Abstract
Background
Neoplasia can be driven by mutations resulting in dysregulation of transcription. In the mesenchymal neoplasm, aggressive fibromatosis, subtractive hybridization identified sterile alpha motif domain 9 (SAMD9) as a substantially down regulated gene in neoplasia. SAMD9 was recently found to be mutated in normophosphatemic familial tumoral calcinosis. In this study, we studied the gene structure and function of SAMD9, and its paralogous gene, SAMD9L, and examined these in a variety of species.
Results
SAMD9 is located on human chromosome 7q21.2 with a paralogous gene sterile alpha motif domain 9 like (SAMD9L) in the head-to-tail orientation. Although both genes are present in a variety of species, the orthologue for SAMD9 is lost in the mouse lineage due to a unique genomic rearrangement. Both SAMD9 and SAMD9L are ubiquitously expressed in human tissues. SAMD9 is expressed at a lower level in a variety of neoplasms associated with β-catenin stabilization, such as aggressive fibromatosis, breast, and colon cancers. SAMD9 and SAMD9L contain an amino-terminal SAM domain, but the remainder of the predicted protein structure does not exhibit substantial homology to other known protein motifs. The putative protein product of SAMD9 localizes to the cytoplasm. In vitro data shows that SAMD9 negatively regulates cell proliferation. Over expression of SAMD9 in the colon cancer cell line, SW480, reduces the volume of tumors formed when transplanted into immune-deficient mice.
Conclusion
SAMD9 and SAMD9L are a novel family of genes, which play a role regulating cell proliferation and suppressing the neoplastic phenotype. This is the first report as far as we know about a human gene that exists in rat, but is lost in mouse, due to a mouse specific rearrangement, resulting in the loss of the SAMD9 gene.
Background
Neoplasia can be driven by a variety of mechanisms. In many cases, oncogenic mutations result in the dysregulation of transcription. This is the case in mutations resulting in β-catenin stabilization in tumors, as stabilized β-catenin activates transcription factors in the Tcf/Lef family. These transcription factors regulate gene expression in a tissue specific manner [1,2]. Aggressive fibromatosis, also known as desmoid tumor, is a locally invasive soft tissue tumor composed of a clonal proliferation of mesenchymal, fibroblast-like, cells [3]. Somatic mutations in either the Adenomatous Polypisis Coli (APC) or β-catenin genes, resulting in the stabilization of β-catenin protein, are present in the majority of lesions. This gives aggressive fibromatosis tumor cells a proliferative advantage through Tcf/Lef dependent transcriptional activation [4,5]. There are a variety of lines of evidences suggesting that differentially expressed genes in aggressive fibromatosis are involved in the development of this tumor. For instance, the differentially expressed genes, COX-2, RHAMM, MMP-3, TIMP-1 and PAI-1 modulate the size or number of aggressive fibromatosis tumor that form [6-9].
To determine how differentially regulated genes contribute to the neoplastic phenotype in aggressive fibromatosis, suppression subtractive hybridization was used to compare gene expression differences between cell cultures derived from an aggressive fibromatosis tumor with an APC mutation and the same cells after transfection of wild-type APC. In this comparison, we found that SAMD9 was one of the most highly differentially regulated genes, and was upregulated with transfection of the wild-type APC. This study focuses on the gene structure and function of SAMD9 and its paralogous gene, SAMD9L.
Results
SAMD9 is differentially regulated in aggressive fibromatosis
Primary cell cultures from an aggressive fibromatosis tumor harbouring a truncating APC mutation were established. These were transfected with either a wild-type APC gene or a control vector, and successful transfection verified using immunohistochemistry. These cell cultures were established for a previous study to determine the role of the interaction between APC and β-catenin in the regulation of cell proliferation in aggressive fibromatosis. The transfection conditions, mutational analysis, and results of proliferation studies have been previously reported [10,11]. RNA from the cells derived from an aggressive fibromatosis tumor with an APC mutation and the same cells after transfection of wild-type APC were compared using suppression subtractive hybridization. Both forward (cells transfected with a control vector compared to cells transfected with wild-type APC) and reverse (cells transfected with wild-type APC compared to cells transfected with a control vector) subtractions were conducted. Clones were identified, and verified using Northern blot. The most differentially expressed clones were sequenced and their identity searched using GenBank.
In this comparison, the most differentially regulated gene in the reverse subtraction was a novel gene that was initially identified on human chromosome 7 using GenBank sequence data. Both 5'- and 3'- rapid amplification of cDNA ends (RACE) using the sequence of the 200 bp clone isolated from suppression subtractive hybridization, as well as analysis of EST in the region were undertaken to identify the full length of this gene. Subsequent to the start of this work, Strausberg et al identified a gene in this region using high through put analysis of EST's and designed the gene sterile alpha motif domain 9 (SAMD9) [12]. During this analysis, we also found a nearby gene, which was also subsequently identified by Strausberg et al as sterile alpha motif domain 9 like (SAMD9L) [12]. Using a search of the DNA sequence database in GenBank, SAMD9 and SAMD9L were found to match human chromosome 7q21.2. SAMD9L is located 5' upstream from the SAMD9 gene (Fig. 1). Both genes are coded by the reverse strand with a head to tail orientation.
Figure 1.
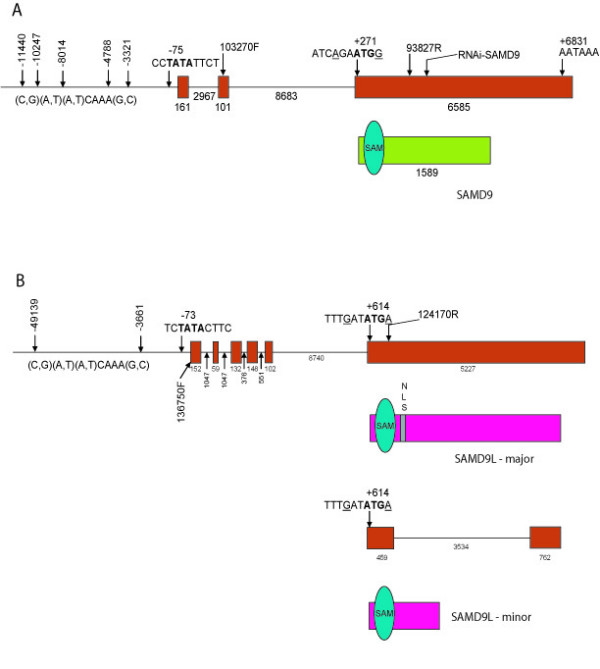
Gene and protein structure of SAMD9 and SAMD9L. A. Diagram of SAMD9 gene structure and predicted protein structure. Exons are shown as red rectangles. The protein is shown in green, with the sterile alpha motif domain as blue oval. B. Diagram of SAMD9L gene structure and predicted protein structure. Exons are shown in red. The protein is shown as a pink box, with two predicted open reading frames of shown. The sterile alpha motif domain is shown as blue oval. A potential nuclear localization domain is shown as grey box.
Structure of SAMD9 and its paralogue – SAMD9L
Compiling all of the sequence data, we located the most 5'- end sequence for SAMD9 in a 5'- RACE clone, and the most 3'- end sequence for SAMD9 in one of the EST clones [GenBank: AA628487] [see Additional file 1]. Using primers corresponding to the sequences of the most 5'- and 3'- ends of the SAMD9 gene, a predicted 7 kb transcript was amplified from an aggressive fibromatosis tumor cDNA. The full-length of this transcript, the longest transcript that we found, was sequenced [GenBank: AF445355]. The cDNA sequence of SAMD9 gene has 6847 bp with a poly-A signal (AATAAA) at 6831 bp downstream from the transcriptional start point (Fig. 1A). Mapping this cDNA sequence to the genomic DNA sequence in the GenBank, the SAMD9 gene is 18505 bp in length and contains three exons (161 bp, 101 bp and 6585 bp in length, respectively). There is a TATA signal (CCTATATTCT) at -75 bp and five predicted Lef/Tcf binding elements [(C,G) (A,T) (A,T)CAAA(G,C)] at -11440 bp, -10247 bp, -8014 bp, -4788 bp and -3321 bp. The clones isolated from 5'- and 3'- RACE of the SAMD9 gene were sequenced. Comparing the 5'- and 3'- RACE sequences to the genomic DNA sequence from GenBank, different transcription initiation sites and polyadenylation sites of SAMD9 are noted, and there are at least two alternative splices.
During the analysis of EST sequences near the initial clone, we identified a second gene,SAMD9L, which is located 12740 bp distal to SAMD9. Using the sequences from the most 5'- and 3'- EST in the region [Celera Database: THC 511215 and THC 513290, respectively] [see Additional file 1], the full-length sequence of the SAMD9L was amplified from an aggressive fibromatosis tumor cDNA, and sequenced [GenBank: AF474973]. The cDNA sequence of SAMD9L gene has 5821 bp with a poly-A signal (AATAAA) at 5659 bp downstream from the transcriptional start point (Fig. 1B). Mapping this cDNA sequence to the genomic DNA sequence in GenBank, the SAMD9L gene is 17583 bp in length, including six exons (152 bp, 59 bp, 132 bp, 148 bp, 102 bp and 5228 bp in length, respectively). There is a TATA signal (TCTATACTTC) at -73 bp and two predicted Lef/Tcf binding elements [(C,G) (A,T) (A,T)CAAA(G,C)] at -49139 bp and -3661 bp.
There are alternatively spliced transcripts in both SAMD9 [GenBank: AF453311] and SAMD9L [GenBank: AY195582-195587, DQ068177] [see Additional file 1]. These alternative splices lead to either inclusion or exclusion of an exon due to the usage of a different 5' donor splice site or a different 3' acceptor splice site. This alternative splicing also changes the internal coding region due to an in-frame insertion or deletion in both SAMD9 and SAMD9L. In the case of SAMD9, non-canonical dinucleotides AT and TC are used as donor and acceptor splice sites for intron 3, and there is an in-frame canonical pair of the donor and acceptor (GT and AG) splice sites near by. This alternative splicing changes the internal coding region due to an in-frame deletion, leading to the exclusion of sequence coding for a putative protein structure domain – sterile alpha motif (SAM) domain near the N-terminus.
The putative protein structures of both genes were determined using the open reading frame finder program on the National Center for Biotechnology Information (NCBI) website [13], and an open reading frame was identified in the last exon of both genes. The two genes share 78% homology at the DNA sequence level in this exon. The putative protein sequences were analyzed using the Conserved Domain search program in NCBI website [14] for SAMD9, the Motif Scan program from Swiss Institute of Bioinformatics [15-17] and the Predict Protein server from European Molecular Biology Laboratory [18] websites for SAMD9L. Both protein products contain a sterile alpha motif (SAM) domain near the N-terminal region. Otherwise, there is no close homology to other well-characterized proteins or protein motifs. Based on a comparison of the SAM domain sequence of SAMD9 to the homology modeling of the RNA-binding Smg SAM domain [19], the SAM domain of SAMD9 lacks the residues that are essential for binding RNA, while it has 98% sequence homology with the Ephrin-B2-receptor SAM domain, which forms homo-oligomerization and provides a platform for the formation of larger protein complexes [20].
SAMD9 and SAMD9L are ubiquitously expressed
The expected 7 kb transcript of SAMD9 was identified by Northern analysis using RNA from different age placentas and a 1729 bp PCR amplified probe encompassing the 3'UTR (Fig. 2A). The expression pattern of SAMD9 in multiple tissues was investigated using a human multiple tissue cDNA panel (Clontech, USA) and RT-PCR. The primers were designed to recognize sequences at 3'UTR of SAMD9. Expression was detected in all human adult, fetal and tumor tissues that were tested, except for fetal brain. Only a very low signal was detected in skeletal muscle (Fig. 2B). Using the sequences at 3'UTR of SAMD9L as primers, SAMD9L was also found to be expressed in all tissues, except for the tumor types, colon cancer (CX-1), breast cancer (GI-101) and pancreatic cancer (GI-103) (Fig. 2B).
Figure 2.
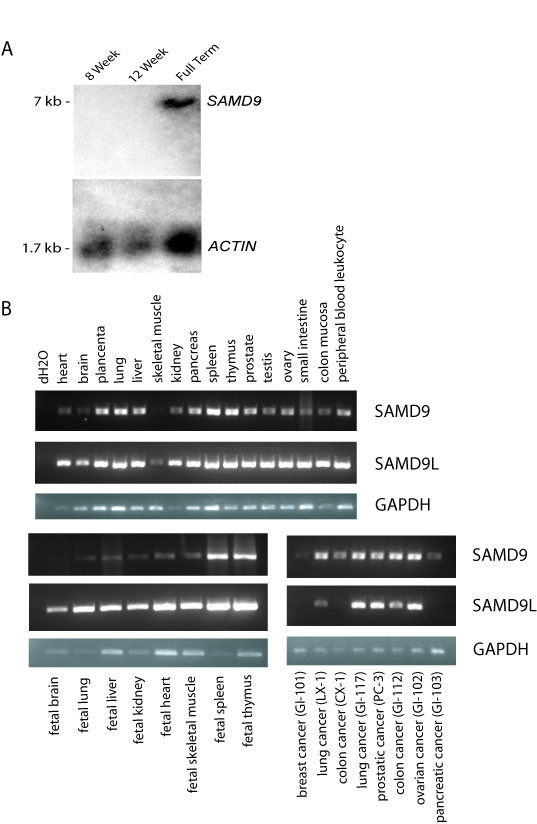
Expression of SAMD9 and SAMD9L in multiple tissue types. A. Northern blot from total RNA of 8 weeks, 12 weeks and full term human placenta probed with 3'UTR of SAMD9. A 7 kb transcript was detected in full term human placenta. B. Human adult, fetal, and tumor multiple tissue cDNA panels were amplified with the sequences at 3'UTR of SAMD9, SAMD9L or GAPDH as primers. SAMD9 was expressed in all human adult, fetal and tumor tissues that were available from human multiple tissue cDNA panels except for fetal brain and breast cancer. It was expressed at a very low level in skeletal muscle and pancreatic cancer. SAMD9L is expressed in all human adult, fetal and tumor tissues that were available from human multiple tissue cDNA panels, except breast cancer, colon cancer, and pancreatic cancer.
SAMD9 and SAMD9L are expressed at lower levels in tumors
To determine the level of expression of SAMD9 in neoplasia, semi-quantitative RT-PCR and real-time quantitative PCR were performed using the sequence at 5'UTR of SAMD9 as a sense primer and a sequence at the beginning of the open reading frame of SAMD9 as an anti-sense primer. Expression was compared between tumor and normal control tissues in cases of aggressive fibromatosis, breast cancer and colon cancer. SAMD9 was expressed at a lower level (about 33% the level for normal control tissues) in aggressive fibromatosis (32.22% ± 29.18% vs 100%, p < 0.05 for real-time PCR, Fig. 3A). Expression of SAMD9 was lower in 20% cases of breast cancer and 35% cases of colon cancer than in the normal control tissues (Fig. 3C). In three cases of colon cancer, there was no RT-PCR product detected. However, there was no significant difference in expression between colon cancers and normal control tissues or breast cancers and normal control tissues when analysed as a group using real-time PCR. Expression studies of SAMD9L were also conducted using a sequence at 5'UTR which is common for all of the splice variants except one, as a sense primer and sequence at the beginning of the open reading frame as an anti-sense primer. There was no statistically significant difference in SAMD9L expression between aggressive fibromatosis or colon cancer and normal control tissues. However, the expression of SAMD9L was lower in breast cancers than in healthy breast epithelial tissues from the same patients (2.81% ± 2.40% vs 100%, n = 10, p < 0.001, Fig. 3B). There was no significant difference in expression of SAMD9L between aggressive fibromatosis samples and normal controls tissues (Fig. 3D). While two genes are expressed at a lower level in tumors, there is a discrepancy between the levels of expression of the two genes in different tumor types. For instance, SAMD9 is expressed at lower levels in aggressive fibromatosis, while SAMD9L is not.
Figure 3.
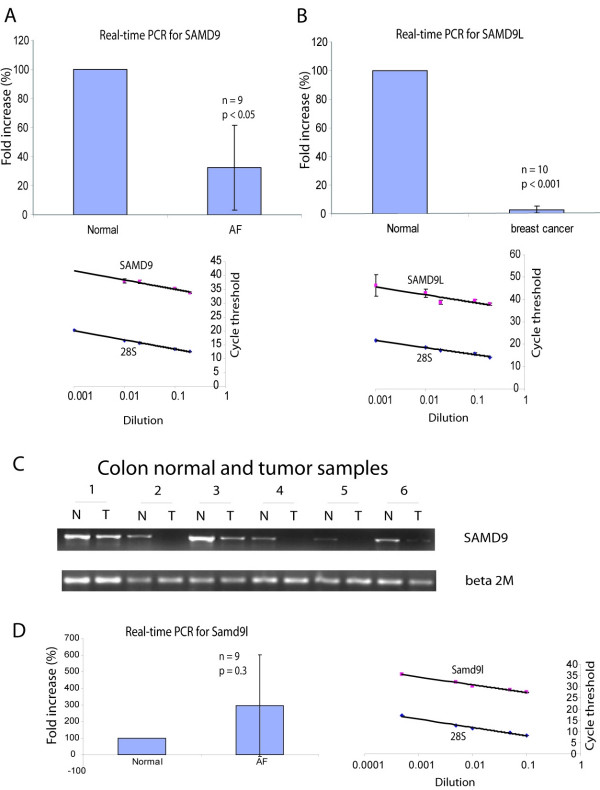
Lower level of expression of SAMD9 and SAMD9L in neoplasia. A. Validation curves for the primer sets of SAMD9 and 28S rRNA, and real time quantitative PCR of SAMD9. SAMD9 and 28S rRNA have similar amplification efficiency since the slope was very similar for both curves. SAMD9 was expressed at a lower level in aggressive fibromatosis (AF) tumors compared to normal fibrous tissues from same patients. B. Validation curves for the primer sets of SAMD9L and 28S rRNA, and real time quantitative PCR of SAMD9L. SAMD9L and 28S rRNA have similar amplification efficiency since the slope was very similar for both curves. SAMD9L was expressed at a lower level in breast cancers compared to normal breast epithelial tissues from same patients.C. Semi-quantitative RT-PCR of SAMD9. SAMD9 was under-expressed in some cases of colon cancer compared to the normal colon mucosa from same patients. Real time PCR data showed no significant difference in expression level between breast and colon cancer and normal controls as a group. D. There was no significant difference in expression of SAMD9L between aggressive fibromatosis samples and normal controls tissues.
SAMD9 is a cytoplasmic protein
In order to investigate the cellular localization of the putative SAMD9 protein, the open reading frame of the SAMD9 gene was cloned, an N-terminal EGFP tag was added and subcloned into a CMV driven vector pLP-EGFP-C1 (Clontech, USA). This vector was transfected into Cos-1 cells, and using an antibody to EGFP, the fusion protein was detected as a 200 kDa band (Fig. 4A). To determine its localization in a variety of cell types, it was transfected into a human fetal fibroblast cell line, MRC-5, and a colon cancer cell line, SW480. The EGFP fusion protein was detected diffusely in the cell cytoplasm (Fig. 4B–F).
Figure 4.
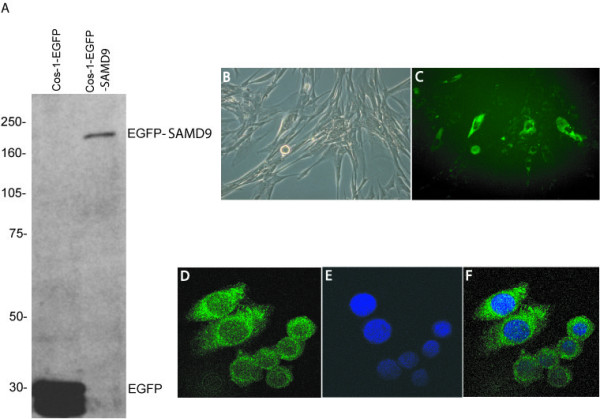
SAMD9 protein is expressed and localized to the cytoplasm. A. Western analysis for the SAMD9-EGFP fusion protein. The SAMD9 fusion protein was detected as a predicted 200 kDa band in Cos-1 cells, using an antibody to GFP. B, C. Expression of SAMD9 putative protein in MRC-5 cells. N-terminal EGFP tagged SAMD9 was detected in the cytoplasm of MRC-5 cells when transiently transfected. B. Light field view. C. Fluorescent microscopy view for FITC. D, E, F. Expression of SAMD9 putative protein in SW480 cells. N-terminal EGFP tagged SAMD9 localized to the cytoplasm of SW480 cells. D. Fusion protein detected under fluorescent wave length for FITC. E. Cell nucleus detected under fluorescent wave length for DAPI. F. Merged view of image D and E.
RNAi for SAMD9 can be used to down regulate its expression
RNA interference was utilized to study the cellular function of SAMD9. RNA interference was designed by searching the public available sequence database to find a 19 bp oligonucleotide sequence specific for SAMD9 and inserting into a pSUPER-RNAi-EGFP expression vector to generate the SAMD9 RNA interference vector (pSUPER-RNAi-SAMD9). One of the sequences is 846 bp after the start of translation, and other is 2371 bp after the start of translation. The pSUPER-RNAi-EGFP expression vector was generated based on pEGFP-C1 (Clontech, USA) and pSUPER [21]. A nucleotide substitution from G to A at the 9th position of this 19 bp oligonucleotide results in abolishing of its specificity to SAMD9 and was used as negative control after subcloned into a pSUPER-RNAi-EGFP expression vector to generate the negative control vector (pSUPER-RNAi-SAMD9-mut9).
SAMD9 expression reduced by RNA interference vector for SAMD9 was tested by co-transfection of an N-terminal EGFP tagged SAMD9 expression vector with either one of the two SAMD9 RNA interference vectors, RNA interference vector with mutated SAMD9 sequence to null its function as negative control, or its control empty vector into Cos-1 cells. The reduction of SAMD9 expression was confirmed using Western blot with anti-GFP (Santa Cruz, USA) for the fusion putative SAMD9 protein. A 90% reduction in expression of SAMD9 fusion protein by RNA interference was detected as measured using densitometry (Fig. 5A). The RNA interference vector that express EGFP was also expressed in SW480 cells along with an N-terminal HA tagged SAMD9 expression vector or its control vector. A 90% reduction in expression of SAMD9 fusion protein by RNA interference was detected (data not shown). Activity of the RNA interference vector for SAMD9 was further verified by the reduction of SAMD9 RNA expression in MRC-5 cells (Fig. 5B).
Figure 5.
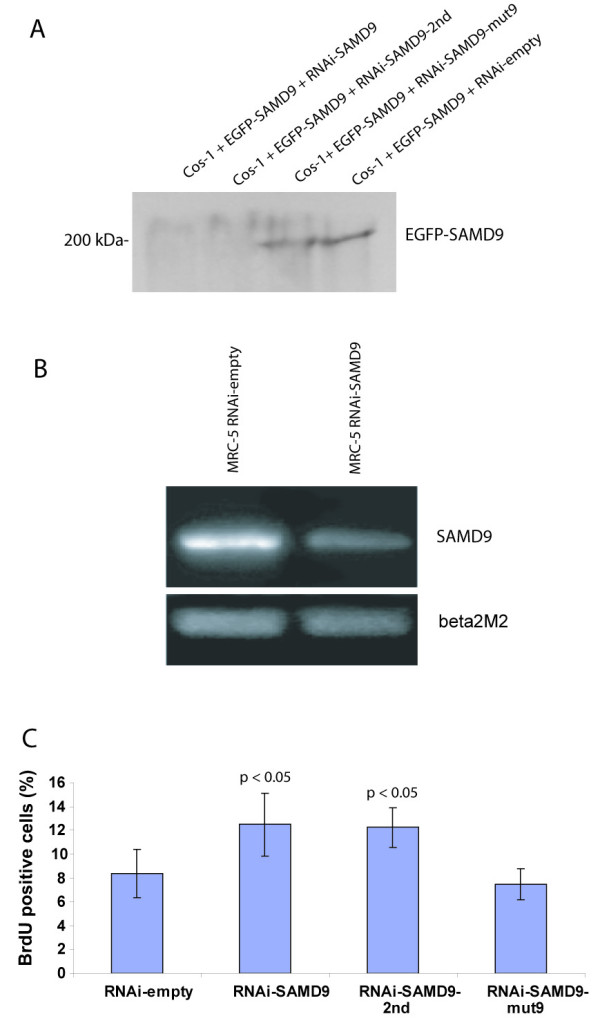
SAMD9 regulates cell proliferation. A. RNAi for SAMD9 is effective in reducing protein expression. A 200 kDa band for the SAMD9 fusion protein was detected in Cos-1 cells after co-transfection of pLP-EGFP-SAMD9 with either pSUPER-RNAi-SAMD9-mut9 or pSUPER-RNAi-EGFP, but it is not detected after co-transfection with either pSUPER-RNAi-SAMD9 or pSUPER-RNAi-SAMD9-2nd. The expression of the EGFP-SAMD9 fusion protein was reduced in Cos-1 cells with RNA interference of SAMD9. B. Semi-quantitative RT-PCR for SAMD9 expression in MRC-5 cells. SAMD9 expression was reduced after RNA interference. C. Proliferation rate (Brdu incorporation percent) increased after RNA interference of SAMD9 in MRC-5 cells.
SAMD9 regulates cell proliferation and apoptosis
Because β-catenin stabilization is known to regulate fibroblast proliferation, motility and invasiveness [11], we studied the effect of down-regulating SAMD9 on these parameters. Proliferation rate, as measured by BrdU incorporation, was increased after RNA interference of SAMD9 in MRC-5 cells (12.5% ± 2.66% with the 1st SAMD9 RNA interference vector, 12.2% ± 1.63% with the 2nd SAMD9 RNA interference vector, and 7.5% ± 1.29% with the control vector, p < 0.01, Fig. 5C). There was not a statistical difference in cell motility as measured by the number of cells passing through the membrane of the culture insert (1.7 ± 0.65 or 2.3 ± 1.31 with RNA interference of SAMD9 vector vs 0.67 ± 0.65 with control vector). Cell invasion was increased, as measured by the number of cells passing through the membrane of the Matrigel invasion chambers, after SAMD9 expression was decreased in MRC-5 cells by RNA interference (16 ± 5.19 with RNA interference of SAMD9 vector vs 5.7 ± 1.73 with control vector, p < 0.05). These studies were also undertaken in Cos-1 cells, and showed a similar reduction in proliferation rate after over-expression of the putative SAMD9 protein in Cos-1 cells (20.7% ± 21.9% vs 76.8% ± 20.2%, p < 0.005). Taken together, this shows that SAMD9 plays a role regulating cell proliferation.
To explore the effect in neoplasia, cells from the SW480 colon cancer cell line were transfected with a SAMD9 expression vector to determine if increasing expression of this gene would suppress the neoplastic behavior in these cells. Upon transfection of the construct, cell proliferation was reduced (25.6% ± 3.3% vs 46.2% ± 9.4%, p < 0.01, Fig. 6A). Cell invasion index calculated as the ratio of the total number of nuclei counted in the membrane for invasion assay to that in the membrane for motility assay was reduced (0.0335 ± 0.0203 vs 0.0779 ± 0.0183, p < 0.05, Fig. 6B), while cell apoptosis, as indirectly measured by relative caspase-3 activity was increased (253.11% ± 45.90% of baseline, p < 0.005, Fig. 6C). The relative caspase-3 activity was also reduced in Cos-1 cells after expression of the SAMD9 siRNA construct (53.76% ± 0.99% of the baseline, p < 0.01). Thus expressing SAMD9 at higher levels causes cell effects which would be expected to suppress the neoplastic phenotype.
Figure 6.
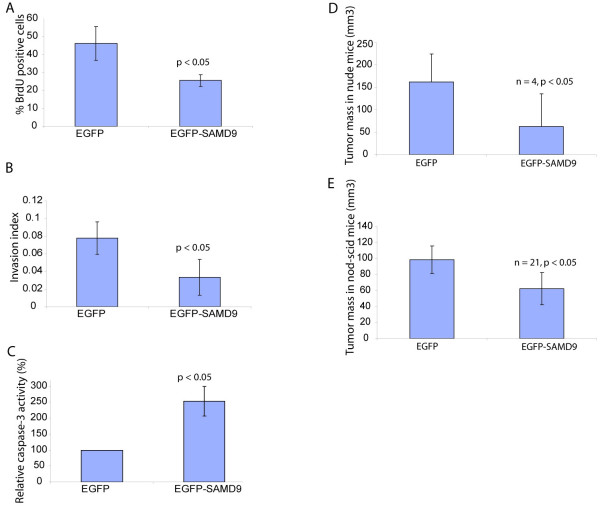
SAMD9 reduces tumor size. A. Increased expression of SAMD9 reduced SW480 cell proliferation rate as detected by Brdu incorporation percent. B. Increased expression of SAMD9 reduced the invasion index of SW480 cells. C. Increased expression of SAMD9 increased caspase activity in SW480 cells. D and E. The tumor volume formed from SW480 xenografts was reduced with increased expression of SAMD9 when transplanted into nude mice (D) or Nod-scid mice (E).
SAMD9 regulates tumor growth
To investigate the role of SAMD9 in tumor growth, the colon cancer cell line, SW480, was transfected with SAMD9 expression vector or its control empty vector. The stably transfected SW480 cells were injected into the immune deficient mice. The tumor volume of the xenograft from SW480 cells over-expressing SAMD9 was reduced in both nude mice (62.5 ± 73.5 mm3 vs 162.5 ± 61.7 mm3, n = 4, p < 0.05, Fig. 6D) and NOD-SCID mice (62.4 ± 20.3 mm3 vs 98.6 ± 17.1 mm3, n = 21, p < 0.01, Fig. 6E). To verify the role of SAMD9 on neoplasia, we performed experiments when the expression of SAMD9 was lowered using RNA interference. The tumor volume of the xenograft from SW480 cells expressing the RNA interference of SAMD9 was increased in nude mice (310 ± 19.6 mm3 vs 270 ± 24 mm3, n = 5, p < 0.05).
Orthologous genes of SAMD9 and SAMD9L
The orthologous genes were identified for SAMD9 and SAMD9L by searching the NR and swissprot databases for best reciprocal matches using human SAMD9 and SAMD9L protein sequences. Genome sequences for all species available were searched by BLAT at the University of California at Santa Cruz genome browser [22]. The results were summarized in Table 1 and the phylogram [see Additional File 1] was constructed using multiple sequence alignment of orthologous genes identified with ClustalW program from European Bioinformatics Institute website [23]. The default parameters from website documentation were used [24]. Based on their sequence similarity, SAMD9 and SAMD9L appear to have originated from a common ancestor by an ancient gene duplication. Since there are orthologous genes of both SAMD9 and SAMD9L in chimpanzees, dogs and rats, but not in chicken, frog or fish species, this duplication event likely occurred after the mammalian radiation. The genomic structures of both SAMD9 and SAMD9L, including the order and orientation of genes within the genomic region and the relative size of the intergenic region, are conserved in all available mammalian genome sequences including opossum, indicating the gene duplication event occurred between 175 to 200 MYA [25]. No evidence for either gene was found in lower eukaryotes such as Drosophila, C. elegans or yeast.
Table 1.
Summary of orthologous genes of SAMD9 and SAMD9L in multiple species
| SAMD9 | SAMD9L | ||
| Species | Accession | Species | Accession |
| Human | GenBank: NP060124.2 | Human | GenBank:NP_689916.1 |
| Chimpanzee | aligns to genomeb | Chimpanzee | GenBank:XP_527818.1 |
| Dog | aligns to genomeb | Dog | GenBank:XP_539422.1 |
| Rat | GenBank:XP_575365.1 | Rat | GenBank:XP_575369.1 |
| Mouse | nonea | Mouse | GenBank:XP_620286.1 |
| Chicken | GenBank:XP_418660.1 | Chicken | nonea |
| Cow | GenBank:XP_592017.1 | Cow | nonea |
| Zebrafish | GenBank:XP_698198.1 | Zebrafish | nonea |
| Xenopus tropicalis | GenBank:AAH91702 | Xenopus tropicalis | nonea |
| Xenopus laevis | GenBank:AAH94169 | Xenopus laevis | nonea |
| Tetraodon nigroviridis | GenBank:CAF92907 | Tetraodon nigroviridis | nonea |
a There is no match in protein or nucleotide database. b There is best reciprocal nucleotide match found in genomic sequence.
SAMD9 is lost in the mouse lineage
There were several overlapping ESTs matching the open reading frame for SAMD9L, and no ESTs corresponding to the open reading frame for SAMD9 in the mouse genome. This was confirmed by bioinformatic searches of all genomic sequences and partial genome sequences (EST libraries). The 168 kb region in human chromosome 7, in which SAMD9 is located, has no match in the mouse genome. A syntenic map of the region was constructed for human, mouse and rat. A break of synteny at the SAMD9 locus was determined in mouse, with distal genes mapping to mouse chromosome 6, and the proximal genes mapping to mouse chromosome 5. The mouse specific rearrangement resulted in new centromere formation at the site of breakage and the subsequent loss of the SAMD9 gene. The SAMD9L region contains numerous gaps in the mouse genome assembly, and an enrichment of mouse specific segmental duplications (Fig. 7, also see Additional file 1). The relatively poor draft quality of the rat genome does not allow for high resolution mapping of this locus, but there is evidence to support the existence of both SAMD9 and SAMD9L genes in rats with no large genomic rearrangements, segmental duplications or breaks in synteny although numerous gaps do exist (Fig. 7, also see Additional file 1). There are no gaps or segmental duplications at this locus in the human genome, determined by searching the Human Genome Segmental Duplication Database.
Figure 7.
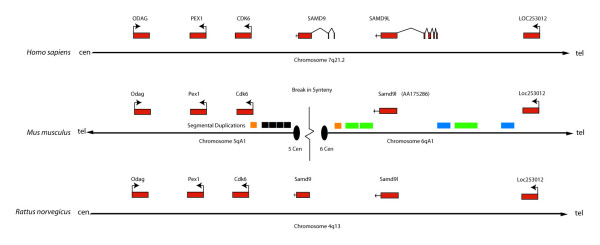
Diagram for the genomic region encompassing the loci of SAMD9 and SAMD9L in human, mouse and rat along with the segmental duplications. Exons are shown as a red box. Segmental duplications are shown as orange, green, blue and grey boxes. Two orange segmental duplications may mediate chromosomal breakage with subsequent deletion of SAMD9, and the centromeres of chromosome 6 and 5 are formed at the sites of breakage. The grey segmental duplications, sharing the sequence among numerous centromeric loci, are accumulated at the centromere of chromosome 5. The green and blue segmental duplications flanking the SAMD9L are mouse specific based on sequence similarity, while the orange and grey segments have occurred at about the same time as the divergence of mouse and rat.
Discussion
Here we identified a novel gene SAMD9 and its paralogue SAMD9L located in human chromosome 7q21.2. These genes are expressed ubiquitously in human tissues. SAMD9 is expressed at a lower level in aggressive fibromatosis and some cases of breast and colon cancer, while SAMD9L is expressed at a lower level in breast cancer, compared to normal control tissues from the same patients. The putative protein of SAMD9 is localized in the cytoplasm. Our in vitro data suggests SAMD9 regulates cell proliferation rate in both normal and tumor cell lines. Over-expression of SAMD9 in SW480 cells reduced the volume of tumors that formed in nude mice and NOD-SCID mice, while the decreased expression in SW480 cells increased the tumor volume formed in nude mice suggesting a role suppressing the neoplastic phenotype.
Very recently, it was found that SAMD9 is mutated in the inherited condition, normophosphatemic familial tumoral calcinosis [26]. In the work of Topaz et al, there is a likely inadvertent error, as when comparing the protein sequences in the publication with the swissProt/NCBI database, it was found that the mouse SAMD9 sequence reported is actually a predicted rat SAMD9 protein sequence [GenBank: XP_575365] and the rat SAMD9 sequence reported is the rat predicted SAMD9L protein sequence [GenBank: XP_001069386]. While conclusions in their manuscript on the species conservation of SAMD9 are incorrect, the occurrence of soft tissue lesions in this inherited condition supports our notion that SAMD9 plays an important role in cell processes that are important in neoplasia.
There are alternatively spliced transcripts in both SAMD9 and SAMD9L. The mechanism for alternative splice of SAMD9 is unusual in that it uses the non-canonical dinucleotides AT and TC as donor and acceptor splice sites for intron 3, and there is an in-frame canonical pair of the donor and acceptor (GT and AG) splice sites near by. Although only the canonical GT-AG, and the non-canonical GC-AG and AT-AC pairs can recruit the splicing machinery effectively, Burset et al suggested that the other non-canonical pairs can function exclusively in association with a canonical pair, which shares its properties with the neighbour, as some kind of parasitic splice sites [27]. This alternative splice results in the in-frame deletion of the sequence coding for the sterile alpha motif (SAM) domain near the N-terminus of SAMD9. Although it is only detected in a quarter of the normal and tumor tissues tested, it has potential to play a dominate-negative role in the functioning of the main common SAMD9 transcript with the SAM domain. The alternative splicing increases the functional complexity of this novel gene, but the function significance of these alternative splicing variants needs further investigation.
While we did not investigate the regulation of expression in SAMD9 and SAMD9L, there are Lef/Tcf binding elements predicted in the genomic sequence upstream of the transcriptional start point for both genes. We also found that the expression of these genes is disregulated in neoplasms driven by β-catenin mediated signaling. Although this suggests SAMD9 may be regulated by β-catenin, formal promoter studies are needed to verify this possibility.
There is an N-terminal sterile alpha motif (SAM) domain in the predicted putative proteins of SAMD9 and SAMD9L. The SAM domain sequence of SAMD9 was compared using the homology modeling of the RNA-binding Smg SAM domain [19]. This showed that the SAM domain lacks the residues that are essential for binding RNA, while it has 98% sequence homology with the Ephrin-B2-receptor SAM domain, which is important in homo-oligomerization and acts as a platform for the formation of larger protein complexes [20]. This raises the possible that SAMD9 may have a similar function participating in protein complexes.
Human chromosome 7q21.2 is located at an evolutionary breakpoint between mouse chromosome 5 and 6, which was localized after human-mouse comparative mapping of the genomic region containing CDK6, i.e., human genes' centromeric to CDK6 map to proximal mouse chromosome 5, while those telomeric to the open reading frame for SAMD9L, map to proximal mouse chromosome 6 [28]. Based on our bioinformatics analysis, there are chimpanzee, dog, and rat orthologous genes for both SAMD9 and SAMD9L. Although there is a mouse ortholog of SAMD9L, SAMD9 is lost in the mouse genome. This is the first report as far as we know about a human gene that exists in rat, but is lost in mouse.
Four human genes, collagen XXI (COL21A1), serine-threonine kinase family member (STK17A), G-protein coupled receptor family member (GPR145) and ras homologue gene family member I (ARH I), located in regions corresponding to evolutionary breakpoints in rodents are absent in both mouse and rat genomes due to an unexpected consequence of evolutionary chromosome rearrangement at the evolutionary breakpoint and lost during evolution [29]. SAMD9 locates in a 168 kb region on human chromosome 7, which has no match in the mouse genome, and at the break in synteny in the mouse genome. But there is no break in synteny at this locus in the rat genome. Mouse-human breakpoint absent in the human-rat comparison suggests the rearrangement is specific to the mouse lineage, i.e. mouse-specific breakpoint. There are no segmental duplications at this locus in human and rat genomes after search of human and rat segmental duplication databases. Fluorescent in situ hybridization (FISH) using a BAC containing mouse EST for SAMD9L (Estm25) revealed a single strong signal at 6A1-A2 in mouse chromosome 6 [28]. Since the sequence of Estm25 maps to the open reading frame of mouse SAMD9L, and it has very high homology with the sequence at the open reading frame of human SAMD9, the FISH result indicates SAMD9 does not exist in mouse.
Here we propose a model, which is mouse specific, for the genomic rearrangement, break in synteny, centromere formation and deletion of SAMD9 although it is very difficult to properly estimate all those events. We highlight a mouse specific rearrangement, resulting in the loss of the SAMD9 gene. Segmental duplications have been shown to mediate chromosomal rearrangements via non-allelic homologous recombination [30], and it has been shown that there is a significant correlation between low copy repeats and break of synteny between human and mouse [31]. Taken together, the existence of the mouse specific rearrangement and mouse specific low copy repeats, it would suggest a causative role, but no direct conclusions can be made. It has also been shown that centromeric regions act as reservoirs for recently duplicated sequence and this may account for the enrichment of low copy repeats at this locus [32]. As shown in Figure 7, in the mouse genome, two segmental duplications, one centromeric to cdk6 and the other centromeric to SAMD9L, may mediate chromosomal breakage with subsequent deletion of SAMD9, and the centromeres of mouse chromosome 6 and 5 are formed at the sites of breakage. There is no evidence of the segmental duplications at this locus in the rat genome after searching the Evan Eichler's rat segmental duplication database [33].
Conclusion
SAMD9 and SAMD9L are expressed ubiquitously in human tissues, but they are expression at lower levels in neoplasia. SAMD9 regulates cell proliferation and apoptosis, and decreases tumor growth of colon cancer cell line in immune deficient mice. This is the first report as far as we know about a human gene that exists in rat, but is lost in mouse, due to a mouse specific rearrangement, resulting in the loss of the SAMD9 gene. Although these novel genes were identify in the study of a relatively rare disorder, aggressive fibromatosis, these genes seem to play a role regulating cell growth in a variety of tumor types.
Methods
Cell lines, human and mouse materials
SW480, MRC-5, Cos-1 and NIH3T3 cell lines were obtained from the American Tissue and Cell Collection. For each experiment in which cell lines were used for a quantitative assay, the experiment was conducted nine independent times, each time on a different day. The RNA populations used for subtractive hybridization were prepared from cells derived from an aggressive fibromatosis tumor with an APC mutation after transfection of wild-type APC or a control vector as previously reported [11]. Cells, tumor and normal control tissues were collected from patients with sporadic aggressive fibromatosis, colon cancer, and breast cancer while the patients were under surgical excision of the tumors. All of the samples were cryopreserved as soon as possible after resection and stored in liquid nitrogen vapor for later nucleic acids extraction. The local ethical approvals for this research were obtained.
Suppression subtractive hybridization
Suppression subtractive hybridization was used to compare the gene expression differences in a cell culture derived from an aggressive fibromatosis tumor with an APC mutation after transfection of a wild type APC to the same cell culture after transfection of a control vector using Clontech PCR-select cDNA subtraction kit (K1804-1, Clontech, CA, USA). Both forward (cDNA from cells transfected with control vector was subtracted by cDNA from cells transfected with wild-type APC) and reverse (cells transfected with wild-type APC was subtracted by cells transfected with control vector) subtractions were conducted. There were eight genes identified in the forward subtraction and 16 genes in the reverse subtraction that were found to be differentially expressed, based on the screening of dot blots containing 196 clones in the forward and reverse subtracted libraries, respectively. SAMD9 was confirmed as being differentially expressed in the reverse subtraction and selected for further analysis in this paper.
Northern analysis
15 μg of total RNA from 8 weeks, 12 weeks and full term human placenta were electrophoresed on 1% RNase-free agarose gel and Northern transferred to a nylon membrane according to according to the method from Sambrook et al [34]. A probe for SAMD9 was generated using PCR with 89729F and 88001R as primers for a 1729 bp fragment from the 3'UTR. The Northern membrane was first probed with the SAMD9, then stripped and probed with the human actin probe as a loading control. The probes were labeled with isotope 32P.
Cloning of the full-length gene
(1) 5'- and 3'- rapid amplification of cDNA ends (RACE)
RACE was performed using SMART™ RACE cDNA synthesis kit (Clontech Inc., USA) according to the manufacture's instruction. The anti-sense (93769R, see Additional file 1) and the sense (93961F, see Additional file 1) primers for the 5'- and 3'- RACE were designed using the sequence of the 200 bp clone of SAMD9 isolated from suppression subtractive hybridization. The amplified transcripts were cloned into a pCR 2.1 vector (Invitrogen, USA), sequenced, and mapped to the DNA sequence in the GenBank.
(2) Long-distance PCR, cloning and sequencing
Sequences corresponding to the 5'- and the 3'- ends of the SAMD9 gene (106430F' and 87926R', see Additional file 1) or SAMD9L gene (136752F' and 119170R', see Additional file 1) were used as primers for long-distance PCR. The amplified transcripts were cloned into a pCR vector (Invitrogen, USA), and the insertion of the transcript was verified by restriction enzyme digestion and sequencing.
PCR with human multiple tissue cDNA panels
Expression of SAMD9 and its paralogous gene SAMD9L were investigated using PCR of human multiple tissue cDNA panels including adult, fetal and tumor tissues (Clontech, USA). The sequences at the 3' UTR of the SAMD9 and SAMD9L genes were used to design the target primers according to the instruction from the manufacture since the DNase pretreated RNA was used for synthesis of those cDNA panels. 88347F and 88001R [see Additional file 1] were primers for SAMD9, while 119622F and 119170R [see Additional file 1] for SAMD9L. GAPDH was used as a control.
Semi-quantitative RT-PCR
The total RNA was extracted from tumor tissue and normal control tissue from aggressive fibromatosis, breast and colon cancers using Trizol RNA reagents (Invitrogen, USA). The anti-sense and sense primers for SAMD9 and human beta-2 microglobulin (beta 2M) were designed using the sequence across the introns, and beta 2M was used as control. For beta 2M, beta 2M-F was used as sense primer and beta 2M-R as anti-sense primer to produce a 112 bp PCR product [see Additional file 1]. For SAMD9, 103270F was used as sense primer and 93871R as anti-sense primer to produce a 711 bp PCR product for SAMD9 with SAM domain coding sequence and a 222 bp PCR product for SAMD9 lacking SAM domain coding sequence [see Additional file 1]. The optimal amplification cycles were set within the linear range, 30 cycles for beta 2M and 37 cycles for SAMD9, after applying series of diluting cDNA samples and subjecting for series of PCR amplification cycles.
The polymerase cycling reactions of paired cDNA samples were conducted at the same time for both target and control genes. The amplified products were electropheresed on a 2% agarose gel and photographed under UV light. The expression of beta 2M was set as control to normalize the level of target expression. The expression level of the target gene SAMD9 was analyzed using computer software ImageQuant and represented as the ratio of its intensity verse the intensity of the beta 2M gene under the condition that the same amount of the cDNA was applied as template for the target gene and the control gene.
Real-time quantitative PCR
Real-time quantitative PCR was undertaken using 28S rRNA (28S) as the control gene. PCR primer pairs for human 28S rRNA were taken from Simpson et al, 2000 [35], for SAMD9, 103270F was used as sense primer and 94424R as anti-sense primer, and for SAMD9L, 133857F was used as sense primer and 124170R as anti-sense primer [see Additional file 1]. Validation curves were carried out for the primer sets using RNA from SW480 cells diluted to 1:5, 1:10, 1:50, 1:100, and 1:1000. Delta delta Ct method was used for setting up the experiment and analysis of the data. An arbitrarily designed threshold was set at 0.2 for all analysis, while the baseline cycles were set for all analysis from 3 to 10 cycles for 28S and from 3 to 30 cycles for SAMD9 or SAMD9L. The threshold cycle, Ct was determined using the analysis software (SDS 2.1, Applied Biosystems). The result was analyzed using the relative quantitative 2-ΔΔ C t method [36]. The expression level of the target gene SAMD9 or SAMD9L in tumor tissues were represented as the fold difference from normal control tissues.
Generation of expression vectors
(1) Long distance PCR amplification of SAMD9 or SAMD9L open reading frame
The sequences flanking the predicted open reading frame of the SAMD9 or SAMD9L gene were used as primers. For SAMD9, 94504F was the sense primer and 89735R was the anti-sense primer, while 124378F as sense primer and 119624R as anti-sense primer for SAMD9L [see Additional file 1]. The predicted open reading frame of the SAMD9 or SAMD9L was subcloned into pDNR-1r donor vector using standard methods [34]. The correct orientation of the insert was confirmed by restriction enzyme digestion and sequencing to generate pDNR-1r-SAMD9-ORF, pDNR-1r-SAMD9L-ORF-major and pDNR-1r-SAMD9L-ORF-minor.
The pDNR-1r-SAMD9-ORF, pDNR-1r-SAMD9L-ORF-major and pDNR-1r-SAMD9L-ORF-minor were subcloned into several acceptor vectors, such as pLP-EGFP-C1 and pLP-CMV-HA (Clontech, USA), using Clontech Creator™ DNA cloning kit (PT3460-2, Clontech, USA) with Cre recombinase through Cre-loxP site-specific recombination to catalyze the transfer of the open reading frame from pDNR-1r donor vector to the acceptor vectors according to the manufacture's instruction.
Western blot analysis with anti-GFP
The total protein from Cos-1 cells transfected with either pLP-EGFP-SAMD9 or its control vector pLP-EGFP were harvested using Nonidet P-40 lysis buffer (1% Nonidet P-40, 50 mM Tris-HCL [pH 7.5], 150 mM NaCl, 2 mM EDTA, 10% glycerol, proteinase inhibitor tablet [1 tablet per 10 mL buffer, Roche, USA]). The protein was electropheresed on 8% sodium dodecyl sulphacrylamide gel and transferred to membrane according to the method from Sambrook et al, 1989. The membrane was then probed with anti-GFP (1:200, Santa Cruz, USA) and secondary antibody (HRP-conjugated monoclonal anti-mouse IgG, 1:2000, Transduction Laboratory, USA). Finally, the membrane was exposed to X-ray film for 20 minutes. The total protein from Cos-1 cells transiently co-transfected with pLP-EGFP-SAMD9 and either one of the two SAMD9 RNA interference vectors (pSUPER-RNAi-SAMD9, pSUPER-RNAi-SAMD9-2nd), RNA interference vector with mutated SAMD9 sequence (pSUPER-RNAi-SAMD9-mut9) to null its function as negative control, or its control empty vector (pSUPER-RNAi-EGFP) were also harvested for Western blot analysis to confirm the knock-down of SAMD9 expression by RNA interference.
Generation of RNA inference vector for SAMD9
After searching of the public available sequence database, a 19 bp oligonucleotide sequence (846 bp after the start of translation) specific for the RNA inference of the SAMD9 expression was found (5'-GTGCATTCGAGAGCCAAGA-3') and subcloned into a pSUPER-RNAi-EGFP expression vector to generate the SAMD9 RNA interference vector (pSUPER-RNAi-SAMD9). A nucleotide substitution from G to A at the 9th position of this 19 bp oligonucleotide results in abolishing of its specificity to SAMD9 to generate the negative control vector (pSUPER-RNAi-SAMD9-mut9). Another 19 bp oligonucleotide sequence (2371 bp after the start of translation) specific for the RNA inference of the SAMD9 expression was found (5'-TACGTACCTGTACTACTCC-3') and also subcloned into a pSUPER-RNAi-EGFP expression vector to generate a second SAMD9 RNA interference vector (pSUPER-RNAi-SAMD9-2nd).
Cell culture, gene transfection, selection, flow cytometry cell sorting, immunoflurescent staining and confocal microscopy
SW480, MRC-5, or Cos-1 cells (ATCC, USA) were cultured in the DMEM medium with 10% fetal calf serum (Invitrogen, USA) in a CO2 incubator at 37°C. Gene transfection was conducted using Fugene 6 reagent (Roche, USA). The transfected cells were either harvested for transient transfection or selected with the addition of 200 ng of G418 (Sigma, USA) for 2 weeks, and sorted by flow cytometry using GFP as marker. The cellular location of SAMD9 was analyzed under a confocal microscope in Cos-1, MRC-5 and SW480 cells transfected with pLP-EGFP-SAMD9. The effectiveness of SAMD9 RNA interference was documented by using confocal microscopy in SW480 cells co-transfected with pLP-CMV-HA-SAMD9 and either the SAMD9 RNA interference vector (pSUPER-RNAi-SAMD9) or its empty control vector (pSUPER-RNAi-EGFP), which were fixed, stained with anti-HA (1:200, Santa Cruz, USA) and Tex-red conjugated secondary antibody (1:100, Molecular Probes, USA).
In vitro assays
(1) Cell proliferation assay
Cells were seeded at the 2.5 × 104 cells/mL on glass cover slips and cultured in DMEM with 10% fetal bovine serum at 37°C. S-phase cells were labelled overnight by supplementation of the cell culture medium with BrdU (Sigma, St louis, MO; 10 μM) and detected by mouse monoclonal anti-BrdU (DAKO, Demark; 1:100) as the published method [37]. After the final step, the glass cover slips were removed from the well and mounted on microscope slides by mounting medium with DAPI to stain the nuclei. The number of S-phase cells and total number of nuclei were counted over five fields at center and four corners on the glass cover slip at 100× magnification. Each experiment was conducted nine times.
(2) Cell motility and invasion assays
Cells were seeded at 5 × 104 cells/mL and cultured with DMEM and 10% fetal bovine serum for 22 hours at 37°C in the BioBoat® culture inserts (BD Bioscience) for motility assay and BioBoat® Matrigel invasion chambers (BD Bioscience) for invasion assay. After the incubation period, cells on the upper surface of the membrane were removed, but cells that migrated to the lower surface of the membrane were fixed to the membrane with 4% paraformaldehyde. The membranes were then excised from the insert and mounted on microscope slides by mounting medium with DAPI to stain the nuclei. The total number of nuclei was counted in each of five fields across the diameter of the member viewed at 400× magnification using UV fluorescence microscopy. The experiments were conducted nine times.
(3) Cell apoptosis assay
Cell apoptosis activity was studied using caspase-3 colorimetric activity assay kit (Chemicon International, USA). The experiments were conducted nine times and relative caspase-3 activity was represented as percentage changes of the treatment group to the control group and calculated by dividing the optical density reading of the treatment group to the control group and multiplying 100.
Xenograft in immune deficient mice
(1) SW480
The flow cytometry sorted SW480 cells transfected with either pLP-EGFP-SAMD9 or its control vector pLP-EGFP were suspended in 1× PBS (120 mM NaCl, 10 mM Na2HPO4, 3 mM KH2PO4, pH 7.3) and injected underneath the skin on the back of nude mice and NOD-SCID mice at a concentration of 2.5 million cells per injection. A similar explant was performed using sorted SW480 cells co-transfected with pLP-CMV-HA-SAMD9 and the SAMD9 RNA interference vector (pSUPER-RNAi-SAMD9) or pLP-CMV-HA-SAMD9 and the empty control vector (pSUPER-RNAi-EGFP). The mice were sacrificed after 1 week of injection and the size of tumors were blindly scored.
(2) MRC-5
The flow cytometry sorted MRC-5 cells transfected with the SAMD9 RNA interference vector (pSUPER-RNAi-SAMD9) or the empty control vector (pSUPER-RNAi-EGFP) were also suspended in 1× PBS and injected underneath the skin on the back of NOD-SCID mice at a concerntration of 5.9 million cells per injection. The mice were sacrificed after 1 week of injection and the size of tumors were blindly scored.
Statistical analysis
The counting process was conducted by a blinded observer. Means and 95% confidence intervals were calculated. The t-test was applied to compare differences.
New sequence accession numbers
GenBank: AF445355, AF474973, AF453311, AY195582-195587, DQ068177.
Authors' contributions
CFL carried out the subtraction hybridization, the initial experiments cloning the genes, generating the expression and siRNA constructs, and drafting the manuscript, JRM carried out the analysis of the genes in different species, and assisted in drafting the manuscript. RYW carried out experiments to determine the expression levels of SAMD9 and SAMD9L in normal and tumor tissues, JR carried out experiments to determine the SAMD9L splice variants, KL carried out gene transfection experiments, CK carried out cell proliferation experiments, RK carried out experiments to detect SAMD9 using Western analysis, SB carried out experiments to detect SAMD9 using Northern analysis, SWS carried out the analysis of the localization on chromosome seven, participated in the design of the study, and assisted with the draft of the manuscript. BAA conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary Results and a summary of Primers used in the cloning of these genes. The data provided represents additional details on the structure and splice variants of SAMD9 and SAMD9L. In addition, there is a table listing the PCR primers used in this work.
Acknowledgments
Acknowledgements
We appreciate the help of Andrew Boright for the bioinformatics analysis. We would like to acknowledge Dr. Irene Andrulis and Dr. Steven Gallinger, The Samuel Lunenfeld Research Institute, University of Toronto for providing us the normal and tumor samples from breast and colon cancer patients; Dr. Reuven Agami, Division of Tumor Biology, The Netherlands Cancer Institute, Amsterdam, Netherlands, for giving us the construct to stable express short interfering RNA in mammalian cells (pSUPER); and Paul Barnfield and Eric Arruda for providing us pSUPER-RNAi-EGFP expression vector. This work was funded by the National Cancer Institute of Canada, the Canadian Institute of Health Research, and the Canadian Research Chairs Program.
Contributor Information
Catherine F Li, Email: catherine.li@utoronto.ca.
Jeffrey R MacDonald, Email: jmacdonald@sickkids.ca.
Robert Y Wei, Email: fibromatosis@yahoo.com.
Jocelyn Ray, Email: jray@mshri.on.ca.
Kimberly Lau, Email: fibromatosis@yahoo.com.
Christopher Kandel, Email: fibromatosis@yahoo.com.
Rachel Koffman, Email: fibromatosis@yahoo.com.
Sherilyn Bell, Email: Sherilyn.Bell@sickkids.ca.
Stephen W Scherer, Email: swScherer@sickkids.ca.
Benjamin A Alman, Email: benjamin.alman@sickkids.ca.
References
- Martin B, Schneider R, Janetzky S, Waibler Z, Pandur P, Kuhl M, Behrens J, von der Mark K, Starzinski-Powitz A, Wixler V. The LIM-only protein FHL2 interacts with beta-catenin and promotes differentiation of mouse myoblasts. J Cell Biol. 2002;159:113–122. doi: 10.1083/jcb.200202075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goruppi S, Chiaruttini C, Ruaro ME, Varnum B, Schneider C. Gas6 induces growth, beta-catenin stabilization, and T-cell factor transcriptional activation in contact-inhibited C57 mammary cells. Mol Cell Biol. 2001;21:902–915. doi: 10.1128/MCB.21.3.902-915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alman BA, Pajerski ME, Diaz-Cano S, Corboy K, Wolfe HJ. Aggressive fibromatosis (desmoid tumor) is a monoclonal disorder. Diagn Mol Pathol. 1997;6:98–101. doi: 10.1097/00019606-199704000-00005. [DOI] [PubMed] [Google Scholar]
- Li C, Bapat B, Alman BA. Adenomatous polyposis coli gene mutation alters proliferation through its beta-catenin-regulatory function in aggressive fibromatosis (desmoid tumor) Am J Pathol. 1998;153:709–714. doi: 10.1016/s0002-9440(10)65614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejpar S, Li C, Yu C, Poon R, Denys H, Sciot R, Van Cutsem E, Cassiman JJ, Alman BA. Tcf-3 expression and beta-catenin mediated transcriptional activation in aggressive fibromatosis (desmoid tumour) Br J Cancer. 2001;85:98–101. doi: 10.1054/bjoc.2001.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon R, Smits R, Li C, Jagmohan-Changur S, Kong M, Cheon S, Yu C, Fodde R, Alman BA. Cyclooxygenase-two (COX-2) modulates proliferation in aggressive fibromatosis (desmoid tumor) Oncogene. 2001;20:451–460. doi: 10.1038/sj.onc.1204107. [DOI] [PubMed] [Google Scholar]
- Tolg C, Poon R, Fodde R, Turley EA, Alman BA. Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor) Oncogene. 2003;22:6873–6882. doi: 10.1038/sj.onc.1206811. [DOI] [PubMed] [Google Scholar]
- Kong Y, Poon R, Nadesan P, Di Muccio T, Fodde R, Khokha R, Alman BA. Matrix metalloproteinase activity modulates tumor size, cell motility, and cell invasiveness in murine aggressive fibromatosis. Cancer Res. 2004;64:5795–5803. doi: 10.1158/0008-5472.CAN-03-3112. [DOI] [PubMed] [Google Scholar]
- Li CF, Kandel C, Baliko F, Nadesan P, Brunner N, Alman BA. Plasminogen activator inhibitor-1 (PAI-1) modifies the formation of aggressive fibromatosis (desmoid tumor) Oncogene. 2005;24:1615–1624. doi: 10.1038/sj.onc.1208193. [DOI] [PubMed] [Google Scholar]
- Alman BA, Li C, Pajerski ME, Diaz-Cano S, Wolfe HJ. Increased beta-catenin protein and somatic APC mutations in sporadic aggressive fibromatoses (desmoid tumors) Am J Pathol. 1997;151:329–334. [PMC free article] [PubMed] [Google Scholar]
- Li C, Bapat B, Alman BA. Adenomatous polyposis coli gene mutation alters proliferation through its beta-catenin-regulatory function in aggressive fibromatosis (desmoid tumor) Am J Pathol. 1998;153:709–714. doi: 10.1016/s0002-9440(10)65614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Sigrist CJA, Le Saux V, Langendijk-Genevaux PS, Bordoli L, Gattiker A, De Castro E, Bucher P, Bairoch A. Recent improvements to the PROSITE database. Nucleic Acids Res. 2004;32:D134–D137. doi: 10.1093/nar/gkh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Applied Bioinformatics. 2002;1:107–108. [PubMed] [Google Scholar]
- Rost B. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol. 2003;10:614–621. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- Thanos CD, Goodwill KE, Bowie JU. Oligomeric structure of the human EphB2 receptor SAM domain. Science. 1999;283:833–836. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- University of California at Santa Cruz genome browser http://genome.ucsc.edu. [DOI] [PMC free article] [PubMed]
- European Bioinformatics Institute website http://www.ebi.ac.uk/clustalw/.
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges B. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Topaz O, Indelman M, Chefetz I, Geiger D, Metzker A, Altschuler Y, Choder M, Bercovich D, Uitto J, Bergman R. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am J Hum Genet. 2006;79:759–764. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Lee-Lin SQ, Green ED. Human-mouse comparative mapping of the genomic region containing CDK6: localization of an evolutionary breakpoint. Mamm Genome. 1999;10:764–767. doi: 10.1007/s003359901088. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J, Bateman JF. Why mice have lost genes for COL21A1, STK17A, GPR145 and AHRI: evidence for gene deletion at evolutionary breakpoints in the rodent lineage. Trends Genet. 2004;20:408–412. doi: 10.1016/j.tig.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet. 2004;Spec No 1:R57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Armengol L, Marques-Bonet T, Cheung J, Khaja R, Gonzalez JR, Scherer SW, Navarro A, Estivill X. Murine segmental duplications are hot spots for chromosome and gene evolution. Genomics. 2005;86:692–700. doi: 10.1016/j.ygeno.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- Tuzun E, Bailey JA, Eichler EE. Recent segmental duplications in the working draft assembly of the brown norway rat. Genome Res. 2004;14:493–506. doi: 10.1101/gr.1907504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor Press; 1989. [Google Scholar]
- Simpson DA, Feeney S, Boyle C, Stitt AW. Retinal VEGF mRNA measured by SYBR green I fluorescence: A versatile approach to quantitative PCR. Mol Vis. 2000;6:178–183. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Tischler AS, Ruzicka LA, Riseberg JC. Immunocytochemical analysis of chromaffin cell proliferation in vitro. J Histochem Cytochem. 1992;40:1043–1045. doi: 10.1177/40.7.1351491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Results and a summary of Primers used in the cloning of these genes. The data provided represents additional details on the structure and splice variants of SAMD9 and SAMD9L. In addition, there is a table listing the PCR primers used in this work.


