Abstract
Hampin, homolog of drosophila MSL1, is a partner of histone acetyltransferase MYST1/MOF. Functions of these proteins remain poorly understood beyond their participation in chromatin remodeling complex MSL. In order to identify new proteins interacting with hampin, we screened a mouse cDNA library in yeast two-hybrid system with mouse hampin as bait and found five high-confidence interactors: MYST1, TPR proteins TTC4 and KIAA0103, NOP17 (homolog of a yeast nucleolar protein) and transcription factor GC BP. Subsequently, all these proteins were used as baits in library screenings and more new interactions were found: tumor suppressor RASSF1C and spliceosome component PRP3 for KIAA0103, ring finger RNF10 for RASSF1C, and RNA polymerase II regulator NELF-C for MYST1. The majority of the observed interactions was confirmed in vitro by pull-down of bacterially expressed proteins. Reconstruction of a fragment of mammalian interactome suggests that hampin may be linked to diverse regulatory processes in the nucleus.
Keywords: Hampin, MSL1, MYST1, interactome, TTC4, NOP17, NELF, RASSF1C, protein interactions
INTRODUCTION
Hampin has been known for a long time as MSL1 protein of Drosophila melanogaster where its mutations are associated with Male-Specific Lethal phenotype [1]. We proposed a different name for its orthologs (hampin [2]) in order to emphasize the poor sequence conservation: even two drosophilid species show only 37% identity (D. melanogaster vs. D. pseudoobscura) and homology between mammalian hampin and D. melanogaster MSL1 is rather weak: only 26% identity is maintained, overwhelmingly in its C-terminal domain called IVpehe [3]. Primary structure of mammalian hampin was determined for the first time as a short partial sequence (Genbank Acc. Number BM945758) and full-length clones were identified later (Genbank Acc. Number BC043039 etc).
Some of the MSL1 and hampin functions accomplished via the IVpehe domain are identical: both human and Drosophila MSL orthologs are involved in multiprotein complexes (MSL) with histone acetyltransferase (HAT) activity [1, 4], that is executed by MYST1/MOF. On the other hand, the drosophila MSL complex, called compensasome, seems to be highly specialized for one peculiar function, X-chromosome dosage compensation in males [1]. It should be noted that the use of compensasome for dosage compensation is known only in drosophila [1, 5]. Also, mammalian MSL complex associates with all chromosomes [4] and its HAT activity is important for maintaining genome integrity, ATM-dependent DNA repair and tumor suppression [6–8].
It is interesting that murine hampin has at least five alternative splice variants [2]. These variants (A–E) contain 616 (GenBank acc. Number NP_082998), 600 (XP_999185), 463 (XP_999167), 370 (BAB29868) and 233 (ABD46887) amino acid residues in its sequence and this diversity is a consequence of use of different exon sets. The longest variant A contains four domains: Pro-rich low complexity and highly variable domain I (absent in hampin D and E variants), short coiled-coil domain IIcc, modestly conserved domain III and highly conservative domain IVpehe (absent in C and E). All the variants were recently shown to be localized in nuclei [9].
RNA interference of either MYST1 or hampin A results in a decrease of histone H4 K16 acetylation [4]. Recombinant MYST1 is specific to H4 K16 in vivo but, in vitro, acetylates also H3 [6] indicating that its specificity may be attuned by other proteins. Apparently, the hampin/MSL1-MYST1/MOF association is conserved from arthropods to vertebrates. Drosophila MSL1 also interacts with MSL3 and MOF proteins via domain IVpehe and with MSL2 via domain IIcc [10]. It is known also that mammalian MYST1 participates in at least two protein complexes that possess histone acetylase activity, - one including proteins hampin A, MSL2, MSL3 and second complex including so-called “NSL”-proteins [11]. Both these complexes are evolutionary conserved, but interactions between their individual components are not well studied.
Therefore, it can be concluded that function both of hampin-MYST1 pair is poorly understood in the global interactome of mammals. To address this question, we searched for new protein-protein interactions of murine hampin and MYST1.
MATERIALS AND METHODS
Two-hybrid system
To construct the bait plasmids, the cDNA fragments corresponding to E266-K616 fragment of mouse hampin A was cloned by PCR into the GAL4 DNA binding domain fusion pGBKT7 vector at Nde I/Sma I sites. Bait-transformed yeast strain AH109 were mated with Y187 cells pretransformed with BD Matchmaker 17day mouse embryo cDNA library in pGADrec (pGAD-T7rec) vector (BD Biosciences) and grown on high stringency selection medium deficient for histidine, adenine, leucine, and tryptophan. Primary colonies were regrown several times on the selection medium with α-X-Gal to select for induction of all three available reporter genes (HIS, ADE and MEL1). Plasmids were rescued from the positive clones by transformation of E .coli and retested to exclude false positives by cotransformation of AH109 cells with either hampin bait or empty pGBK plasmid. Plasmids clones passed all these tests were subjected to sequence determination.
Dual-tag system and protein expression
Plasmids pHPMLQ, pCBDQ and pD1HTHQ were prepared by cloning fragments encoding T166-S371 (HPML), K1057-V1205 (CBD) fragments of human Plasma Membrane Calcium ATPase (hPMCA4b) and M1-K394 of E. coli nicotinamide nucleotide transhydrogenase (NNT) in pQE42 vector (Qiagen) at BamH I/ Bgl II sites (instead of DHFR coding sequence). These coding DNA sequences were amplified using PCR from pO plasmid (for hPMCA4b fragments) or from E.coli genomic DNA as template (for D1HTH). All other full-length ORFs were amplified by RT-PCR from various mouse cDNAs using specific primers and standard PCR conditions with the exception of hampin A and C: they required melting temperature to be set at 98 C. Murine ORFs for hampin A, C and RASSF1C were cloned into pD1HTHQ at Bgl II and Hind III sites, mouse KIAA0103 protein was cloned in pHPMLQ vector at Bgl II and Sal I sites. MYST1 - in pCBDQ at Bgl II / Sma I sites, NELF-C - at Bgl II and Sph I sites of pD1HTHQ. Full-length TTC4 ORF as well as a fragment corresponding to TPR-containing M1-R310 fragment (termed 51HIN) and mouse NOP17 full-length ORF were cloned into pQE30 vector (Qiagen) at Bgl II-Bam HI and Hind III sites. His6-tagged NOP17, 51HIN, D1HTH, CBD-MYST1, and CBD were expressed at +37 C for 30 min in presence of 1mM IPTG. Hampin fusions with D1HTH, His6-tagged TTC4, D1HTH-RASSF1C and D1HTH-NELFC were expressed for one hour at +19 C to avoid extensive degradation of the proteins. HMPL-tagged mouse KIAA0103 protein was expressed for 4 hours at +37 C.
In vitro binding assays
To prepare soluble cell extracts, E.coli cells were lysed in TE buffer (10mM Tris, 1 mM EDTA, pH 8) containing 1 mg/ml lysozyme, protease inhibitor cocktail (Sigma P2714), 1 mM PMSF for 30 min on ice, then sonified and centrifuged for 15000 rpm 15 min (Beckman JA-20 rotor). For co-immunoprecipitation, E.coli extracts were mixed with 1 μl of ascites fluid and incubated for one hour at room temperature. Then 3 μl Protein G-sepharose was added and incubated for one hour on rotary shaker, washed three times with “wash” buffer (25mM Hepes pH 7.6, 0.5% NP40, 400 mM NaCl, 1mM MgCl2, 1mM CaCl2) and eluted by 50 μl of electrophoresis sample loading buffer (2xSLB). For co-immunoprecipitation experiments with HPML-tagged KIAA0103 protein, the protein was purified in denaturing conditions and dialyzed against PBS. For pull-down experiments on calmodulin-sepharose, protein extracts were mixed in the same way except that 20 μl calmodulin-sepharose instead of Protein G-sepharose was used and 2 mM CaCl2. was included in all buffers. For D1HTH-NELFC pull-down on CBD-MYST1, 10 mM MgCl2 was also added. For pull-down from myeloma cells, nuclear extract from myeloma cell line X63AG8.653 was prepared as described [4] except that proteins were extracted using PBS containing 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS (RIPA). The nuclear extracts were incubated overnight with either 51HIN or D1HTH-hampin immobilized on 20 μl Ni-NTA-agarose (Sigma N3158) at +4°C, washed three times and eluted using 2xSLB buffer. Western blotting was performed as described earlier [12] using chemiluminescent substrates ECL+ (Amersham Biosciences) or Immun-Star (Bio-Rad).
Antibodies and immunocytochemistry
His6-tagged and HPML-tagged KIAA0103 proteins were purified as described [13]. Rabbit polyclonal antibodies against hampin were obtained using His6-tagged fragment K259-A463 of hampin C as described in [2]. His6-tagged 51HIN was used for immunization of rabbits and mouse, whereas His6-tagged NOP17 was used only for immunization of mouse. Monoclonal and polyclonal antibodies to D1HTH domain (2G10) and monoclonal antibodies 2D8 against first cytoplasmic loop and 3C12 against calmodulin-binding domain of hPMCA4b were described earlier [14] (N.B.P., unpublished data). Mouse 3T3 fibroblasts and the myeloma cells were grown in DMEM containing 10% fetal bovine serum. For immunofluorescence, 3T3 fibroblasts grown onto glass tissue slides were fixed in methanol at −20°C, blocked for one hour in 5% normal bovine serum (Difco), incubated with the antibodies (1:1000 dilution for rabbit and 1:200 for mouse antibodies, 50 mins at RT) washed with TBST (tris buffered saline with 0.1% tween-20), then incubated with anti-rabbit (Alexa Fluor 555 nm) or anti-mouse (Alexa Fluor 594 nm) secondary antibodies (1:1000 dilution), washed with TBST, counterstained with DAPI (Molecular Probes), mounted in Anti-Fade medium (Molecular Probes) and images collected on a fluorescent microscopy equipped with a digital camera.
Results
Search of hampin protein interactions in two-hybrid system
To find new protein partners of hampin, we screened a 17 day mouse whole embryo cDNA library using yeast two-hybrid method with E266-K616 fragment of mouse hampin A as bait. Using high stringency selection conditions and verification of the clones in order to exclude obvious false positives, we identified several clones encoding five different proteins: histone acetyl transferase MYST1, two tetratricopeptide repeat proteins – mouse KIAA0103 protein and TTC4, C2H2 Zn finger protein GC BP, and a novel protein homologous to yeast NOP17 (Table 1). To widen the interactome map centered on hampin, we screened the same library for novel interactors of these five proteins. No positive clones were identified in case of TTC4 and NOP17. Screening for interactors of KIAA0103 gave RASSF1C and PRP3. RASSF1C attracted our attention because of its role in oncogenesis: we chose it for a further screen and obtained one new clone, identified as RNF10 (RING-finger protein 10). Screening of interactions for MYST1 gave NELF-C/TH1L and also one clone encoding hampin A itself (coordinates Y437-K616).
TABLE 1.
Identification of protein-protein interactions from two-hybrid library screenings
| Bait | Interactor | GenBank number | Amino acid coordinates | Length; Domains | Known or hypothetical functions of the interactors |
|---|---|---|---|---|---|
| Hampin | KIAA0103/TTC35 | NP_080012 | Full length | 297; TPR2 | None |
| TTC4 | NP_082485 | Full length | 386; TPR | Putative co-chaperone and tumor supressor | |
| GC BP
NOP17 |
NP_666371
NP_083682 |
P83-C terminus
Full length |
619; C2H2 Zn finger
290; NOP17 |
Transcription factor, regulates IL12 p35 gene in macrophages
Putatively involved in pre-rRNA processing |
|
| MYST 1 | NP_080646 | G39-C terminus | 458; MYST; C2HC Zn Finger; chromodomain; | Histone acetyl transferase | |
| KIAA0103 | RASSF1C | NP_062687 | Full length | 270; RA | Tumor suppressor |
| PRP3 | NP_081817 | E291-C terminus | 683; PWI | Splic ing factor | |
| MYST1 | NELF-C/TH1L | NP_065605 | Full length | 592 | Negative regulator of RNAPII transcription elongation and A-raf protein kinase |
| Hampin A | AK_014463 | Y437-K616 | 616 | Component of histone acetyl transferase complex | |
| RASSF1C | RNF10 | NP_057907 | G372-C terminus | 805 | None |
Confirmation of interactions in vitro
To assess reliability of the interactions identified by the two-hybrid screenings, we tested them in vitro. These experiments were also aimed at investigating whether the interactions are direct and whether they require any modifications specific to eukaryotes. For this purpose, we expressed His6-tagged proteins NOP17 and TTC4 in E .coli. Expression of full-length hampin A and C was not successful either as His6-tagged proteins or as fusions with several other proteins, such as DHFR (results not shown). After these attempts, we decided to employ C-terminal fusions with nicotinamide nucleotide transhydrogenase domain I (D1HTH), and two domains of hPMCA4b (HPML or CBD). With these domains, we developed a new dual tag system useful for immunoprecipitation and pull-down. This system relies on available precipitating monoclonal antibodies against the tags (described in Methods). Finally, we successfully expressed in E. coli soluble full-length hampins A and C, RASSF1C and soluble NELF-C as C-terminal fusions with D1HTH, whereas MYST1 was expressed as C-terminal fusions with CBD. Expression of KIAA0103 was successful in the case of HMPL fusion.
Fig. 1 illustrates confirmation of identified interactions in vitro. Known interaction between hampin A and MYST1 was confirmed using our dual-tag system (see Supplementary Fig. S1.). Interactions of hampin with His-tagged TTC4 and HPML-tagged KIAA0103 proteins were tested by co-immunoprecipitation from E. coli extracts with D1HTH-tagged hampin A and C (Fig. 1A) or, in case of NOP17 protein, with D1HTH-tagged hampin A only (Fig. S2). It was shown that TTC4 co-precipitates with both hampin isoforms whereas KIAA0103 - only with hampin A. It would be also interesting if the observed interactions can be confirmed with endogenous proteins; D1HTH-tagged hampin A and C were used for pull-down of myeloma extract. NOP17 was detected in the eluate from immobilized hampin A, and was lacking in the eluate from hampin C (Fig. 1B). All these results indicate that domain IVpehe of hampin is required for binding NOP17 and KIAA0103 whereas domain III is necessary for interaction with TTC4.
Figure 1. Confirmation of observed interactions using in vitro-based methods.
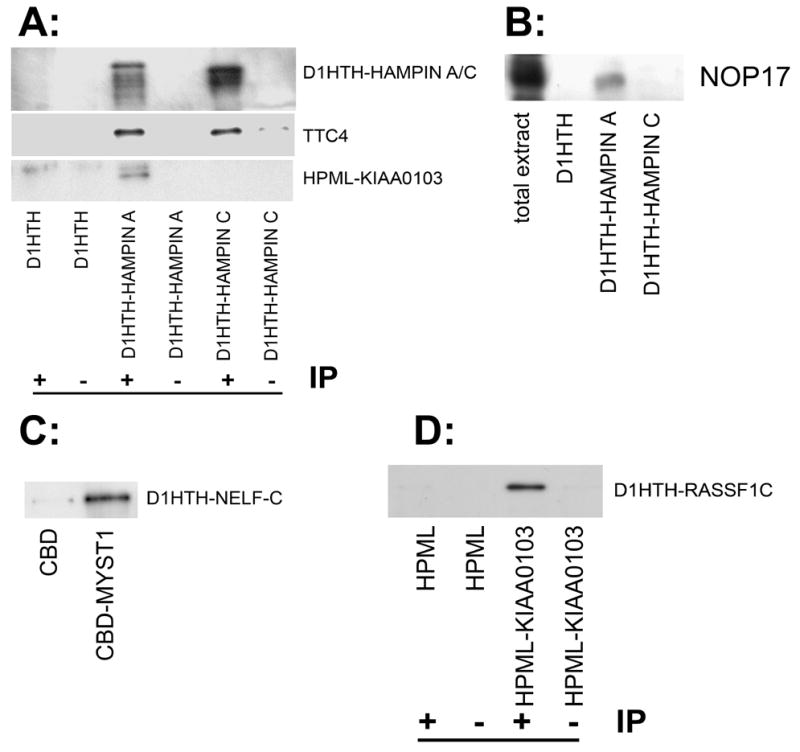
A: Results of co-immunoprecipitation (IP) of overexpressed D1HTH-hampin isoforms A and C His6-tagged proteins TTC4 and HPML-tagged KIAA0103 from E. coli extracts using monoclonal antibody to D1HTH (marked “+”). Co-precipitations with D1HTH polypeptide or with an unrelated antibody (“−“) were used as controls. Western-blotting was done using rabbit anti-hampin, anti-TTC4 and anti-HMPL antibodes (from top to bottom, respectively). Multiple bands of hampin isoforms A and C (upper panel) with lower molecular weights represent products of partial proteolytic degradation during bacterial expression. B: Pull-down of endogenous NOP17 protein from mouse myeloma nuclear extract on D1HTH-tagged hampin isoforms A and C, and D1HTH itself (negative control) immobilized on Ni-NTA-sepharose. Eluates from Ni-NTA agarose were stained with the help of mouse antibodies against NOP17. C: Pull-down of bacterially expressed D1HTH-tagged NELF-C on CBD-tagged MYST1 or CBD (negative control). Recombinant D1HTH-NELF-C was incubated together with CBD-MYST1 and then pulled down using calmodulin-sepharose. Beads were washed, and eluates were probed with monoclonal antibody 2G10 specific to D1HTH. D: Co-immunoprecipitation of D1HTH-tagged RASSF1C with HPML-tagged KIAA0103 from bacterial extracts. Proteins HPML or HPML-tagged KIAA0103 were precipitated with monoclonal anti-HMPL antibody 2D8 from bacterial extract containing D1HTH-RASSF1C. Eluates from protein-G-sepharose were analyzed using rabbit antibodies against D1HTH.
Interaction MYST1-NELF-C was confirmed by pull-down from E. coli extracts using our dual-tag system (Fig. 1C) and interaction KIAA0103-RASSF1C was confirmed using co-immunoprecipitaion from E. coli extracts (Fig.1D).
In all cases when we were able to bacterially express protein pairs, we successfully detected the protein-protein interactions by pull-down/co-immunoprecipitation. This provides an estimate that interactions identified in this work by the yeast two-hybrid system have an excellent rate or true positives.
Demonstration of nuclear localization of new hampin-interacting proteins
It is known that hampin is localized exclusively in nuclei [4, 9]. However, TTC4 and NOP17 are almost unstudied; therefore we characterized them in more detail. We performed immunofluorescence experiments and showed that mouse proteins TTC4 and NOP17 are also localized predominantly in nuclear compartments (Fig. 2 and Fig. S3). Nuclear localization of NOP17 and TTC4 provides additional support for the existence of protein complexes with both TTC4 and NOP17 associated with hampin.
Figure 2. Subcellular localization of endogenous proteins TTC4 and NOP17 in 3T3 fibroblasts.
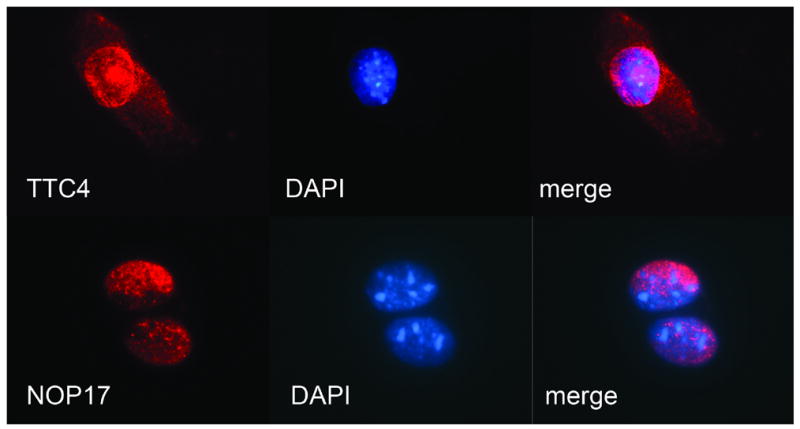
Mouse 3T3 fibroblasts were labeled with polyclonal antibodies against TTC4 (top) or NOP17 (bottom) followed by secondary antibodies conjugated with Alexa Fluor (red fluorescence). Nuclei counterstained with DAPI (blue fluorescence). From left to right: antibody labeling, DAPI and merged images.
Discussion
In this work, the interaction of mouse hampin/MSL1 with MYST1/MOF histone acetyl transferase has been found using yeast two-hybrid system, in accord with recent independent reports of Smith et al and [4] and Mendjan et al [11] where cultured human cells were used. This interaction requires domain IVpehe of hampin and is conserved from insects to mammals despite poor overall sequence conservation of hampin/MSL1. MYST1/MOF can be regarded as a catalytic subunit of the MSL complex, which function is acetylation of histone H4 at Lys16.
Functions of other identified hampin interactors are largely unknown. For example, the only available information about KIAA0103 is its presence in inner nuclear membrane [15]. Structurally, it belongs to TPR proteins; TPR is a repeat structure of typically 34 amino acids that is present in a large number of functionally diverse proteins [16]. Another TPR protein interacting with hampin is TTC4. Its gene was reported to be frequently damaged in breast cancer [17] and melanoma [18] (the latter is contradictory [19]). Its Drosophila homologue, Dpit47, has an interesting function as a nuclear co-chaperone of Hsp90 protein that interacts with DNA polymerase α [20]. However, our results indicate that TTC4 binds both hampin A and C isoforms and, therefore, the hampin-TTC4 interaction apparently occurs through variable domain III.
No studies on mammalian NOP17 have been published to date. Its yeast homologue NOP17p is known to be located in nucleoli where it interacts with snoRNP protein NOP58p, exosome component Prp43p (homologous to mammalian OIP-2) and NOP53p. These proteins are involved in pre-rRNA processing [21].
GC BP is a C2H2-Zn-Finger transcription factor that suppresses interleukine-12 p35 gene in macrophages during phagocytosis of apoptotic cells [22]. GC BP stands for GC - binding protein because it binds a GC motif in the IL-12 p35 promoter. The GC BP gene is expressed in all tissues, therefore GC BP may have a broad physiological role not limited to regulation of interleukine-12.
Subsequent screenings using the hampin interactors as baits revealed several new interesting protein-protein interactions, for example, between MYST1 and NELF-C/TH1L. NELF-C/TH1L is a component of negative elongation factor (NELF) that is involved in promoter-proximal pausing by RNA polymerase II [23]. Also, NELF-C/TH1L negatively regulates protein kinase A-raf [24]. In this context, the interaction MYST1-NELF-C may link acetylation of H4 K16 to diverse processes including regulation of transcription elongation and A-raf signaling.
The identified binding of KIAA0103 to tumor suppressor RASSF1C [25] is especially interesting since it may reveal a liaison between H4 K16 acetylation and oncogenesis. Recently, RASSF1C was shown to localize to nuclei via anchoring to Daxx and to an unknown protein [26]. Hypothetically, RASSF1C translocation to nuclei may involve KIAA0103 and, indirectly, hampin A.
Our results together with published data allowed reconstruction of a mammalian nuclear interactome fragment centered on hampin (Fig. 3). Genes encoding homologs of KIAA0103, NOP17 and TTC4 exist in genomes from yeast to mammals, whereas true GC BP orthologs can be identified only in vertebrates. Therefore, many of these interactions may be well conserved. However, high-throughput screening of drosophila interactome yielded no high-confidence protein interactors for MSL1 [27]. Therefore, hampin appears to possess a functional profile more diverse than that of drosophila MSL1. This complexity may be even higher since hampin splice variants differ in domain composition.
Figure 3. Fragment of mammalian interactome map including already known interactions and those identified in this work.
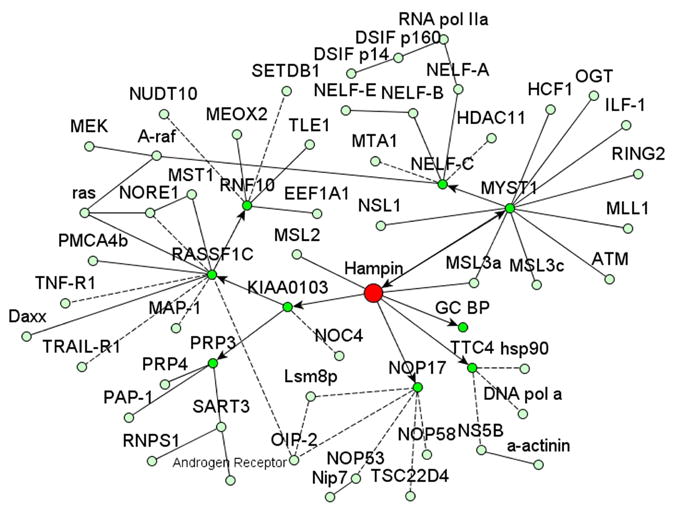
Prepared using yEd software (yWorks). Hampin is shown in red; interactions identified in this work are shown by arrows; other interactions were taken from databases HPRD, BioGRID, BOND, DIP and PPI; interrupted lines stand for unconfirmed interactions (from high throughput screenings or inferred from homology).
Supplementary Material
Figure S1. Confirmation of in vitro interaction between proteins MYST1 and hampin A. D1HTH-tagged hampin A or D1HTH itself were pulled down on CBD and CBD-tagged MYST1 bound to calmodulin-sepharose. Eluates from calmodulin-sepharose were stained with monoclonal antibodies against CBD and D1HTH polypeptides.
Figure S2. Additional in vitro based confirmations of interactions between proteins hampin A, TTC4 and NOP17. Results of co-immunoprecipitation (IP) from E. coli extracts of overexpressed D1HTH-hampin A and His6-tagged proteins TTC4 and NOP17 using monoclonal antibody to D1HTH (marked “+”). Western-blotting was done using monoclonal antibody to D1HTH, mouse anti-NOP17 antibody and rabbit anti-TTC4.
Figure S3. Subcellular localization of endogenous proteins SPCA1, hampin, TTC4, NOP17 and SERCA2 in 3T3 fibroblasts. Mouse 3T3 fibroblasts were labeled with primary rabbit polyclonal antibodies against hampin, TTC4, and mouse antibodies against NOP17 and SERCA2 (negative control for mouse serum). Fibroblasts were stained using secondary antibodies conjugated with Alexa Fluor (red) and counterstained using DAPI (blue) as nuclear marker.
Acknowledgments
We are grateful to I. Berezkin, N. Bystrov, S. Volkov, O. Molokoyedova, Y. Makarova, and especially to Dr. V. Baklaushev for excellent help. Supported by RFBR grant 04-04-49413 and NIH grants HL-36573.
Abbreviations
- PBS
phosphate buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Gen. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 2.Dmitriev RI, Pestov NB, Korneenko TV, Gerasimova AV, Zhao H, Modianov NN, Kostina MB, Shakhparonov MI. Tissue specificity of alternative splicing products of mouse mRNA encoding new protein hampin homologous to the Drosophila MSL-1 protein. Bioorg Khim. 2005;31:363–371. doi: 10.1007/s11171-005-0045-1. [DOI] [PubMed] [Google Scholar]
- 3.Marin I. Evolution of chromatin-remodeling complexes: comparative genomics reveals the ancient origin of "novel" compensasome genes. J Mol Evol. 2003;56:527–539. doi: 10.1007/s00239-002-2422-1. [DOI] [PubMed] [Google Scholar]
- 4.Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiz MF, Esteban MR, Donoro C, Goday C, Sanchez L. Evolution of dosage compensation in Diptera: the gene maleless implements dosage compensation in Drosophila (Brachycera suborder) but its homolog in Sciara (Nematocera suborder) appears to play no role in dosage compensation. Genetics. 2000;156:1853–1865. doi: 10.1093/genetics/156.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. Involvement of human MOF in ATM function. Mol Cell Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 9.Dmitriev RI, Pestov NB, Korneenko TV, Shakhparonov MI. Intracellular location of hampin isoforms. Dokl Biochem Biophys. 2006;408:130–132. doi: 10.1134/s1607672906030069. [DOI] [PubMed] [Google Scholar]
- 10.Scott MJ, Pan LL, Cleland SB, Knox AL, Heinrich J. MSL1 plays a central role in assembly of the MSL complex, essential for dosage compensation in Drosophila. EMBO J. 2000;19:144–155. doi: 10.1093/emboj/19.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Pestov NB, Korneenko TV, Adams G, Tillekeratne M, Shakhparonov MI, Modyanov NN. Nongastric H-K-ATPase in rodent prostate: lobe-specific expression and apical localization. Am J Physiol Cell Physiol. 2002;282:C907–C916. doi: 10.1152/ajpcell.00258.2001. [DOI] [PubMed] [Google Scholar]
- 13.Pestov NB, Gusakova TV, Kostina MV, Shakhparonov MI. Phage mimotopes for monoclonal antibodies against plasma membrane Ca2+-ATPase. Bioorg Khim. 1996;22:567–573. [PubMed] [Google Scholar]
- 14.Bizouarn T, Fjellstrom O, Axelsson M, Korneenko TV, Pestov NB, Ivanova MV, Egorov MV, Shakhparonov M, Rydstrom J. Interactions between the soluble domain I of nicotinamide nucleotide transhydrogenase from Rhodospirillum rubrum and transhydrogenase from Escherichia coli: Effects on catalytic and H1-pumping activities. Eur J Biochem. 2000;267:3281–3288. doi: 10.1046/j.1432-1327.2000.01358.x. [DOI] [PubMed] [Google Scholar]
- 15.Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci U S A. 2001;98:11943–11948. doi: 10.1073/pnas.211201898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Su G, Roberts T, Cowell JK. TTC4, a novel human gene containing the tetratricopeptide repeat and mapping to the region of chromosome 1p31 that is frequently deleted in sporadic breast cancer. Genomics. 1999;55:157–163. doi: 10.1006/geno.1998.5633. [DOI] [PubMed] [Google Scholar]
- 18.Poetsch M, Dittberner T, Cowell JK, Woenckhaus C. TTC4, a novel candidate tumor suppressor gene at 1p31 is often mutated in malignant melanoma of the skin. Oncogene. 2000;19:5817–5820. doi: 10.1038/sj.onc.1203961. [DOI] [PubMed] [Google Scholar]
- 19.Irwin N, Walker G, Hayward N. Lack of TTC4 mutations in melanoma. J Invest Dermatol. 2002;119:186–187. doi: 10.1046/j.1523-1747.2002.18181.x. [DOI] [PubMed] [Google Scholar]
- 20.Crevel G, Bates H, Huikeshoven H, Cotterill S. The Drosophila Dpit47 protein is a nuclear Hsp90 co-chaperone that interacts with DNA polymerase alpha. J Cell Sci. 2001;114:2015–2025. doi: 10.1242/jcs.114.11.2015. [DOI] [PubMed] [Google Scholar]
- 21.Gonzales FA, Zanchin NI, Luz JS, Oliveira CC. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J Mol Biol. 2005;346:437–455. doi: 10.1016/j.jmb.2004.11.071. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity. 2004;21:643–653. doi: 10.1016/j.immuni.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, Endoh M, Yamada T, Handa H. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W, Shen X, Yang Y, Yin X, Xie J, Yan J, Jiang J, Liu W, Wang H, Sun M, Zheng Y, Gu J. Trihydrophobin 1 is a new negative regulator of A-Raf kinase. J Biol Chem. 2004;279:10167–10175. doi: 10.1074/jbc.M307994200. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Wang F, Protopopov A, Malyukova A, Kashuba V, Minna JD, Lerman MI, Klein G, Zabarovsky E. Inactivation of RASSF1C during in vivo tumor growth identifies it as a tumor suppressor gene. Oncogene. 2004;23:1–9. doi: 10.1038/sj.onc.1207789. [DOI] [PubMed] [Google Scholar]
- 26.Kitagawa D, Kajiho H, Negishi T, Ura S, Watanabe T, Wada T, Ichijo H, Katada T, Nishina H. Release of RASSF1C from the nucleus by Daxx degradation links DNA damage and SAPK/JNK activation. EMBO J. 2006;25:3286–3297. doi: 10.1038/sj.emboj.7601212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Confirmation of in vitro interaction between proteins MYST1 and hampin A. D1HTH-tagged hampin A or D1HTH itself were pulled down on CBD and CBD-tagged MYST1 bound to calmodulin-sepharose. Eluates from calmodulin-sepharose were stained with monoclonal antibodies against CBD and D1HTH polypeptides.
Figure S2. Additional in vitro based confirmations of interactions between proteins hampin A, TTC4 and NOP17. Results of co-immunoprecipitation (IP) from E. coli extracts of overexpressed D1HTH-hampin A and His6-tagged proteins TTC4 and NOP17 using monoclonal antibody to D1HTH (marked “+”). Western-blotting was done using monoclonal antibody to D1HTH, mouse anti-NOP17 antibody and rabbit anti-TTC4.
Figure S3. Subcellular localization of endogenous proteins SPCA1, hampin, TTC4, NOP17 and SERCA2 in 3T3 fibroblasts. Mouse 3T3 fibroblasts were labeled with primary rabbit polyclonal antibodies against hampin, TTC4, and mouse antibodies against NOP17 and SERCA2 (negative control for mouse serum). Fibroblasts were stained using secondary antibodies conjugated with Alexa Fluor (red) and counterstained using DAPI (blue) as nuclear marker.


