Abstract
Mutations in the nuclear structural protein lamin A cause the premature aging syndrome Hutchinson-Gilford progeria (HGPS). Whether lamin A plays any role in normal aging is unknown. We show that the same molecular mechanism responsible for HGPS is active in healthy cells. Cell nuclei from old individuals acquire defects similar to those of HGPS patient cells, including changes in histone modifications and increased DNA damage. Agerelated nuclear defects are caused by sporadic use, in healthy individuals, of the same cryptic splice site in lamin A whose constitutive activation causes HGPS. Inhibition of this splice site reverses the nuclear defects associated with aging. These observations implicate lamin A in physiological aging.
Mutations in the lamin A gene (LMNA) are responsible for the premature aging disease Hutchinson-Gilford progeria syndrome (1-3). The most prevalent HGPS mutation (heterozygous Gly608→Gly608 with C changed to T) leads to a splicing defect and consequent generation of a truncated, dominant gain-of-function lamin A isoform (2, 3). HGPS patient cells have various defects in nuclear structure and function (4-6). They are characterized by dysmorphic nuclear shape (4), increased DNA damage (5), and down-regulation of several nuclear proteins, including the heterochromatin protein HP1 and the LAP2 group of lamin A-associated proteins (6). Furthermore, HGPS cells have altered histone modification patterns, including reduced heterochromatinspecific trimethylation of Lys9 on histone H3 (Tri-Me-K9H3) (6).
It has not been clear how HGPS relates to normal aging and whether lamin A plays any role in the physiological aging process (7, 8). To address these questions, we determined whether HGPS-like nuclear defects occurred in cells from normally aged individuals. Multiple skin fibroblast cell lines from old (81 to 96 years) individuals consistently showed nuclear aberrations similar to those seen in HGPS cells (Fig. 1, A to D) (6). Whereas most cells from young individuals (3 to 11 years) showed robust staining for HP1, LAP2s, and Tri-Me-K9H3, a significant subpopulation of nuclei in cells from old individuals had reduced signals, similar to previous observations in HGPS cells (Fig. 1A) (6). Quantitative single-cell analysis demonstrated that the distributions of marker levels were significantly different in cells from young compared with old individuals (Fig. 1, B and C) (P < 0.001). In cells from old individuals, between 40 and 90% (average of 61 ± 23%) of cells showed a reduction of Tri-Me-K9H3 (9), whereas this fraction was 20 to 32% (average of 25 ± 3.8%) in cells from young individuals (Fig. 1, B to D). The percentage of cells with reduced HP1 and Tri-Me-K9H3 in cells from old donors was comparable to that found in HGPS patient cells, although a stronger overall reduction was observed in HGPS cells, with many cells showing nearly no signal (Fig. 1, B to C). Similar differences were found for LAP2s.
Fig. 1.
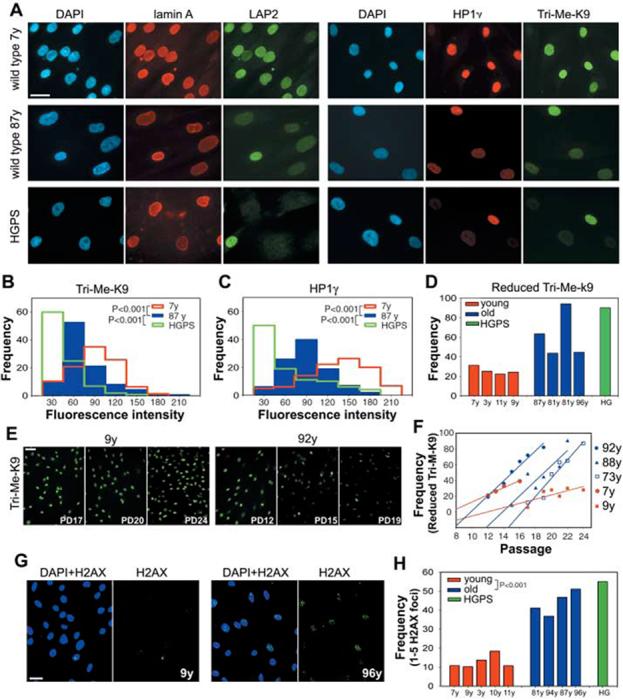
Nuclear abnormalities in cells from old individuals. (A) Immunofluorescence microscopy on primary dermal fibroblasts from young (7 y) and old (87 y) healthy individuals and a HGPS patient. Scale bar, 10 μm. DAPI, 4′,6′-diamidino-2-phenylindole. Intensity distributions of the average fluorescent signal for (B) Tri-Me-K9H3 and (C)HP1γ in fibroblasts from healthy individuals of indicated age and a HGPS patient. (D) Quantitation of cells showing reduced amounts of Tri-Me-K9 in passage-matched population doublings (PD) 19 cell lines from healthy individuals of indicated age and a HGPS patient (9). (E and F) Reduction of Tri-Me-K9 H3 over cell passage. Best linear fits are shown. (G and H)Increased DNA damage in passage-matched (PD20) cell lines from healthy individuals of indicated age and a HGPS patient detected by antibody to H2AX. Scale bar, 20 μm. N > 200.
The nuclear defects in HGPS patient cells have previously been shown to accumulate during passage in culture (4). Cell lines from both young and old donors exhibited increased defects with prolonged passage (Fig. 1, E and F). However, defects accumulated much more rapidly in cells from old donors than in cells from young individuals (Fig. 1F and fig. S1). A further hallmark of HGPS cells is an increased amount of unrepaired DNA damage as indicated by the presence of foci containing phosphorylated histone H2AX (5). Similar to HGPS patient cells, the percentage of nuclei with phospho-H2AX foci was significantly higher in cells from old individuals (43 ± 6%) compared with young individuals (12 ± 3%; P < 0.001) (Fig. 1, G and H). The accumulation of H2AX foci in aging human cells is consistent with similar recent observations in baboons and mice (10, 11). We conclude that cells from HGPS patients and normally aged individuals share several common nuclear defects.
The vast majority of HGPS cases are caused by the constitutive activation of a cryptic splice site in exon 11, which leads to aberrant removal of the 3′-terminal 150 nucleotides of this exon (2, 3). The resulting Δ150 LMNA mRNA gives rise to a dominant gain-of-function lamin A isoform containing an internal deletion of 50 amino acids (Δ50 lamin A) (2, 3). Because the cryptic splice site in the wild-type LMNA gene is highly homologous to a generic consensus splice donor site (Fig. 2A), we asked whether this cryptic splice site is also used by default in cells from healthy individuals. Reverse transcription polymerase chain reaction (RT-PCR) using primers in exon 9 and 12 amplified a minor truncated product in cells from healthy individuals of all ages (Fig. 2, B and C). DNA sequencing confirmed the identity of the product. Furthermore, all cell lines tested expressed the Δ150 LMNA isoform when detected with a pair of primers specific for the aberrant splice junction (Fig. 2, B and D). Transfection of a splicing reporter in multiple cell lines from both young and old donors confirmed the sporadic use of the cryptic splice site in the wild-type LMNA transcript (Fig. 2, F and G). Quantitative RT-PCR analysis using the splice junction primers demonstrated that use of the cryptic splice site was about 50-fold lower in cells from healthy individuals than in HGPS cells and did not increase with age (Fig. 2H). Use of the cryptic splice site is not a cell-culture artifact, because Δ150 LMNA mRNA is detectable in unprocessed human tissues (Fig. 2I). Similar to cultured cells, no significant difference in the amount of Δ150 LMNA mRNA was observed in tissues from either young or old individuals (Fig. 2I).
Fig. 2.
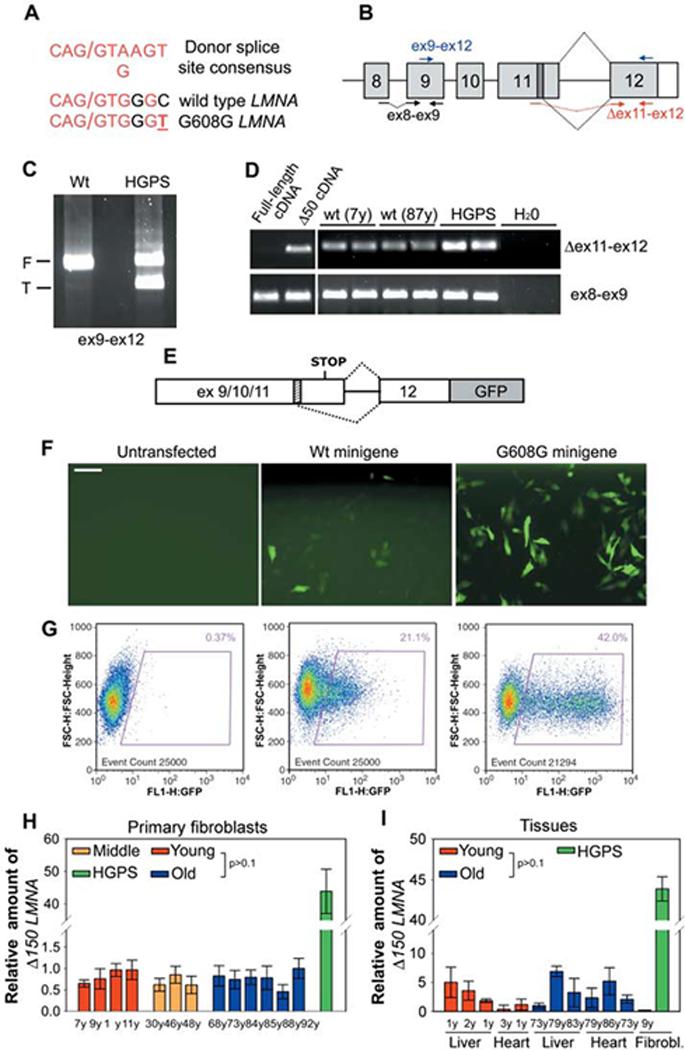
Sporadic use of exon 11 LMNA cryptic splice site in cells from healthy individuals. (A)Comparison between the consensus donor splice sequence, the wild-type LMNA cryptic splice site in exon 11, and the mutated cryptic splice site in HGPS patients. Forward slash indicates the splice junction. (B)Schematic representation of splicing events (dotted black lines) involving LMNA exon 11 and exon 12. The cryptic splice site (dark box) and the position of different primer sets used for RT-PCR are indicated. RT-PCR analysis of wild-type and HGPS fibroblasts using (C) primers detecting both the full-length (F) and the truncated (T) LMNA isoforms or (D) primers specific for Δ150 LMNA (Δ ex11-ex12). Control ex8-ex9 primers detect all LMNA transcripts. Samples were analyzed in duplicate. Plasmids containing either full-length or truncated lamin A cDNA were used as controls for specificity of Δ150 LMNA detection. (E) Schematic representation of LMNA splicing reporter (9). The cryptic splice site (dashed box) is indicated. (F) Immunofluorescence microscopy and (G) fluorescence-activated cell sorting analysis of fibroblasts from a healthy individual transiently transfected with the indicated reporter minigene or untransfected. Scale bar, 60 μm. Quantitative RT-PCR analysis of (H) wild-type fibroblasts and (I) tissues from healthy individuals of indicated age and a HGPS patient. Δ150 LMNA RNA detected by using Δex11-ex12 primers relative to total LMNA RNA detected by using ex8-ex9 primers is shown.
The occurrence of lamin A aberrant splicing in wild-type cells suggests that Δ50 lamin A is present in cells from healthy individuals. A truncated protein of the expected molecular weight was detected in wild-type cells by antibodies against lamin A and C (lamin A/C) upon extraction of soluble proteins and enrichment for membrane-associated proteins (Fig. 3A and fig. S2). Analysis of liver tissues confirmed the presence of Δ50 lamin A in old individuals (Fig. 3B). Consistent with the absence of any increase in Δ150 LMNA mRNA levels, the truncated protein did not significantly accumulate during aging (Fig. 3A). However, a striking change in lamin A/C localization was detected in cells from old donors (Fig. 3, C to E). Whereas in cells from young individuals a substantial fraction of lamin A/C was present throughout the nucleoplasm, this fraction was almost completely absent in cells from old donors, and the vast majority of lamin A accumulated at the nuclear rim (Fig. 3, C and D). This distribution was similar to that in HGPS cells, where the presence of the mutant lamin A protein leads to the accumulation of wild-type lamin A at the nuclear periphery (Fig. 3C) (6). Importantly, lack of nucleoplasmic lamin A/C strongly correlated with reduced amounts of Tri-Me-K9H3 and HP1 at the single-cell level (Fig.3D).Thus, as in HGPS patients, cells from old individuals express the Δ150 LMNA mRNA, and lamin A is aberrantly localized in the cell nucleus.
Fig. 3.
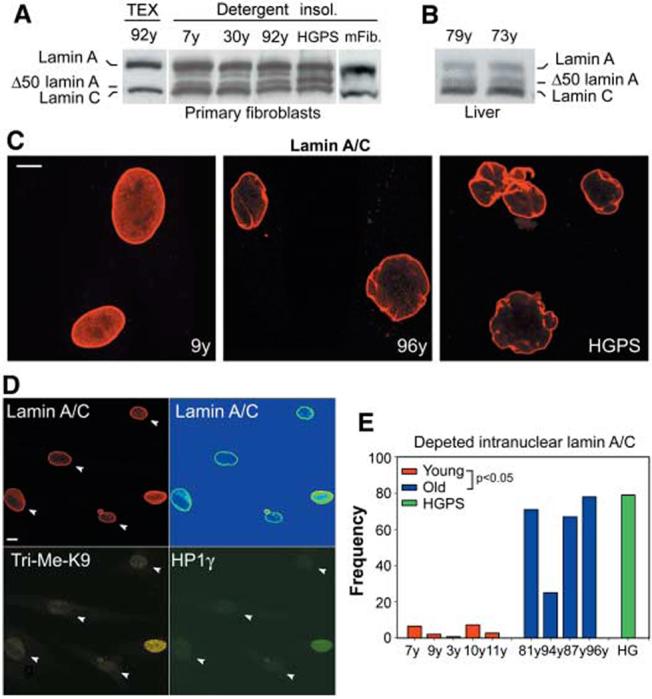
Accumulation of lamin A/C at the lamina in cells from old individuals. Western blot analysis of total protein extract and detergent insoluble fraction of fibroblasts from (A)healthy individuals of indicated age, a HGPS patient, and mouse fibroblasts and (B) from liver from healthy individuals probed with antilamin A/C. Δ50 lamin A is not detected in mouse fibroblasts. A fourth protein consistent with Δ10 lamin A (15) is also detected. Equal loading was verified by Coomassie staining of the blot. (C and D) Immunofluorescence confocal microscopy on passage-matched (PD21) fibroblasts. (D) Co-staining of fibroblasts from an old individual. Scale bar, 10 μm. Lamin A/C signal shown in intensity pseudocolors in upper right image. Arrowheads indicate nuclei with depleted nucleoplasmic lamin A/C and reduced amounts of HP1γ and Tri-Me-K9H3. (E) Quantitation of percentage of cells showing depletion of nucleoplasmic lamin A/C (average fluorescence intensity ratio nucleoplasm/lamina < 0.2) in passage-matched (PD21) cell lines from healthy individuals of indicated age and a HGPS patient.
To directly demonstrate that the production of the Δ50 lamin A protein by sporadic use of the cryptic splice site in LMNA exon 11 was responsible for the observed changes in nuclear architecture in cells from aged individuals, we inhibited the cryptic splice site in cells from old donors by using a previously characterized oligonucleotide-based approach (6). A morpholino oligonucleotide targeted to the cryptic splice site (Exo11) was introduced by repeated electroporation into cells from old individuals. We previously demonstrated that the Exo11 oligonucleotide efficiently inhibits the use of the cryptic splice site in vivo (6). Treatment of multiple cell lines from old individuals with Exo11 reversed their aberrant cellular phenotypes (Fig. 4). Fluorescent signals of HP1 and Tri-Me-K9H3 increased in cells treated with Exo11 compared with the same cells treated with a scrambled control oligonucleotide (Fig. 4A). Quantitation of the distribution of HP1 and Tri-Me-K9H3 confirmed the restoration of amounts similar to those seen in cells from young individuals at similar passages (Fig. 4, B and C) (P < 0.001). The reversibility of nuclear defects upon blocking of the cryptic splice site in LMNA directly demonstrates that lamin A is causal in generating the age-related nuclear defects. A smaller, but statistically significant, effect was also seen in cells from young individuals at later passages, further supporting a correlation between age and lamin A-dependent nuclear abnormalities (Fig. 4, B and C).
Fig. 4.
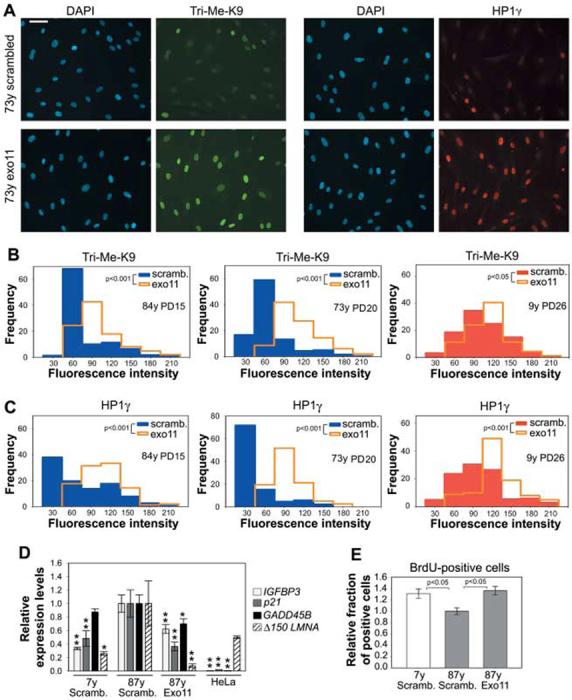
Nuclear defects in cells from old individuals are caused by use of the LMNA exon 11 cryptic splice site. (A) Immunofluorescence microscopy on fibroblasts from an old healthy individual after treatment with either Exo11 oligonucleotide or a control scrambled oligonucleotide. Scale bar, 60 μm. (B and C) Quantitation of phenotypic rescue upon treatment with Exo11 oligonucleotide. (D) Quantitative RT-PCR analysis of HeLa cells and cell lines from young and old donors upon treatment with either Exo11 oligonucleotide or a control scrambled oligo-nucleotide. Values represent averages ± SD from a representative experiment. Statistical significances of the differences compared with mock-treated cells from the old donor are indicated. One asterisk indicates P < 0.1. Two asterisks indicate P < 0.05. (E) Quantitation of BrdU-positive cells in cell lines from young and old donors upon treatment with either Exo11 oligonucleotide or a control scrambled oligonucleotide. Values represent averages ± SD from three independent experiments.
Organismal aging has been linked to activation of p53-dependent signaling pathways and initiation of the senescence program in a premature aging mouse model (12). To test whether lamin A plays a role in the activation of p53-dependent signaling pathways, we probed the status of p53 target genes after elimination of Δ50 lamin A from cells of old individuals using the Exo11 oligonucleotide. Inhibition of lamin A aberrant splicing resulted in significant down-regulation of p21, IGFBP3, and GADD45B compared with that of mock-treated cells (Fig. 4D). The status and response of p53 target genes were variable among cell lines. Consistent with the reduction in p53 activation, upon elimination of Δ50 lamin A from old cells, the fraction of 5-bromo-2′-deoxyuridine (BrdU)-positive proliferating cells increased by 30% and was similar to that in mock-treated young cells (Fig. 4E) (P < 0.05).
The sum of our observations implicates lamin A in physiological aging, showing that the same molecular mechanism responsible for the premature aging disease HGPS acts at a low level in healthy cells. The observation that HGPS and physiological aging share a common cellular and molecular basis strongly supports the notion that HGPS mimics at least some aspects of physiological aging and suggests that lamin A participates in the aging process in healthy individuals. Accelerated aging in HGPS might thus reflect an exaggerated lamin A-dependent mechanism, which normally contributes to physiological aging.
The fact that inhibition of the LMNA aberrant splicing event reverses the age-related defects in nuclear structure demonstrates that these abnormalities are caused by the Δ50 isoform of lamin A. Δ50 lamin A lacks an internal proteolytic processing site that is normally used as part of the lamin A maturation process but which is removed during the aberrant splicing event (13). The uncleaved pre-lamin A intermediate accumulates at the nuclear rim (14), where it is thought to exert its dominant effects (12, 14). We show here that sporadic use of the cryptic splice site in exon 11 in healthy individuals leads to the production of the same pre-lamin A intermediate. Our observation that neither the amount of aberrantly spliced LMNA mRNA nor the amount of Δ50 lamin A increases during aging suggests that the nuclear defects are due to the prolonged presence of Δ50 lamin A in the nucleus. Aged cells might be more sensitized to the presence of the aberrant lamin A isoforms and less able to neutralize the negative effects of Δ50 lamin A, possibly due to the existence of a p53-dependent checkpoint, which senses structural abnormalities of the nuclear lamina and links those to the activation of the senescence program (12). Given our finding that several nuclear defects in aged cells are reversible upon inhibition of the aberrant splicing event in LMNA, it will be interesting to determine whether other cellular features of aging respond to such treatment and whether organismal aging can be modulated by interference with lamin A.
References and Notes
- 1.Hutchison CJ. Nat. Rev. Mol. Cell Biol. 2002;3:848. doi: 10.1038/nrm950. [DOI] [PubMed] [Google Scholar]
- 2.De Sandre-Giovannoli A, et al. Science. 2003;300:2055. doi: 10.1126/science.1084125. published online 17 April 2003 (10.1126/science.1084125). [DOI] [PubMed] [Google Scholar]
- 3.Eriksson M, et al. Nature. 2003;423:293. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman RD, et al. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8963. [Google Scholar]
- 5.Liu B, et al. Nat. Med. 2005;11:780. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 6.Scaffidi P, Misteli T. Nat. Med. 2005;11:440. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrew W. Adv. Gerontol. Res. 1964;18:87. [PubMed] [Google Scholar]
- 8.Haithcock E, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16690. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Materials and methods are available as supporting material on Science Online.
- 10.Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Science. 2006;311:1257. doi: 10.1126/science.1122446. published online 2 February 2006 (10.1126/science.1122446). [DOI] [PubMed] [Google Scholar]
- 11.Sedelnikova OA, et al. Nat. Cell Biol. 2004;6:168. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 12.Varela I, et al. Nature. 2005;437:564. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 13.Young SG, Fong LG, Michaelis S. J. Lipid Res. 2005;46:2531. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Yang SH, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10291. [Google Scholar]
- 15.Machiels BM, et al. J. Biol. Chem. 1996;271:9249. doi: 10.1074/jbc.271.16.9249. [DOI] [PubMed] [Google Scholar]
- 16.We thank D. Donato for help with statistical analysis, C. Baker for design of specific primer pairs, M. Sinensky for advice on metabolic labeling of farnesylated lamin A, and K. Wilson and T. Jenuwein for providing reagents. Fluorescence imaging was performed at the NCI Fluorescence Imaging Facility. This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.


