Abstract
Complex microbial communities reside within the intestines of humans and other vertebrates. Remarkably little is known about how these microbial consortia are established in various locations within the gut, how members of these consortia behave within their dynamic ecosystems, or what microbial factors mediate mutually beneficial host–microbial interactions. Using a gnotobiotic zebrafish–Pseudomonas aeruginosa model, we show that the transparency of this vertebrate species, coupled with methods for raising these animals under germ-free conditions can be used to monitor microbial movement and localization within the intestine in vivo and in real time. Germ-free zebrafish colonized with isogenic P. aeruginosa strains containing deletions of genes related to motility and pathogenesis revealed that loss of flagellar function results in attenuation of evolutionarily conserved host innate immune responses but not conserved nutrient responses. These results demonstrate the utility of gnotobiotic zebrafish in defining the behavior and localization of bacteria within the living vertebrate gut, identifying bacterial genes that affect these processes, and assessing the impact of these genes on host–microbial interactions.
Keywords: Danio rerio, establishment of a gut microbiota, flagellar motility, host–microbial symbiosis and mutualism, Pseudomonas aeruginosa
Starting at birth, we are colonized by communities of microorganisms that establish residency on our external and internal surfaces. These resident microbes outnumber our human cells by an order of magnitude, and their aggregate genomes (microbiome) specify important physiologic traits that are not encoded in our own genome (1). The vast majority of these microbes reside in our intestine: most of our 10–100 trillion gut-dwelling microbes belong to the domain Bacteria, although members of Archaea (Euryachaeota and Crenarchaeota) and Eukarya are also represented (2–6). Over the last 50 years, experiments comparing mice and rats raised in the absence of any microorganisms [germ-free (GF)] to those colonized with members of gut microbial communities have revealed that the microbiota plays an integral role in many aspects of intestinal and extraintestinal host biology, ranging from postnatal development of the gut's blood and lymphatic vascular systems (7, 8) to the proliferative activity of intestinal epithelial cells (9, 10), metabolism of ingested xenobiotics (1, 11), regulation of energy balance (12–14), maturation of the innate and adaptive immune systems (15–18), heart size (19), and behavior (e.g., locomotor activity) (14).
The notion that each of us is a supraorganism, composed of microbial and human parts, focuses attention on the question of how our microbial communities are assembled (20). Understanding the dynamic patterns of microbial entry into and movement within their gut habitats is critical for deciphering how different species establish and maintain a presence in the intestinal ecosystem, and how they interact with their host and other microbial community members. Fluorescence in situ hybridization and confocal and electron microscopic analyses have provided static rather than dynamic views of the positioning of microbial cells within the mammalian intestine, and in vivo bioluminescence analyses do not permit resolution of individual microbial cells.
The zebrafish (Danio rerio) possesses several key attributes that make it a distinctively powerful model organism for addressing these questions. First, the zebrafish digestive tract is structurally similar to that of mammals, with proximal-distal specification of functions and multiple self-renewing epithelial cell lineages (21, 22). Second, comparisons of GF zebrafish and those colonized with a microbiota harvested from the intestines of conventionally raised (CONV-R) zebrafish or mice have revealed a broad range of host processes that are impacted by the gut microbiota and that are conserved between mammals and fish (23–25). Moreover, individual bacterial representatives of the zebrafish and mouse gut microbiotas have been identified that can provoke evolutionarily conserved host responses in gnotobiotic zebrafish (23–25). Third, this vertebrate species and its gut are optically transparent from the time of fertilization through the onset of adulthood. This unusual feature provides an opportunity to make real-time in vivo observations of microbial–microbial and microbial–host interactions. Because zebrafish larvae can be grown in a 96-well plate format, their transparency could also be used to conduct genetic and chemical screens for host and/or microbial factors that mediate host–microbial interactions. Finally, with the development of methods for rearing zebrafish under GF conditions, reciprocal transplantations of gut communities from normal mouse and zebrafish donors into GF zebrafish and mouse recipients have revealed that differences in the normal gut communities of these vertebrates arise in part from distinct selective pressures imposed within their respective gut habitats. These experiments also revealed a striking degree of conservation of host responses to the different microbiotas (24).
We have used a simplified system, consisting of GF zebrafish colonized with the Gram-negative γ-proteobacterium Pseudomonas aeruginosa, to define the mechanisms by which members of the microbiota elicit these conserved host responses. P. aeruginosa is best known as an opportunistic pathogen. However, it has several characteristics that facilitate its use as a model mutualist in this system. Pseudomonads are common members of the fish gut microbiota (23–28) as well as the gut microbiota of some mammals (e.g., the African zebra and others; R. E. Ley and J.I.G, unpublished observations) [see supporting information (SI) Materials and Methods and Fig. 4 for a 16S rRNA sequence-based tree of zebrafish Pseudomonads and their relationship to P. aeruginosa]. Although Pseudomonads are rare members of the intestinal microbiota of healthy humans, their representation is increased in certain pathologic states, notably inflammatory bowel diseases (29–31). In an initial survey, 10 different bacterial species representative of the zebrafish or mouse gut microbiota were tested for their ability to elicit the innate immune and nutrient metabolic responses produced when a complete microbiota is introduced into GF zebrafish hosts. In this survey, P. aeruginosa was the most potent inducer of these responses (24) (see SI Fig. 5). Finally, in addition to the large body of knowledge that exists about P. aeruginosa biology, valuable genetic resources are available, including a finished genome sequence for strain PAO1 (32), deep draft genome assemblies for several other strains (PA14, C3719, 2192, PA7, and PACS2), and saturation-level sequenced transposon insertion libraries for strains PAO1 (33) and PA14 (34).
In the present study, we take advantage of the transparency of zebrafish and these genetic resources to demonstrate a linkage between motility/flagellar function and regulation of conserved innate immune responses.
Results
Real-Time in Vivo Imaging of Microbial Consortia and Individual Bacterial Species in the Transparent Intestine of Gnotobiotic Zebrafish.
As noted above, the transparency of the zebrafish provides opportunities for exploring the movement as well as localization of microbes within their intestinal habitat through real-time microscopy of live whole-mount zebrafish. CONV-R zebrafish typically hatch from the GF environment within their protective chorions at 3 days postfertilization (dpf). This hatching event coincides with the anterior digestive tract achieving full patency (21, 35). Fluorescence in situ hybridization has revealed that the zebrafish digestive tract is colonized by bacteria as early as 4 dpf (25); however, the timing and route of initial colonization remained unclear.
Therefore, we first colonized GF zebrafish at 3 dpf with a normal zebrafish microbiota harvested from adult CONV-R zebrafish (a process called conventionalization) and then imaged their digestive tracts at different time points. In vivo bright-field microscopy of the gut microbiota in these conventionalized (CONVD) animals revealed a striking amount of microbial movement within their intestinal lumen (SI Movies 1 and 2), although the activity of individual microorganisms was difficult to monitor.
Upon exposure to P. aeruginosa, 3-dpf GF zebrafish are colonized at densities similar to the conventional zebrafish gut microbiota (104-105 cfu per gut at 6 dpf; SI Table 1) and elicit host responses that are conserved across vertebrate hosts (see below and ref. 24). To facilitate real-time in vivo microscopic observation of individual microbial cells, we introduced a plasmid that allows constitutive expression of the gene encoding GFP under the control of the trc promoter (pMF230) (36) into P. aeruginosa strain PAO1 to create PAO1 pMF230. GF 3-dpf zebrafish exposed to 104 cfu of P. aeruginosa PAO1 pMF230/ml gnotobiotic zebrafish medium (GZM) were initially colonized with a small cohort of bacteria that was readily seen as early as 3.5 dpf (Fig. 1 A and D and SI Movie 3). Because the anus does not achieve patency until ≈4 dpf (21, 35), our findings establish that the anterior digestive tract becomes colonized within just a few hours after its lumen first opens.
Fig. 1.
Gut bacteria display diverse behaviors within the intestines of gnotobiotic zebrafish. (A and D) Whole-mount preparation of a live 3.5-dpf zebrafish colonized since 3 dpf with GFP-expressing P. aeruginosa PAO1 (PAO1 pMF230) demonstrates the transparency of the developing zebrafish intestine. Brightfield microscopy of the anterior intestine (segment 1, A) shows the intestinal lumen (lum) and the adjacent intestinal epithelium (ep). Fluorescence time-lapse microscopy of the same field (D) shows the movements of individual bacteria over the course of 10 frames, or 4 sec (D extracted from SI Movie 3). The locations of individual bacteria in the first (1) and the last (10) frames are numbered accordingly. (B and E) Brightfield (B) and fluorescence time-lapse (E) microscopy of the same field from a live 6-dpf zebrafish, colonized since 3 dpf with PAO1 pMF230, shows increasing bacterial density and behavioral complexity in the midintestine (junction of segments 1 and 2) over the course of 10 frames or 2.6 sec (E extracted from SI Movie 4). Note that the intestines shown in D and E both contain bacteria that are nonmotile in association with the host epithelium or luminal contents (yellow), whereas other bacteria exhibit high rates of motility in both ascending (distal to proximal; red tracks) and descending (green tracks) directions. Note that ascending and descending bacteria were tracked for only the first several frames because they quickly moved out of the focal plane; the first and last frames over which bacteria were tracked are numbered. (C and F) Brightfield (C) and fluorescence time-lapse (F) microscopy of a live 4.5-dpf zebrafish colonized since 3 dpf with DsRed-expressing E. coli MG1655 (MG1655 pRZT3) showing movement of luminal bacteria (green tracks) in the midintestine (segment 1). Over the course of 14 frames or 14 sec (F extracted from SI Movie 5), some bacteria appear adherent to the epithelium or luminal structures (yellow track), whereas most bacterial motion is synchronous and attributed to intestinal motility (green tracks). Anterior is to the left, and dorsal is to the top in all images. (Scale bars: 20 μm.)
The size of this monocomponent community increased rapidly over the course of the next 2.5 days. As in CONVD zebrafish, bacteria in 6-dpf P. aeruginosa monoassociated zebrafish were observed along the entire proximal-distal length of the intestine (e.g., SI Movie 4). Individual bacteria displayed a range of behaviors, from intimate association with the intestinal epithelium, to incorporation into large multicellular structures in the luminal space, to rapid movement of planktonic cells through the lumen (Fig. 1 B and E and SI Movie 4). In 6-dpf hosts, individual bacteria were observed moving at speeds as high as 24 μm/sec within the lumen (equivalent to ≈12 body lengths per sec). This movement is likely the result of flagella-mediated swimming motility (see below).
A central challenge for members of a gut microbiota is to avoid washout from its continuously perfused ecosystem. Static scanning electron microscopic studies in the gnotobiotic mouse intestine indicate this can be achieved by bacterial attachment to nutrient platforms consisting of partially digested food particles, exfoliated fragments of mucus, and shed epithelial cells (37, 38). From an engineering perspective, these platforms represent well settling particles, analogous to those that prevent microbial washout from human-made bioreactors (37).
Our gnotobiotic zebrafish provided a dynamic view of the interactions of bacteria with such luminal contents. Similar to the microbiota in CONV-R and CONVD zebrafish, PAO1 pMF230 was observed interacting with large slowly moving luminal structures (SI Movie 2 and data not shown) that were distributed along the length of the gut lumen; similar masses were observed in GF animals, indicating that their formation does not depend upon microbes. Individual bacterial cells could be seen intermittently contacting the surface of these masses (SI Movie 2).
To determine whether bacteria reside within these structures, we fixed P. aeruginosa PAO1 monoassociated 6-dpf zebrafish en bloc and processed them for transmission electron microscopy (TEM): this en bloc fixation was designed to minimize disruption of the in vivo spatial relationships among microbes, other gut contents, and host cells. Transverse TEM sections through the zebrafish intestine revealed that the luminal masses contained many intact bacteria mixed with other gut contents, including mucus-like material and large electron-dense lamina (Fig. 2 A and B). Consistent with our real-time in vivo imaging results, bacterial cells were also observed outside these luminal aggregates in close juxtaposition to the host epithelium (Fig. 2 A). TEM disclosed that in CONV-R, CONVD, and P. aeruginosa-monoassociated zebrafish, actively dividing and nondividing bacterial cells were closely associated with epithelial cells in the intact mucosa and in the luminal structures (Fig. 2C plus data not shown) (23).
Fig. 2.
TEM of gut bacteria in gnotobiotic zebrafish. Transverse sections are shown that include segments 1 and 2 of the intestine of a 6-dpf zebrafish colonized since 3 dpf with P. aeruginosa strain PAO1. (A) Bacteria are clustered together in the luminal space, and some remain close to the host epithelium (arrowhead in A). (B and C) Bacteria (arrowheads) are also observed in association with unidentified electron-dense laminated objects in the lumen (arrows in B) and undergoing fission (C). (Scale bars: A, 3 μm; B, 1 μm; C, 500 nm.)
Together, these findings show that P. aeruginosa appears to recapitulate the range of movements, as well as the locations occupied by members of the intestinal microbiota. To investigate whether the behavior of P. aeruginosa in this system was characteristic of other γ-Proteobacteria, we colonized 3-dpf GF zebrafish with Escherichia coli MG1655 carrying a plasmid that directs constitutive expression of red fluorescent protein (DsRed) under the control of the lac promoter (MG1655 pRZT3). Strain MG1655 pRZT3 displayed significantly less motility than strain PAO1 pMF230 in 6-dpf zebrafish digestive tracts (Fig. 1 E and F and SI Movies 4 and 5), even though its density of colonization was not significantly different from P. aeruginosa (SI Table 1). In contrast, in vitro assays revealed that E. coli MG1655 has higher rates of swimming motility than P. aeruginosa PAO1 in soft agar (SI Fig. 6), suggesting that the zebrafish gut environment influences motility in these bacterial species.
Characterization of P. aeruginosa as a Model Zebrafish Mutualist.
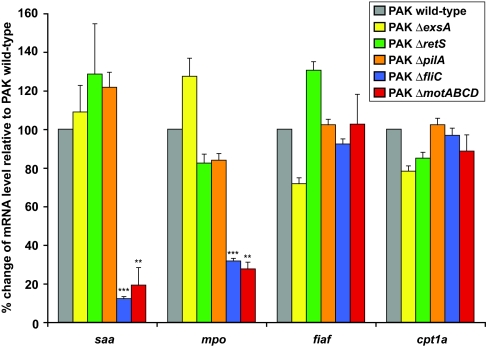
P. aeruginosa strains generally express one of two flagellin proteins (type-a and -b flagellin) that differ by 35% in amino acid sequence (39). To determine whether the motility phenotype and other effects on the host were specific to type-b strains such as PAO1, we tested a well characterized P. aeruginosa strain that expresses type-a flagellin (strain PAK) (40). Both strains colonized the digestive tracts of GF zebrafish to similar densities (SI Table 1) and were highly motile in vivo (SI Movies 3 and 4 and data not shown). Moreover, both strains elicited evolutionarily conserved nutrient and innate immune responses: quantitative RT-PCR (qRT-PCR) assays conducted on RNA extracted from whole 6-dpf monoassociated zebrafish indicated they suppressed expression of fiaf [also known as angptl4; encodes a secreted inhibitor of lipoprotein lipase (12, 41)] and carnitine palmitoyltransferase 1a (cpt1a; involved in mitochondrial oxidation of fatty acids) and induced expression of serum amyloid a (saa; an acute-phase protein) and myeloperoxidase (mpo; a granulocyte-specific biomarker of the innate immune response to the normal gut microbiota) (23, 24) (Fig. 3 and SI Table 2).
Fig. 3.
The impact of P. aeruginosa flagellar mutants on host responses in gnotobiotic zebrafish. Expression levels of serum amyloid a (saa), myeloperoxidase (mpo), fasting-induced adipose factor (fiaf), and carnitine palmitoyltransferase 1a (cpt1a) were assessed by qRT-PCR by using RNA extracted from whole 6-dpf larvae colonized since 3 dpf with P. aeruginosa PAK wild-type strain, PAK exsA deletion mutant (PAK ΔexsA), PAK retS deletion mutant (PAK ΔretS), PAK pilA deletion mutant (PAK ΔpilA), PAK fliC deletion mutant (PAK ΔfliC), or PAK motABCD deletion mutant (PAK ΔmotABCD). Data from biological duplicate pools (6–17 animals per pool) were normalized to 18S rRNA levels. Normalized mRNA levels in colonized fish were referenced against age-matched GF controls (ΔΔCt method), and the results are expressed as mean percent change relative to the PAK wild-type strain ± SEM. ∗∗∗, P < 0.0001; ∗∗, P < 0.001 compared with wild-type, based on a two-tailed Student's t test.
Animal models of P. aeruginosa infection and disease have identified specific factors that this bacterium uses for virulence. A multicomponent Type III secretion system (TTSS) functions to translocate effector proteins into host cells. Strains PAO1 and PAK both secrete three effectors by the TTSS (ExoS, ExoT, and ExoY); these toxins target various host signaling pathways, leading to disruption of the actin cytoskeleton and cytotoxicity (42). The TTSS is required for colonization and virulence in a range of mouse models of infection (43–45).
To test whether these gene products play a role in P. aeruginosa colonization of the zebrafish gut, we exposed GF 3-dpf zebrafish to the PAK strain carrying a deletion for the TTSS master transcriptional regulator exsA (46). In many of the pathogenesis models of P. aeruginosa infection, this mutant exhibits decreased virulence and colonization (43, 47, 48). In contrast, the wild-type and exsA mutant strains achieved comparable densities of colonization within the intestines of 6-dpf zebrafish (SI Table 1), indicating that the TTSS and its secreted toxins are not essential for colonization. In addition, qRT-PCR analysis showed that the exsA mutant strain was capable of regulating expression of fiaf, cpt1a, saa, and mpo in a manner that was not significantly different from its wild-type parent strain (Fig. 3).
P. aeruginosa uses the RetS hybrid two-component system to coordinately activate TTSS expression and repress exopolysaccharide production (49). Similar to exsA mutants, deletion of retS leads to loss of the TTSS. In addition, these mutants overproduce exopolysaccharides implicated in biofilm formation (49). To ascertain whether overproduction of these exopolysaccharides influenced P. aeruginosa–zebrafish interactions, we introduced an isogenic PAK retS mutant strain into 3-dpf GF hosts. As with the TTSS mutant, neither colonization density nor the response of host innate immune and nutrient biomarker genes differed between the mutant and wild-type strains when measured at 6 dpf (Fig. 3 and SI Table 1).
P. aeruginosa utilizes several surface appendages for colonization and virulence, including a single polar flagellum and Type IV pili. To test the role of bacterial movement on host responses, we examined isogenic PAK strains with loss-of-function mutations in fliC and pilA (40, 50). The flagellar apparatus of P. aeruginosa is assembled through an intricate regulatory process, concluding with synthesis and assembly of FliC protein into the flagellar filament (40, 51). Transmembrane ion gradients provide energy for physical rotation of the filament by the flagellar motor (51). The motor consists of two structures: the rotor (the switch that determines direction of rotation) and the stator (the stationary component through which the rotor turns). Because FliC is the major structural component of the flagellar filament, the fliC mutant fails to assemble an intact flagellar filament (40, 51, 52). The pilA mutant, which is missing the major pilin structural subunit, does not exhibit pili-dependent twitching or swarming motility (53).
Both mutants achieved a normal density of colonization in the zebrafish gut (SI Table 1). However, the fliC strain did not show the highly motile phenotype characteristic of wild-type P. aeruginosa in this habitat or in vitro (SI Fig. 6 and data not shown), suggesting that swimming motility is a primary method of locomotion in vivo. qRT-PCR assays revealed that, like wild-type P. aeruginosa, the fliC and pilA mutants were able to suppress expression of the nutrient metabolic biomarkers fiaf and cpt1a in 6-dpf larvae (Fig. 3). Unlike the wild-type or three other mutant strains, fliC-deficient bacteria failed to elicit a significant increase in expression of innate immune response biomarkers saa or mpo (reference controls, GF 6-dpf animals; Fig. 3).
The attenuated host immune responses to fliC mutants could be caused by absence of FliC protein, the absence of an intact flagellar filament, and/or the absence of flagellar function. To help distinguish among these possibilities, we colonized 3-dpf GF zebrafish with a P. aeruginosa PAK strain that carries deletions of the motAB and motCD genes (ΔmotABCD), encoding the bacterium's two flagellar stators. PAK motABCD mutants assemble an intact flagellar filament that contains the FliC protein, but the assembled filament is nonmotile (SI Fig. 6) because of a failure of filament rotation (54, 55). As with the fliC mutant, we found that the motABCD mutant was able to colonize 6-dpf zebrafish at densities comparable to the isogenic wild-type strain (SI Table 1) and to recapitulate its elicited nutrient responses: thus, flagellar motility is not required for these host reactions to colonization (Fig. 3). Also similar to fliC mutants, innate immune responses to motABCD mutants in 6-dpf zebrafish larvae were significantly attenuated (Fig. 3). Therefore, the ability of P. aeruginosa to evoke these conserved innate immune responses to a complete gut microbiota is due in part to flagellar function, rather than exclusively to the presence of an intact flagellar filament and its component proteins such as FliC.
Discussion
For P. aeruginosa and other bacteria, flagella perform several nonexclusive functions that can impact bacterial colonization and host responses in the digestive tract. First, flagella mediate swimming motility that can facilitate interactions with and invasion of host cells (56), as well as chemotaxis toward preferred habitats and nutrient sources (57) [e.g., studies of V. cholerae mutants indicate bacterial motility and chemotaxis are important virulence determinants in the (mouse) intestine (58)]. Second, flagella can act as adhesins that bind bacteria to host epithelial cells independent of their role in motility (59, 60). Third, flagella can serve as a secretion apparatus for virulence factors (61, 62), a role that may be played by P. aeruginosa flagella (63). Finally, flagellin can serve as a major immunostimulatory antigen recognized by Toll-like receptor 5 (TLR5) homologs in both fish and mammals (64–66). Activation of mammalian TLR5 triggers NF-κB-dependent proinflammatory signaling pathways that stimulate production of acute-phase proteins and neutrophil chemoattractants (67, 68). P. aeruginosa flagella can also bind the glycolipid asialoGM1, leading to TLR2-dependent activation of similar signaling pathways (69). Recent studies have revealed that flagellin from other bacterial species can also be detected by TLR5-independent mechanisms (70).
Our observations suggest that flagellar function, including the swimming motility observed by real-time in vivo microscopy, is an important component of host–bacterial interaction in this system. We hypothesize that flagella-dependent swimming motility promotes physical interaction between P. aeruginosa and the host epithelium, where the presence of surface-attached antigens (including the flagellum itself) and other bacterial products can be monitored by the host. Although it remains possible that flagella-dependent immune responses are ultimately stimulated by FliC acting as an antigen, the attenuated immune response to the flagellated but nonmotile motABCD mutant shows that flagella motor function is required for this process to occur. These observations set the stage for future experiments that further dissect how dynamic interactions between P. aeruginosa and the gut epithelium mediate the observed flagellar-motility-dependent host response in zebrafish.
Our results demonstrate the utility of using gnotobiotic zebrafish for defining and monitoring microbial behavior and localization within the living vertebrate gut and for identifying bacterial genes that affect host–microbial interactions. As such, this genetically pliable host provides an opportunity to explore how habitat influences the establishment of a microbiota, and how microbial dynamics in vivo affect host biology. Although P. aeruginosa is often used as a model opportunistic pathogen, our study indicates that it can also serve as a model mutualist, capable of colonizing the gut of gnotobiotic zebrafish and eliciting nutrient metabolic and innate immune responses that have been conserved during the ≈400 million years since fish and mammals diverged from their last common ancestor. The combined advantages of P. aeruginosa (genome sequence, saturation-level insertion libraries, and genetic tools) and gnotobiotic zebrafish (conservation of metabolic and immune responses to a microbiota with mammals, amenability to high-throughput genetic and chemical screens and the ability to directly observe the gut and its microbial inhabitants in a living vertebrate) offer an opportunity to systematically decipher the foundations of host–microbial mutualism in the gut and perhaps to apply the findings to our own species.
Materials and Methods
Animal Husbandry.
All experiments using zebrafish were performed by using protocols approved by the Animal Studies Committees of Washington University and the University of North Carolina at Chapel Hill.
Zebrafish gametes were expressed manually from CONV-R adults (C32 inbred strain), fertilized in vitro, and embryos derived as GF according to established protocols (23). GF zebrafish were reared under a 14-h light cycle in sterile vented tissue culture flasks (Becton Dickinson, Sparks, MD) at an average density of 1.3 individuals per milliliter of GZM (GZM components are defined in ref. 23). Animals were maintained at 28.5°C in an air incubator. Fish were fed daily beginning at 3 dpf with a sterilized solution containing 0.1 mg of ZM000 fish food (ZM Ltd., Winchester, United Kingdom) per milliliter of GZM. A 90% water change was performed before each daily feeding, starting at 3 dpf. GF zebrafish were monitored routinely for sterility by using culture-based methods (23).
Colonization and in Vivo Imaging.
At the time of hatching at 3 dpf, we exposed GF zebrafish reared in sterile vented tissue culture flasks to (i) an unfractionated gut microbiota harvested directly from CONV-R adult C32 donors, (ii) P. aeruginosa PA01 containing pMF230 [harbors GFP under the control of a constitutive trc promoter (36); supplied by Michael Franklin, Montana State University, Bozeman, MT], (iii) E. coli MG1655 containing pRZT3 (DsRed under the control of a constitutive lac promoter; a gift from Wilbert Bitter, Vrije University Medical Centre, Vrije, The Netherlands), (iv) wild-type P. aeruginosa PAK or the isogenic ΔfliC strain carrying pSMC21 [a derivative of pSMC2 (71), harboring GFP under the control of a constitutive lac promoter; provided by Matthew Wolfgang, University of North Carolina, Chapel Hill], or (v) isogenic wild-type or mutant P. aeruginosa PAK strains without plasmids (supplied by Matthew Wolfgang and Reuben Ramphal, University of Florida, Gainesville, FL; plus Stephen Lory, Harvard University, Boston, MA). Bacterial strains were grown overnight at 37°C in Luria–Bertani broth before inoculation. Microbes were introduced at a density of 104 cfu/ml GZM. A complete list of bacterial strains and plasmids used can be found in SI Table 1.
Monoassociated and age-matched CONVD zebrafish were imaged at various times after exposure to bacteria by using the following protocol. Animals were anesthetized in 0.2 mg/ml Tricaine (Sigma, St. Louis, MO), placed on a 40 × 22-mm glass coverslip, and imbedded in low-melting-point 1% NuSieve GTG agarose (FMC Bioproducts, Philadelphia, PA) containing 0.2 mg/ml Tricaine anesthetic. After the agarose quickly solidified, animals were viewed by using an Axiovert 200M inverted fluorescence microscope and Axiovision 4.1 software (Zeiss, Thornwood, NY).
Supplementary Material
Acknowledgments
We are grateful to Edward Flynn for zebrafish husbandry; to Jaime Dant and Howard Wynder for help with TEM; to Adam Schreck for video editing; and to Fredrik Bäckhed, Eric Martens, Justin Sonnenburg, and Matthew Wolfgang for helpful suggestions. This work was supported in part by the Ellison Medical Foundation, the W. M. Keck Foundation, and National Institutes of Health Grants DK30292, DK62675, and DK73695.
Abbreviations
- GF
germ-free
- CONV-R
conventionally raised
- dpf
days postfertilization
- CONVD
conventionalized
- GZM
gnotobiotic zebrafish medium
- TEM
transmission electron microscopy
- TTSS
Type III secretion system
- qRT-PCR
quantitative RT-PCR.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702386104/DC1.
References
- 1.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller TL, Wolin MJ. Syst Appl Microbiol. 1986;7:223–229. [Google Scholar]
- 3.Rieu-Lesme F, Delbes C, Sollelis L. Curr Microbiol. 2005;51:317–321. doi: 10.1007/s00284-005-0036-8. [DOI] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R, Gottschalk G, Thauer RK. J Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Stappenbeck TS, Hooper LV, Gordon JI. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bäckhed F, Crawford PA, O'Donnell D, Gordon JI. Proc Natl Acad Sci USA. 2007;104:606–611. doi: 10.1073/pnas.0605957104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lesher S, Walburg HE, Jr, Sacher GA., Jr Nature. 1964;202:884–886. doi: 10.1038/202884a0. [DOI] [PubMed] [Google Scholar]
- 10.Khoury KA, Floch MH, Hersh T. J Exp Med. 1969;130:659–670. doi: 10.1084/jem.130.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, et al. Proc Natl Acad Sci USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 14.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 16.Macpherson AJ, Uhr T. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 17.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon HA, Wostmann BS, Bruckner-Kardoss E. Proc Soc Exp Biol Med. 1963;114:301–304. doi: 10.3181/00379727-114-28658. [DOI] [PubMed] [Google Scholar]
- 20.Ley RE, Peterson DA, Gordon JI. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 21.Ng AN, de Jong-Curtain TA, Mawdsley DJ, White SJ, Shin J, Appel B, Dong PD, Stainier DY, Heath JK. Dev Biol. 2005;286:114–135. doi: 10.1016/j.ydbio.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Rawls JF, Samuel BS, Gordon JI. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Dev Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Cahill MM. Microb Ecol. 1990;19:21–41. doi: 10.1007/BF02015051. [DOI] [PubMed] [Google Scholar]
- 27.Huber I, Spanggaard B, Appel KF, Rossen L, Nielsen T, Gram L. J Appl Microbiol. 2004;96:117–132. doi: 10.1046/j.1365-2672.2003.02109.x. [DOI] [PubMed] [Google Scholar]
- 28.Romero J, Navarrete P. Microb Ecol. 2006 doi: 10.1007/s00248-006-9037-9. [DOI] [PubMed] [Google Scholar]
- 29.Graham DY, Yoshimura HH, Estes MK. J Lab Clin Med. 1983;101:940–954. [PubMed] [Google Scholar]
- 30.Wei B, Huang T, Dalwadi H, Sutton CL, Bruckner D, Braun J. Infect Immun. 2002;70:6567–6575. doi: 10.1128/IAI.70.12.6567-6575.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spivak J, Landers CJ, Vasiliauskas EA, Abreu MT, Dubinsky MC, Papadakis KA, Ippoliti A, Targan SR, Fleshner PR. Inflamm Bowel Dis. 2006;12:1122–1130. doi: 10.1097/01.mib.0000235833.47423.d7. [DOI] [PubMed] [Google Scholar]
- 32.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 33.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, et al. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace KN, Pack M. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- 36.Nivens DE, Ohman DE, Williams J, Franklin MJ. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnenburg JL, Angenent LT, Gordon JI. Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 38.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 39.Spangenberg C, Heuer T, Burger C, Tummler B. FEBS Lett. 1996;396:213–217. doi: 10.1016/0014-5793(96)01099-x. [DOI] [PubMed] [Google Scholar]
- 40.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. Mol Microbiol. 2003;50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 41.Sukonina V, Lookene A, Olivecrona T, Olivecrona G. Proc Natl Acad Sci USA. 2006;103:17450–17455. doi: 10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbieri JT, Sun J. Rev Physiol Biochem Pharmacol. 2004;152:79–92. doi: 10.1007/s10254-004-0031-7. [DOI] [PubMed] [Google Scholar]
- 43.Holder IA, Neely AN, Frank DW. Burns. 2001;27:129–130. doi: 10.1016/s0305-4179(00)00142-x. [DOI] [PubMed] [Google Scholar]
- 44.Lee VT, Smith RS, Tummler B, Lory S. Infect Immun. 2005;73:1695–1705. doi: 10.1128/IAI.73.3.1695-1705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vance RE, Rietsch A, Mekalanos JJ. Infect Immun. 2005;73:1706–1713. doi: 10.1128/IAI.73.3.1706-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfgang MC, Lee VT, Gilmore ME, Lory S. Dev Cell. 2003;4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 47.Smith RS, Wolfgang MC, Lory S. Infect Immun. 2004;72:1677–1684. doi: 10.1128/IAI.72.3.1677-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolfaghar I, Evans DJ, Ronaghi R, Fleiszig SM. Infect Immun. 2006;74:3880–3889. doi: 10.1128/IAI.01891-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 50.Kagami Y, Ratliff M, Surber M, Martinez A, Nunn DN. Mol Microbiol. 1998;27:221–233. doi: 10.1046/j.1365-2958.1998.00679.x. [DOI] [PubMed] [Google Scholar]
- 51.Macnab RM. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 52.Soscia C, Hachani A, Bernadac A, Filloux A, Bleves S. J Bacteriol. 2007 doi: 10.1128/JB.01677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strom MS, Lory S. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 54.Arora SK, Neely AN, Blair B, Lory S, Ramphal R. Infect Immun. 2005;73:4395–4398. doi: 10.1128/IAI.73.7.4395-4398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toutain CM, Zegans ME, O'Toole GA. J Bacteriol. 2005;187:771–777. doi: 10.1128/JB.187.2.771-777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Neil HS, Marquis H. Infect Immun. 2006;74:6675–6681. doi: 10.1128/IAI.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ottemann KM, Lowenthal AC. Infect Immun. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butler SM, Camilli A. Proc Natl Acad Sci USA. 2004;101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arora SK, Ritchings BW, Almira EC, Lory S, Ramphal R. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giron JA, Torres AG, Freer E, Kaper JB. Mol Microbiol. 2002;44:361–379. doi: 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- 61.Young GM, Schmiel DH, Miller VL. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fleiszig SM, Arora SK, Van R, Ramphal R. Infect Immun. 2001;69:4931–4937. doi: 10.1128/IAI.69.8.4931-4937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 65.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 66.Tsujita T, Tsukada H, Nakao M, Oshiumi H, Matsumoto M, Seya T. J Biol Chem. 2004;279:48588–48597. doi: 10.1074/jbc.M407634200. [DOI] [PubMed] [Google Scholar]
- 67.Schleimer RP, Sha Q, Vandermeer J, Lane AP, Kim J. Clin Exp All Rev. 2004;4:176–182. [Google Scholar]
- 68.Prince A. Am J Respir Cell Mol Biol. 2006;34:548–551. doi: 10.1165/rcmb.2006-0022SF. [DOI] [PubMed] [Google Scholar]
- 69.Adamo R, Sokol S, Soong G, Gomez MI, Prince A. Am J Respir Cell Mol Biol. 2004;30:627–634. doi: 10.1165/rcmb.2003-0260OC. [DOI] [PubMed] [Google Scholar]
- 70.Neish AS. Am J Physiol. 2007;292:G462–G466. doi: 10.1152/ajpgi.00274.2006. [DOI] [PubMed] [Google Scholar]
- 71.Bloemberg GV, O'Toole GA, Lugtenberg BJ, Kolter R. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.