippocampal region considering age, treatment and time after restraint. The regional analyses revealed effects in CA1, CA3 and DG (see Fig. 5, Panels A–C). In all of these areas, an age effect was detected (F(2, 231)= 6.33, p=0.05–0.002 depending on the area). A treatment by time interaction was observed in CA1 (F(4, 231)= 2.59, p=0.04), whereas age by time interactions were detected in CA3 (F(8, 231)=2.11, p=0.04) and DG (F(8, 231)=1.98, p=0.05).
Fig. 5.
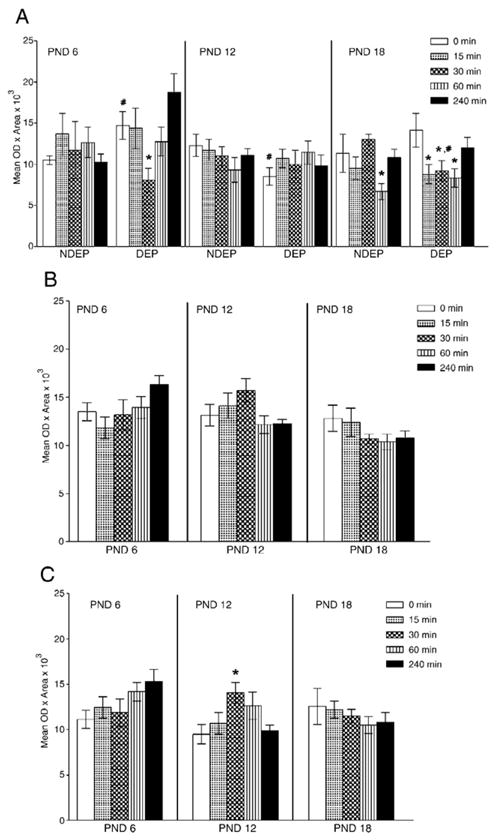
Densitometric analyses of CRH mRNA gene expression in hippocampal CA1 (Panel A), CA3 (Panel B) and dentate gyrus (Panel C) subregions. Panel A depicts the results with all variables tested, whereas Panels B and C show the response to acute restraint across ages (data collapsed across the DEP treatment variable). Post hoc analysis is represented as *p<0.05, significantly different from corresponding basal level (time 0); #p<0.05, significantly different from NDEP time point.
2.4.2. CA1 region
No significant ontogenic changes were detected in CA1 (see NDEP, time 0 across all ages). The age effect detected was linked to the maternal deprivation treatment and the response to the restraint stress (treatment by time interaction: F(4, 231)=2.59, p=0.04). Twelve- and 6-day-old animals had a regulation of CRH mRNA levels following 24 h of maternal deprivation (Fig. 5, Panel A—compare time 0, NDEP and DEP PND 12 animals). Significant CRH mRNA changes after restraint were observed in the NDEP 18-day-old animal and the DEP 6 and 18 days old. See Fig. 5, Panel A for post hoc comparisons across time.
2.4.3. CA3 region
Although an age effect was detected in the CA3 region (F(2, 231)=6.85, p=0.001), no treatment, ontogenic progression or restraint effects were detected. An age by time interaction was observed (F(8, 231)=2.11, p=0.04) linked to the response at 30 min and 240 min. See Fig. 5, Panel B.
2.4.4. Dentate gyrus
Similarly, a treatment effect was not detected in the DG. An age by time interaction was found (F(8. 231)=1.98, p=0.05). The effect on CRH mRNA levels following restraint was seen in the 12-day-old animal only (time: F(4, 89)=2.68, p=0.04). See post hoc comparisons in Panel C, Fig. 5.
2.4.5. CRH binding protein
The CRHbp mRNA analyses revealed limited findings. Specifically, CA1 was the only region that yielded significant mRNA changes. In this region, there was an age effect (age—ontogeny: F(2, 178)=6.73, p=0.001) without any interactions. Post hoc analysis revealed that the 18-day-old animals had low CRHbp mRNA levels when compared to 6- and 12-day-old animals (data not shown).
2.4.6. CRH receptor 1
Distinct hippocampal CRHr1 changes are presented in Fig. 6, Panels A, B and C. A four-way ANOVA revealed significant regional effects (F(3, 968)=1460.04, p<0.0001). Further analyses performed for each different hippocampal region considering age, treatment and time after restraint revealed effects in CA1, CA3 and DG that were parallel to the CRH mRNA changes. In all of these areas an age effect was detected (CA1: F(2, 242)= 147.96–6.11, p<0.0001–0.002). A treatment by time interaction was observed in CA1 (F(4, 242)=3.47, p=0.009), whereas age by time interactions were detected in CA3 and DG (CA3—F(8, 242)= 2.48, p=0.01; DG—F(8, 242)=3.77, p=0.0003).
Fig. 6.
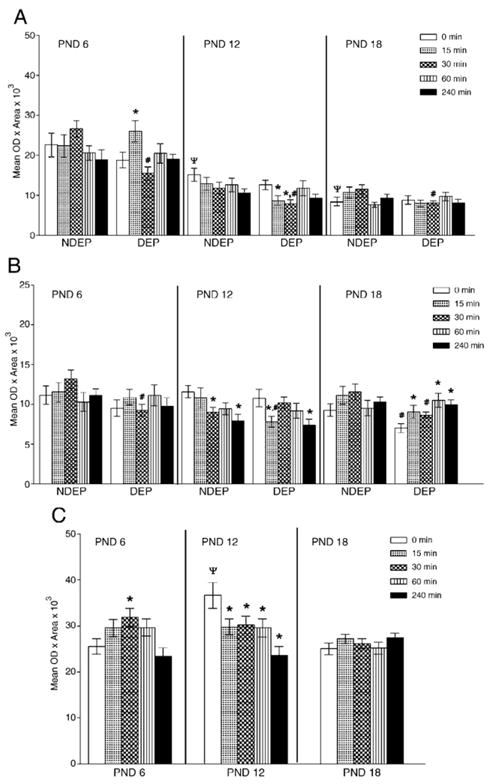
Densitometric analyses of CRH r1 mRNA gene expression in hippocampal CA1 (Panel A), CA3 (Panel B) and dentate gyrus (Panel C) subregions. Panels A and B depict the results with all variables tested, whereas Panel C shows the response to acute restraint across ages (data collapsed across the DEP treatment variable). Post hoc analysis is represented as *p<0.05, significantly different from corresponding basal level (time 0); #p<0.05, significantly different from NDEP time point, ψp<0.05, significantly different from PND 6 time point.
2.4.7. CA1 region
A significant decrease of CRHr1 levels was observed in the CA1 region as the animal matured (NDEP, 0 min: F(2, 23)=9.97, p=0.0008). Maternal deprivation did not alter this pattern (compare, time 0, NDEP vs. DEP). A restraint effect was detected in the DEP 6-day-old (F(4, 41)=3.26, p=0.02) and 12-day-old (F(4, 37)=2.6, p=0.05) animals. Post hoc comparisons can be viewed in Fig. 6, Panel B.
2.4.8. CA3 region
The CRH r1 mRNA levels did not change as the animal mature in the CA3 region. However, DEP caused a down-regulation of CRHr1 gene expression in the 18-day-old animal (compare time 0 NDEP vs. DEP PND 18; F(1, 14)=5.96, p=0.03). An acute restraint effect was observed in the 12-day-old NDEP (F(4, 42)= 2.98, p=0.03) and DEP pups (F(4, 37)=3.00, p=0.03), as well as the 18-day-old DEP animals (F(4, 44)=3.85, p=0.009). See Fig. 6, Panel B for post hoc comparisons.
2.4.9. Dentate gyrus
An age effect on CRHr1 expression was noted in this region (F (2, 242)=6.11, p=0.003), with the 12-day-old animal having the highest gene expression compared to the other ages (NDEP time 0, PND 12—F(2, 51)=10.01, p=0.0002). Though lacking a significant DEP treatment effect, an age by time interaction (F (8, 242)=3.77, p=0.0003) was detected in granular cells of the DG. An acute restraint effect was detected in the 6-day-old (time: F(4, 86)=3.24, p=0.02) and 12-day-old (time: F(4, 84)= 4.76, p=0.002) animals. Post hoc comparisons can be viewed in Panel C, Fig. 6.
In summary, the CA1, A3 and DG regions have prominent findings. A stable expression of CRH mRNA is seen these regions, in all ages studied. In contrast, CRH bp and CRHr1 mRNA expression change with age in a site-specific fashion. Similarly, these patterns are differentially affected by restraint leading to suppression, enhancement, or no change of CRH, CRH binding protein and CRH receptor expression in a region-specific manner.
3. Discussion
The primary goal of this study was to investigate CRH, CRH r1 and CRH binding protein expression in extra-hypothalamic brain regions in 6-, 12- and 18-day-old rat pups. A second goal was to ascertain the modulation of these molecules by maternal deprivation. Also examined were the changes in gene expression following restraint in both non-deprived and deprived pups at different stages of development. The present report provides evidence of the developmental trajectories of specific extra hypothalamic brain structures and the sensitivity of these to stimuli during the neonatal period. Three main findings are derived from this study: (1) the developmental gene expression progression of CRH, CRHr1 and CRH bp is unique in each of the structures studied; (2) the genomic changes observed were not always influenced by maternal deprivation, in fact deprivation enhanced, suppressed or had no consequence on the underlying ontogenic progression of these CRH-related molecules; and (3) there is a temporal response of CRH, CRHbp and CRHr1 mRNA levels following the 15-min restraint stressor that was dependent on the anatomical area and age of the animal.
We examined several brain areas that are important for LHPA axis regulation or that have abundance of CRH neurons or rich reciprocal CRH connections. These areas included cortex, amygdala, septum and hippocampus. We included the septum because it is a conspicuous, integrated part of the limbic system with reciprocal CRH interconnections to rostral, deep and caudal brain. Caudal brain modulates a number of behavioral and physiological processes that overlap direct or indirect CRH mediated actions, namely, learning and memory, anxiety, fear, aggression, as well as autonomic regulation of water/food intake and humoral immune responses (Jakab et al., 1995). Our first finding is that the developmental gene expression progression of CRH, CRHr1 and CRH bp is unique in each of the structures studied. This can be assessed by comparing the time 0 point (basal) mRNA levels of the NDEP animal at all ages and in all structures. We found a general pattern in which the second postnatal week of life appears to be a pivotal time when transitions into increments or decrements of gene expression are seen for all molecules within all structures studied. Although our experimental design does not include ages between day 6 and day 12, our data are consistent with the literature both in terms of ontogenetic development and neuroplasticity. For example, Dent and co-workers described CRH mRNA decreases within the PVN of 12-day-old animals, with further decreases observed on PND 18 (Dent et al., 2000a). Similarly, the expression of arginine vasopressin (AVP) in the PVN increases at PND 12 (Dent et al., 2000a). Avishai-Eliner and co-workers have also demonstrated that CRH receptor mRNA increases in the cortex of 12-day-old animals (Avishai-Eliner et al., 1996). These changes are coincidental with marked organization of the CNS postnatal processes of dendritic arborization extension and pruning, axonal lengthening and neurogenesis that would be specific to striatal, cerebellum, hippocampus and layers IV–II in neocortex (Bayer et al., 1995). The expression changes, be it increases or decreases, may be linked to these processes inherent to the developmental pattern within CRH-related circuits.
There are additional data in the literature, which suggest that the second week of life is a critical period of neuroplasticity. For example, Avishai-Eliner and collaborators have shown that the permanent down-regulation of hypothalamic CRH-mRNA levels observed in rats handled daily from postnatal days 3 to 14 was evident by postnatal day 9 and was sustained through postnatal days 23 and 45, i.e., beyond puberty (Avishai-Eliner et al., 2001; Fenoglio et al., 2006). These animals exhibit a behavioral profile of decreased anxiety and HPA responses in adulthood. Similarly, Moriceau's and co-workers studies indicate that postnatal day 10 is a time when the developmental shift from preference learning to aversion learning occurs (Moriceau et al., 2004; Schulkin et al., 2005). This shift correlates with the functional development of the amygdala and elevation of corticosterone levels in the developing rat as it emerges from the SHRP.
The genomic changes that we observed were not always related to maternal deprivation status, in fact deprivation enhanced, suppressed or had no consequence on the underlying ontogenic progression of these CRH-related molecules. However, focusing on the CRH and CRHr1 mRNA response after 24 h deprivation, we observe a consistent decrease in CRHr1 mRNA with a decrease or no change of CRH mRNA levels in a site- and age-specific manner (see Table 1, maternal deprivation and DEP in Fig. 2, Panel C; Fig. 5, Panel B). The fact that we observe a down-regulation of CRHr1 at the end of the deprivation period suggests that the CRH receptor 1 is sensitive to CRH release triggered by maternal deprivation within the CRH circuit. It may, however, seem paradoxical that the CRH mRNA levels are suppressed rather than enhanced in these structures. However, it has been established that there is a robust modulation of key HPA axis molecules at different time points as the 24-h maternal deprivation progresses (Schmidt et al., 2004). The activation of the key elements of the HPA axis, namely CRH mRNA in PVN, plasma ACTH and corticosterone, occurs between 4 and 8 h of maternal absence. In contrast, negative feedback mechanisms suppress these molecules during the second half of the deprivation period. It is clear from elegant pharmacological studies by Brunson and co-workers that at least in cortex and hippocampus both occupancy of the CRH r1 receptor by its ligand and activation of post-receptor signaling pathways are required for modulation of CRH r1 in the developing animal (Brunson et al., 2002). The down-regulation of CRHr1 mRNA levels observed in specific structures in our study is likely to have resulted from CRH activation, although at the end of the maternal deprivation period there are no discernable changes on the gene expression of the ligand (CRH). Our CRHr1 gene expression data in cortex is in agreement with others who found a decrease in CRHr1 binding levels in homogenates derived from cortex of 10-day-old animals 4 h after administration of intracerebral CRH (Brunson et al., 2002). However, this same study revealed an enhancement of CRHr1 message RNA indicating a dissociation of gene and protein expression. The CRHr1 mRNA up-regulation observed by Brunson et al. in 10-day-old animals was distinct to superficial cortical layers and not at the deeper cortical layers (V–VI). Despite this enhancement, they found decreased binding capacity of CRHr1 in homogenates derived from cortex indicating that the net effect was decreased receptor expression. The decreased binding may reflect either an overall decreased cortical expression of the CRH receptor when all cortical layers are considered or a reduced translation of the receptor protein at this site (Xu et al., 2001). It is also possible that the physiological approach in our study (compared to the pharmacological one utilized by Brunson and co-workers) and the time frame after the challenge (end of 24 h after maternal deprivation vs. 4 h after intra ventricular CRH infusion) contributes to our findings.
We observed a temporal response of CRH, CRHbp and CRHr1 mRNA levels following the 15-min restraint stressor that was dependent on the anatomical area and age of the animal. In those specific regions that genomic changes were observed the response was both rapid and sustained, or the initial alteration in gene expression showed a recovery to its initial starting point. Again, the response was not limited to the maternally deprived animal. For example, 15 min of restraint caused a general pattern of decrease of CRHr1 mRNA in cortex, amygdala and hippocampus at virtually all ages. CRH gene expression was also found to decrease in cortex and amygdala. These changes in expression are relatively rapid, occurring within 15 to 30 min of the restraint challenge and not exclusive to the deprived pup. In view of this short time frame and the pattern of corticosterone release observed at the different ages (Cook, 2002), it is possible that CRH and CRHr1 are indirectly regulated by glucocorticoids in these structures. In the adult animal, neither corticosterone treatment, adrenalectomy nor immobilization stress alters the gene expression of CRH r1 in the amygdala even though regulation is detected in the PVN and AP (31). Of interest is the fact that restraint, particularly in 12- and 18-day-old animals increased the septum, CRH, CRHbp and CRHr1 mRNA levels. This general pattern of up-regulation is similar to what is observed in PVN in both adult and developing animals (Dent et al., 2000b; Makino et al., 1995). These findings provide evidence that there is sensitivity and specificity of the extra-hypothalamic CRH system to environmental manipulation during the neonatal period. The overall impression is of a dynamic system responding to developmental and environmental demands.
Amygdala CRH gene expression increased in both DEP and NDEP 12- and 18-day-old animal. The significance of this overall increased CRH expression in the amygdala may be related to the role of the amygdala CRH in fear-related responses. In the adult the amygdala is a critical part of the neural circuit involved in the expression of fear-related behaviors (Schulkin et al., 2005). However, in the infant there are stages in development when the neonate exhibits a sensitive period for learning an odor preference when confronted with an odor stimulus that has been paired with a stimulus that elicits fear behavior in the adult animal (electric shock). Thus, an odor–shock pairing results in a preference for the conditioned odor during this sensitive period. In contrast after this sensitive period (day 10) the odor-shock pairing results in a robust odor aversion. In the sensitive period, the learning of an odor preference does not involve amygdala activation, whereas in the post-sensitive period amygdala activation is required for the learning of the odor-shock aversion (Moriceau and Sullivan, 2005). Thus a hypo-functional amygdala during infancy appears to prevent the pup from learning aversions to the caregiver. The results of the current study indicated that early in development (day 6), which would correspond to the sensitive period, CRH, CRH bp and CRHr1 gene expression is significantly lower than at day 12 and day 18. The increase in the mRNA levels of these molecules would suggest a transition from a hypo-functional to a hyper-functional amygdala. As previously mentioned, Moriceau et al. (2006) have demonstrated that elevating corticosterone during the sensitive period induces odor-aversion learning whereas depleting cortico-sterone by adrenalectomy maintains preference learning in post-sensitive period pups. Further, if corticosterone is administered directly to the amygdala odor shock aversion is learned in sensitive period pups, but not if the corticosterone is delivered to non-HPA-related brain areas. The relationship between corticosterone and levels of amygdala CRH has been well established in the adult (Schulkin et al., 2005). However, the acute stress induced elevation of amygdala CRH does not appear to be glucocorticoid dependent (Cook, 2002). Our results do not suggest that CRH gene expression is altered by acute restraint. In the NDEP 12-day-old pups CRH mRNA levels are suppressed by 60 min. There is presently little known about the relationship between corticosterone levels and amygdala CRH expression during development.
In conclusion, the present report supports the presence of complex regulatory effects of development and environmental manipulation on CRH, CRHr1 and CRHbp gene expression in cortical and limbic regions. The data once again challenges the basic assumption of a stress hypo-responsiveness in the infant rat. Within extra hypothalamic regions, such as amygdala, hippocampus and septum distinct patterns of CRH, CRHr1 and CRHbp regulation are evident in the non-deprived animal subjected to the immediate effect of physical restraint. We also observe modulation of these molecules under prolonged maternal deprivation when compared to acute restraint. Such modulation may have important repercussions on morphological organization during development and events leading to neuronal protection in the stressed animal.
4. Experimental procedures
4.1. Animals
The tissue samples utilized in this study were collected from infant rats of a research study that has been the subject of a previously published report (Dent et al., 2000a). The infant rats were the hybrid progeny of Sprague–Dawley females and Long–Evans males (Charles River, Chicago, IL, USA). The day of birth was termed day 0 and on day 1 the litters were culled to 10 pups (5 males and 5 females). Culling the litters served two purposes: (1) male/female pairs could be assigned to each of the five treatment groups (see below); and (2) a litter size of ten pups controls for malnutrition (number of pups does not exceed average number of nipples available for lactation). From the time of culling until the start of testing the animals were not handled, nor were the cages disturbed in any manner. Animals were housed with their dams in transparent polycarbonate cages (Nalgene, 24 cm×45 cm×20 cm). Further, litters were maintained in an environment of constant temperature (25±2 °C) and on 12-h light/dark cycle. Access to food and water was ad libitum. Animal care was in compliance with the NIH Guide for the Care and Use of Laboratory Animals and was approved by the University of Delware IACUC.
A total of thirty litters were used in the study (5 litters for each age and treatment condition). Among each litter, one pair of pups (1 male and 1 female) served as a control group, with the other 4 pairs each representing one of four different time points used to examine the stress response to restraint (see details below). Genetic diversity and control for maternal factors was thus ensured with this procedure. The time course of the endocrine response (plasma adrenocorticotropic hormone and corticosterone), hypothalamic CRH and arginine vasopressin gene expression following restraint of these animals has been previously reported (Dent et al., 2000a).
4.2. Deprivation procedure and stress paradigm
Male and female pups were examined at two postnatal times within the SHRP (PND 6 and 12) and one age outside the SHRP (PND 18). Non-deprived pups (NDEP) served as comparison subjects. These pups were left undisturbed with their mothers until the time of testing. Deprived pups refer to those litters where the dam was removed and the pups remained in the home cage for a period of 24 h before testing (on PND 5, 11 and 17, respectively). The cages holding the deprived pups were placed on an electric heating pad (30–33 °C) in a room adjacent to, and maintaining the same temperature and lighting conditions of the main animal colony room.
During the deprivation period food and water were not available, a necessary requirement to observe a hormonal response (Rosenfeld et al., 1993). At the start of testing one pair of pups (1 male and 1 female) within a given litter were immediately sacrificed to serve as controls. The remaining eight pups in each litter were then individually restrained in plastic, cylindrical restrainers. The size of the plastic restrainer used was specific to the age of the pup. For PND 6, 12 and 18 the restrainer width was 2.5, 3 and 4 cm, respectively. For each age and treatment group, one pair of pups from each litter (1 male and 1 female) was restrained for 15 min and sacrificed immediately thereafter. A second set of pups was restrained for 30 min and sacrificed afterwards. The remaining two sets of pups were kept in restraint for 30 min and sacrificed either 1 or 4 h after the start of restraint.
4.3. Tissue processing and in-situ hybridization
Animals were sacrificed via rapid decapitation at their respective time points following restraint. The animal brains were rapidly removed and frozen in 2-methylbutane at −40 °C. Brains were stored at −80 °C until processing tissue for in-situ hybridization. Six brains (3 male and 3 female) obtained from five different litters for each age, treatment and time point were coronally sectioned (16 μ m) and subsequently mounted on Superfrost slides (VWR Scientific, West Chester, PA, USA). 35S-UTP-labeled riboprobes were utilized for the histochemical localization of corticotropin-releasing hormone (CRH) mRNA, CRH receptor 1 (r1) and CRH binding protein (bp) were performed. The CRH cRNA probe was produced using a 353 base-pair fragment derived from a rat cDNA clone including the peptide region of the rat CRH gene, exon 2 (courtesy of R. Thompson). A pBluescript (SK+) plasmid containing 415-bp was used to produce CRHr1 probe. PCR amplification between transmembrane domain II and VII of the CRHr1 receptor produced the CRHr1 fragment. The CRHbp probe was synthesized from a 585 base-pair restriction fragment of CRH bp cDNA cloned into the PstI site of pBluescript SK (courtesy of R. Thompson). The riboprobes were synthesized utilizing the SP6, T3 or T7 transcription systems (Invitrogen, Carlsbad, CA, USA) in a standard labeling reaction mixture consisting of 1 μ g linearized plasmid, 5× of the appropriate transcription buffer (Invitrogen, Carlsbad, CA, USA), 125 μ Ci-35S-UTP (Amersham Biosciences, Arlington Heights, IL, USA), 150 μ M of NTP's (Roche Diagnostics Corp., Indianapolis, IN, USA), 12.5 mM dithiothreitol (DTT) (Sigma-Aldrich, St. Louis, MO, USA), 20 U RNAse inhibitor (Invitrogen, Carlsbad, CA, USA) and 6 U of the respective polymerase (Invitrogen, Carlsbad, CA, USA). The reaction mixture was incubated at 37 °C for 90 min, treated with DNAse I RNA (Roche Diagnostics Corp., Indianapolis, IN, USA), free for 15 min at room temperature, followed by filtration over a Sephadex G50-50 Quick Spin Column (Roche Diagnostics Corp., Indianapolis, IN, USA) to separate labeled probe from free nucleotides.
The hybridization procedure has been previously published (Vázquez et al., 2003). Briefly, tissue sections (2 brain sections per slide) were transferred from storage at −80 °C to room temperature 4% paraformaldehyde solution. After 1 h of fixation, slides were rinsed in isotonic phosphate buffered saline and then treated with proteinase K (1 μ g/ml in 100 mM Tris/HCL, pH 8.0) (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 10 min. Afterwards, tissue sections were subjected to serial washes of 1 min in purified water, 10 min in 0.1 M triethanolamine (pH 8.0, plus 0.25% acetic anhydride; Roche Diagnostics Corp., Indianapolis, IN, USA) and 5 min in 2× SSC (0.3 mM NaCl, 0.03 sodium citrate, pH 7.2, Roche Diagnostics Corp., Indianapolis, IN, USA). Tissue sections then underwent graded alcohol dehydration, air-dried, then hybridized with 1.0×106 counts per minute (cpm) 35S-UTP-labeled riboprobe in a total volume of 25 μ l hybridization buffer [50% formamide, 10% dextran sulfate, 3× SSC, 50 mM sodium phosphate buffer (pH 7.4), 1× Denhardt's solution, 0.1 mg/ml yeast tRNA and 10 mM dithiothreitol]. Brain sections were coverslipped and incubated overnight at 55 °C. On the following day, sections were washed for 5 min in 2× SSC and then treated for 60 min at 37 °C with RNAse A (200 μ g/ml in 10-mM Tris/HCl, pH 8.0), containing 0.5 NaCl (Sigma-Aldrich, St. Louis, MO, USA). Finally, sections were washed successively for 5 min in 2× SSC, 5 min in 1× SSC, 60 min at hybridization temperature in 0.5× SSC, 5 min at room temperature in 0.5× SSC and then dehydrated in graded ethanol dilutions. The slides were placed on Kodak MR X-ray film at room temperature for signal detection, with exposure times dependent upon probe under consideration.
4.4. Data analyses
All numerical data represent values as means±standard error of mean (SEM). Statistical significance was set at p<0.05. Data for each age were analyzed using analysis of variance (ANOVA) that considered brain region, age, sex, DEP treatment and time of sacrifice at rest (0 min) and following restraint (15, 30, 60, 240 min). When no differences were found with a particular variable, the data were collapsed across this variable. A post hoc analysis followed the ANOVA using Fisher protected least significance difference (Fisher PLSD). Auto-radiograms from the in situ hybridization were analyzed by computer-assisted optical densitometry (NIH Image). To control for film non-linearity, 14C-methylmethacrylate standards were used. The mean value of 4 to 6 regional measures (depending on the structure) was taken to represent the final value for each animal. Five to six animals were ultimately studied for each age, treatment and time point.
Acknowledgments
This research was supported by the National Institute of Mental Health MH62099 to JFL and MH-45006 to SL; National Institute of Health HD/DK37431 to DMV. The authors would like to thank Mr. Carl Flink and Mercy Pawar for their help with data collection, data analysis and assistance in the preparation of the photomicrographs.
References
- Avishai-Eliner S, Yi SJ, Baram TZ. Developmental profile of messenger RNA for the corticotropin-releasing hormone receptor in the rat limbic system. Brain Res Dev Brain Res. 1996;91:159–163. doi: 10.1016/0165-3806(95)00158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avishai-Eliner S, Eghbal-Ahmadi M, Tabachnik E, Brunson KL, Baram TZ. Down-regulation of hypothalamic corticotropin-releasing hormone messenger ribonucleic acid (mRNA) precedes early-life experience-induced changes in hippocampal glucocorticoid receptor mRNA. Endocrinology. 2001;142:89–97. doi: 10.1210/endo.142.1.7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA, Atltman J. Neurogenesis and neuronal migration. In: Paxinos G, editor. The Rat Nervous System. Academic Press; San Diego, CA: 1995. pp. 1041–1076. [Google Scholar]
- Bhatnagar S, Dallman M. Neuroanatomical basis for facilitation of hypothalamic–pituitary–adrenal responses to a novel stressor after chronic stress. Neuroscience. 1998;84:1025–1039. doi: 10.1016/s0306-4522(97)00577-0. [DOI] [PubMed] [Google Scholar]
- Brunson KL, Grigoriadis DE, Lorang MT, Baram TZ. Corticotropin-releasing hormone (CRH) downregulates the function of its receptor (CRF1) and induces CRF1 expression in hippocampal and cortical regions of the immature rat brain. Exp Neurol. 2002;176:75–86. doi: 10.1006/exnr.2002.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers DT, Lovenberg TW, Grigoriadis DE, Behan DP, De Souza EB. Corticotrophin-releasing factor receptors: from molecular biology to drug design. TIPS. 1996;17:166–172. doi: 10.1016/0165-6147(96)81594-x. [DOI] [PubMed] [Google Scholar]
- Charney DS, Deutch A. A functional neuroanatomy of anxiety and fear: implications for the pathophysiology and treatment of anxiety disorders. Crit Rev Neurobiol. 1996;10:419–446. doi: 10.1615/critrevneurobiol.v10.i3-4.70. [DOI] [PubMed] [Google Scholar]
- Cook CJ. Glucocorticoid feedback increases the sensitivity of the limbic system to stress. Physiol Behav. 2002;75:455–464. doi: 10.1016/s0031-9384(02)00650-9. [DOI] [PubMed] [Google Scholar]
- Dent GW, Okimoto DK, Simith MA, Levine S. Stress-induced alterations in corticotropin-releasing hormone and vasopressin gene expression in the paraventricular nucleus during ontogeny. Neuroendocrinology. 2000a;71:333–342. doi: 10.1159/000054554. [DOI] [PubMed] [Google Scholar]
- Dent GW, Smith MA, Levine S. Rapid induction of corticotropin-releasing hormone gene transcription in the paraventricular nucleus of the developing rat. Endocrinology. 2000b;141:1593–1598. doi: 10.1210/endo.141.5.7455. [DOI] [PubMed] [Google Scholar]
- Eckart K, Jahn O, Radulovic J, Radulovic M, Blank T, Stiedl O, Brauns O, Tezval H, Zeyda T, Spiess J. Pharmacology and biology of corticotropin-releasing factor (CRF) receptors. Recept Channels. 2002;8:163–177. [PubMed] [Google Scholar]
- Eghbal-Ahmadi M, Hatalski CG, Lovenberg TW, Avishai-Eliner S, Chalmers DT, Baram TZ. The developmental profile of the corticotropin releasing factor receptor (CRF2) in rat brain predicts distinct age-specific functions. Brain Res Dev Brain Res. 1998;107:81–90. doi: 10.1016/s0165-3806(98)00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio KA, Chen Y, Baram TZ. Neuroplasticity of the hypothalamic–pituitary–adrenal axis early in life requires recurrent recruitment of stress-regulating brain regions. J Neurosci. 2006:2434–2442. doi: 10.1523/JNEUROSCI.4080-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TS, Bingaman EW. The amygdala: corticotropin-releasing factor, steroids and stress. Crit Rev Neurobiol. 1996;10:155–168. doi: 10.1615/critrevneurobiol.v10.i2.10. [DOI] [PubMed] [Google Scholar]
- Herman JP, Cullinan WE. Neurocircuitry of stress: central control of the hypothalamo–pituitary–adrenocortical axis. Trends Neurosci. 1997;20:78–84. doi: 10.1016/s0166-2236(96)10069-2. [DOI] [PubMed] [Google Scholar]
- Herman JP, Prewitt CM, Cullinan WE. Neuronal circuit regulation of the hypothalamo–pituitary–adrenocortical stress axis. Crit Rev Neurobiol. 1996;10:371–394. doi: 10.1615/critrevneurobiol.v10.i3-4.50. [DOI] [PubMed] [Google Scholar]
- Jakab RL. Septum. 2. Academic Press, Inc.; San Diego: 1995. pp. 405–431. [Google Scholar]
- Lewis K, Li C, Perrin MH, Blount A, Kunitake K, Donaldson C, Vaughan J, Reyes TM, Gulyas J, Fischer W, Bilezikjian L, Rivier J, Sawchenko PE, Vale WW. Identification of urocortin III, an additional member of the corticotropin-releasing factor (CRF) family with high affinity for the CRF2 receptor. Proc Natl Acad Sci U S A. 2001;98:7570–7575. doi: 10.1073/pnas.121165198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Schulkin J, Smith MA, Pacak K, Palkovits M, Gold PW. Regulation of corticotropin-releasing hormone receptor messenger ribonucleic acid in the rat brain and pituitary by glucocorticoids and stress. Endocrinology. 1995;136:4517–4525. doi: 10.1210/endo.136.10.7664672. [DOI] [PubMed] [Google Scholar]
- Makino S, Shibasaki T, Yamauchi N, Nishioka T, Mimoto T, Wakabayashi I, Gold PW, Hashimoto K. Psychological stress increased corticotropin-releasing hormone mRNA and content in the central nucleus of the amygdala but not in the hypothalamic paraventricular nucleus in the rat. Brain Res. 1999;850:136–143. doi: 10.1016/s0006-8993(99)02114-9. [DOI] [PubMed] [Google Scholar]
- Moriceau S, Sullivan RM. Neurobiology of infant attachment. Dev Psychobiol Suicidal Behav. 2005;47:20–242. doi: 10.1002/dev.20093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriceau S, Roth TL, Okotoghaide T, Sullivan RM. Corticosterone controls the developmental emergence of fear and amygdala function to predator odors in infant rat pups. Int J Dev Neurosci. 2004;22:415–422. doi: 10.1016/j.ijdevneu.2004.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriceau S, Wilson D, Levine S, Sullivan RM. Dual circuitry for odor-shock conditioning during infancy. Neuroscience, J. 2006;26:6737–6748. doi: 10.1523/JNEUROSCI.0499-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld P, Ekstrand J, Olson E, Suchecki D, Levine S. Maternal regulation of adrenocortical activity in the infant rat: the effects of feeding. Dev Psychobiol. 1993;26:261–277. doi: 10.1002/dev.420260504. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Enthoven L, van Woezik JHG, Levine S, De Kloet ER, Oitzl S. The dynamics of the hypothalamic–pituitary–adrenal axis during maternal deprivation. J Neuroendocrinol. 2004;16:52–57. doi: 10.1111/j.1365-2826.2004.01123.x. [DOI] [PubMed] [Google Scholar]
- Schulkin J, Morgan MA, Rosen JB. A neuroendocrine mechanism for sustaining fear. Trends Neurosci. 2005;28:629–635. doi: 10.1016/j.tins.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Seasholtz AF, Burrows HL, Karolyi IJ, Camper SA. Mouse models of altered CRH-binding protein expression. Peptides. 2001;22:743–751. doi: 10.1016/s0196-9781(01)00387-4. [DOI] [PubMed] [Google Scholar]
- Vazquez DM, Morano MI, Lopez JF, Watson SJ, Akil H. Short-term adrenalectomy increases glucocorticoid and mineralocorticoid receptor mRNA in selective areas of the developing hippocampus. Mol Cell Neurosci. 1993;4:455–471. doi: 10.1006/mcne.1993.1057. [DOI] [PubMed] [Google Scholar]
- Vazquez DM, Van Dours H, Ko LS, Ko AH. Regulation of the glucocorticoid and mineralocorticoid receptor mRNA in the hippocampus of the maternally deprived infant rat. Brain Res. 1996;731:79–90. doi: 10.1016/0006-8993(96)00465-9. [DOI] [PubMed] [Google Scholar]
- Vázquez DM, Eskandari R, Phelka A, López JF. The impact of maternal deprivation on brain corticotropin releasing hormone circuits: prevention of CRH receptor-2 mRNA changes by desipramine treatment. Neuropsychopharmacology. 2003;28:898–909. doi: 10.1038/sj.npp.1300126. [DOI] [PubMed] [Google Scholar]
- Walker CD, KAnand JS, Plotsky PM. Development of the hypothalamic–pituitary–adrenal axis and the stress response. In: McEwen BS, editor. Handbook of Physiology: Coping with the Environment. Oxford Univ. Press; New York: 2001. pp. 237–270. [Google Scholar]
- Xu G, Rabadan-Diehl C, Nikodemova M, Wynn P, Spiess J, Aguilera G. Inhibition of corticotropin releasing hormone type-1 receptor translation by an upstream AUG triplet in the 5′ untranslated region. Mol Pharmacol. 2001;59:485–492. doi: 10.1124/mol.59.3.485. [DOI] [PubMed] [Google Scholar]


