Abstract
Objectives: Warfarin is a commonly prescribed anticoagulant drug used to prevent thromboses that may arise as a consequence of orthopedic and vascular surgery or underlying cardiovascular disease. Warfarin is associated with a notoriously narrow therapeutic window where small variations in dosing may result in hemorrhagic or thrombotic complications. To ultimately improve dosing of warfarin, we evaluated models for stable maintenance dose that incorporated both clinical and genetic factors.
Method: A model was constructed by evaluating the contribution to dosing variability of the following clinical factors: age, gender, body surface area, and presence or absence of prosthetic heart valves or diabetes. The model was then sequentially expanded by incorporating polymorphisms of cytochrome P450 (CYP) 2C9; vitamin K 2,3 epoxide reductase complex, subunit 1 (VKORC1); gamma carboxylase; factor VII; and apolipoprotein (Apo) E genes.
Results: Of genetic factors evaluated in the model, CYP2C9 and VKORC1 each contributed substantially to dose variability, and together with clinical factors explained 56% of the individual variability in stable warfarin dose. In contrast, gamma carboxylase, factor VII and Apo E polymorphisms contributed little to dose variability.
Conclusion: The importance of CYP2C9 and VKORC1 to patient-specific dose of warfarin has been confirmed, while polymorphisms of gamma carboxylase, factor VII and Apo E genes did not substantially contribute to predictive models for stable warfarin dose.
Keywords: Bleeding, Coagulation, Vitamin K
Warfarin is an inexpensive, commonly prescribed anticoagulant that inhibits activity of vitamin K dependent blood coagulation factors. Warfarin is prescribed for the treatment of deep venous thrombosis and pulmonary embolism and also prophylactically for prevention of thromboembolic events.1–3
Despite its proven efficacy and low cost, warfarin is underutilized among individuals who could benefit from this therapy due to the high rate of adverse events, the lack of precise warfarin dosing parameters and the need to monitor patients on a regular basis.4–6 Strategies to more reliably achieve optimal anticoagulation and maintain patients in therapeutic range would increase the use of warfarin by practitioners and offer the potential to improve patient safety and reduce adverse events.
Warfarin is a racemic mixture of S-warfarin and R-warfarin. S-warfarin is approximately 5 times more potent than R-warfarin. Metabolism of S-warfarin occurs through the cytochrome P450 (CYP) 2C9 enzyme, while metabolism of R-warfarin occurs through CYP1A2 and CYP3A4. Three prominent genetic variants have been identified in CYP2C9, resulting in reduced enzymatic activity for warfarin metabolism. The *2/*2 homozygous mutant leads to a reduction to approximately 12% of CYP2C9 activity and the *3/*3 homozygous mutant has <5% of wild type CYP2C9 activity.7 Additionally, dosing variability observed among subjects with at least one CYP2C9 *1 allele may be partly attributable to genetic polymorphisms in the C1 subunit of the vitamin K 2,3 epoxide reductase complex (VKORC1).8–13 This enzyme has recently been identified as the rate-limiting step in the warfarin sensitive, vitamin K-dependent gamma carboxylation system.14 Because the initiation of warfarin treatment is problematic and is dependent on complex interactions between physiological, environmental and genetic factors, we previously developed a warfarin-dosing algorithm designed to more accurately initiate warfarin therapy.15 That algorithm predicts an initiation dose that more closely approximates stable dosing requirements by formally incorporating the contributions of genetic variants in CYP2C9. In this study, we extend our earlier model to incorporate variability in stable dose contributed by VKORC1, and we investigate the putative contribution of known polymorphisms in gamma carboxylase, factor VII and apolipoprotein (Apo) E.
Materials and Methods
Participating Subjects
Subjects were seen at Marshfield Clinic, a multi-specialty group practice in Wisconsin. Almost all subjects were Caucasian and most were of German decent.16 Most subjects were recruited from the anticoagulation management service (AMS). Physician-approved protocols were used to provide a standardized approach to patient assessment and warfarin dosing. The AMS staff communicates with the anticoagulation medical director and the patient’s referring physician regarding changes in warfarin dose.
Patients were excluded from this study if they were known to have underlying conditions that influence drug dosage, such as cancer, renal or hepatic insufficiency, or congestive heart failure. Seven patients taking CYP2C9 inducer medications were excluded.
The majority of patients (533/570) were recruited for a prior study of the impact of CYP2C9 genotype on warfarin dosing, and data from 453 of these participants were reported previously.15 Additional patients (37/570) were recruited for a prospective study,17 which evaluated the predictive model developed in the first study in a randomized controlled pilot trial. A subset of the study subjects (501/570) subsequently consented to additional genetic testing to evaluate other promising genetic markers. All study subjects were consented and enrolled according to institutional protocols approved by the Institutional Review Board of Marshfield Clinic. Age, gender, body surface area, clinical attributes such as presenting condition (e.g., valve replacement), presence or absence of diabetes, anticoagulation data and prescription medications were abstracted from the clinical records of patients.
Genetic Testing
The CYP2C9 assay was performed as previously described.15 For VKORC1, we tested single nucleotide polymorphism (SNP) rs8050894 that is described in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/index.html). We performed this testing prior to publications that tested other SNPs in VKORC1. This SNP corresponds to position 6853 of GenBank sequence AY587020 and is a variant that tags the major haplotypes present in VKORC1.12 A validated TaqMan assay for detection of this polymorphism was purchased from Applied Biosystems (Foster City, CA), catalog number C_2847860_10.
Testing of Apo E was performed using TaqMan genotyping assays purchased from Applied Biosystems. These assays tested for the SNPs that encode Arg to Cys amino acid substitutions at position 112 in rs429358 and position 158 in rs7412 (catalog numbers C_3084793_20 and C_904973_10, respectively). Possible haplotypes include E2=Cys(112), Cys(158); E3=Cys(112), Arg(158); E4= Arg(112), Arg(158). These data were summarized for statistical analysis as the number of E4 alleles.
Testing of factor VII was done to determine if variants at positions −402 and −401 of its promoter region affect warfarin dosing.18,19 Since these positions are adjacent to each other, it is difficult to design direct assays for these positions where the testing of one polymorphism would not be hindered by variation at the adjacent polymorphism. To overcome this testing problem, we assayed the SNP at position 1704 rs762635, which is in complete linkage disequilibrium with position 1972 (aka −402) rs570317, and the SNP at position 2456 rs6039 that tags the common haplotypes for position 1973 (aka −401) rs7981123 through linkage disequilibrium with a discordance of 2%.20 Custom designed TaqMan assays were used for these SNP analyses (a TaqMan assay for rs6039 is now commercially available from Applied Biosystems under catalog number C_783182_10). The two SNPs were considered separately as potential predictors of warfarin metabolism for statistical analyses.
To test the microsatellite polymorphism in gamma carboxylase (rs10654848), we performed polymerase chain reaction (PCR) amplification followed by electrophoresis through a 6% acrylamide gel and analysis on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Primers used for amplification were 5′-agtcagctcctcagacaaca and 5′-cagctaggcgtcggtggca. Thermal cycling conditions were 95°C for 5 min, followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 1 min with a final extension at 72°C for 6 min. Allele sizes of 10, 11, 13 and 14 were confirmed by sequencing PCR products and were used as standards for gel electrophoresis. For statistical analysis, repeat size was summarized by the maximum allele size up to 14 with alleles of 10,13 or 11,13 classified as 13 (e.g., 10,10 was classified as 10; and 10,14 was classified as 14).
Statistical Analysis
The clinical parameters considered here were those reported previously:15 age, gender, body surface area, treatment indication (heart valve or not), and presence or absence of diabetes. Most of our subjects had an INR target range of 2 to 3, with the exception of heart valve patients for whom the range is generally 2.5 to 3.5. This association implies that the heart valve parameter in our models is to some degree a surrogate for INR target range.
Because not all study subjects consented to additional genetic testing, and because genetic test results are sometimes inconclusive for any given DNA sample, fewer than 570 observations were available for analyses of genetic data other than CYP2C9. In all analyses, all informative cases were included, and a consistent model fitting strategy was applied. Whenever models involving different predictor variables were compared, the same study subjects were used in each analysis. For example, when comparing a model that included clinical parameters and CYP2C9 to a model with only clinical parameters, all 570 subjects were included, but only 431 subjects with valid VKORC1 results were used to evaluate the effects of adding VKORC1 to the set of clinical and CYP2C9 predictors.
All models were fit with the log of stable warfarin dose as the response. Clinically, it was felt that the effect of warfarin was best reflected by a proportional (multiplicative) scale.21 This implies, for example, that the adverse effects of 15 mg/week in a patient needing 10 mg/week would be equivalent to the adverse effects of 60 mg/week in another patient needing 40 mg/week, since in each case the dose is 50% higher than needed. Statistically, model assumptions of constant variance and normality were observed to be better met under models fit to the log of dose.
Although there is ample evidence in the literature of an important association between CYP2C9 and stable warfarin dose, even this large study did not include a sufficient number of subjects to reliably estimate the independent impact of clinical and genetic predictors of stable dose in those with no wild type CYP2C9 alleles (e.g., no *1 alleles). Therefore, we conducted multiple regression analyses only on the *1 genotypes and used the observed mean dose (log scale) as the predicted value for those with only non-wild CYP2C9 alleles (*2/*2, *2/*3, and *3/*3). Accordingly, our models that include genetic variables are composites of multiple regression and simple mean-based estimation approaches.
We employed a modified step-down approach in developing our regression models by first estimating a “full” model. Full models included all potential clinical and genetic predictors pertinent to that model, plus all two-factor interactions and quadratic terms for age and body surface area. Terms were then cautiously excluded from this full model only if they contributed very little. The regression models were finalized after evaluating residuals and excluding cases with large residuals as outliers (|Studentized Residual| >3). At most, 5 cases (<1% of the cohort) were excluded as outliers in the genetic models.
In order to evaluate potential improvements to dosing based solely on clinical factors like age and body size, a series of genetic models was evaluated. Since subject numbers were too limited to consider all genetic factors simultaneously, models were built sequentially by modeling dose with and without each of the individual genetic terms under consideration. The evidence in the literature is strongest for CYP2C9 and VKORC1; therefore, these were the first factors considered. The explanatory power for competing models was assessed graphically and by using the adjusted R-squared (R2adj) statistic which is based on the squared difference between the observed and predicted dose (log scale). The R2adj statistic measures the proportion of the total variability explained by the model but with adjustment for the number of parameters in the model. The statistic was directly calculated to use all cases under analysis by pooling across both parts of the composite models and by applying the final regression model to any outliers dropped when estimating the parameters in the regression models.
Results
Table 1 ▶ summarizes the basic patient characteristics for the 570 study subjects. The majority were male (56%); 20% had diabetes; and the primary indications for warfarin were arrhythmias (52%) and heart valves (20%). Table 2 ▶ lists the genotype frequencies for the different genes tested in the study.
Table 1.
Characteristics of the 570 study subjects.
| Median (range) | |
| Dose (mg/wk) | 30.0 (4.5 – 122.5) |
| Age (years) | 71.6 (27.9 – 94.1) |
| Body surface area (m2) | 1.97 (1.41 – 2.78) |
| N(%) | |
| Male | 319 (56.0) |
| Diabetes | 113 (19.8) |
| Indication for warfarin | |
| Atrial arrhythmias | 299 (52.5) |
| Prosthetic heart valves | 114 (20.0) |
| Thromboembolic disease | 94 (16.5) |
| Other (arthroplasty, stroke, etc.) | 63 (11.1) |
Table 2.
Overall genotype frequencies.
| Gene | Frequency | Percent |
| CYP2C9 (n=570) | ||
| *1/*1 | 373 | 65.4 |
| *1/*2 | 109 | 19.1 |
| *1/*3 | 65 | 11.4 |
| *2/*2 | 9 | 1.6 |
| *2/*3 | 11 | 1.9 |
| *3/*3 | 3 | 0.5 |
| VKORC1 (n=431) | ||
| CC | 52 | 12.1 |
| CG | 211 | 49.0 |
| GG | 168 | 39.0 |
| ApoE E4 count (n=352) | ||
| 0 | 259 | 73.6 |
| 1 | 83 | 23.6 |
| 2 | 10 | 2.8 |
| Gamma carboxylase CAA repeats (n=331) | ||
| 10 | 82 | 24.8 |
| 11 | 51 | 15.4 |
| 12 | 5 | 1.5 |
| 13 | 122 | 36.9 |
| 14 | 71 | 21.5 |
| Factor VII | ||
| F71704AC-104 (n=387) | ||
| LL | 230 | 59.4 |
| LH | 138 | 35.7 |
| HH | 19 | 4.9 |
| F72456GA-186 (n=387) | ||
| LL | 6 | 1.6 |
| LH | 82 | 21.2 |
| HH | 299 | 77.3 |
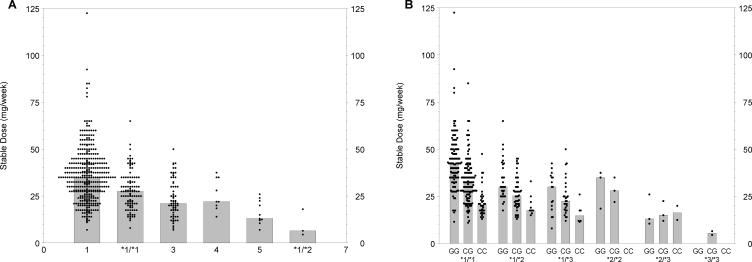
We, and others, have previously demonstrated that CYP2C9 haplotypes significantly contribute to variation in warfarin stable dose,15,22–29 as defined by a therapeutic International Normalized Ratio (INR). Figure 1A ▶ depicts this variability for our study cohort. Individuals who are homozygous wild type (*1/*1) on average require more warfarin than patients with one or more variant alleles, with the median dose for wild type patients at 35 mg/week and the median dose for *3/*3 patients less than one-fifth as high (6.5 mg/week). Within each CYP2C9 group, there remains substantial variation that is unexplained.
Figure 1.
Stable coumadin dose by genotype. Distribution of weekly stabilizing warfarin dose in relation to (A) CYP2C9 and (B) CYP2C9 and VKORC1 genotypes. For (B) CYP2C9 genotype is labeled as *1/*1, *1/*2, *1/*3, *2/*2, *2/*3 and *3/*3. Within each CYP2C9 genotype, the VKORC1 genotype is shown as GG, GC or CC.
Recently, VKORC1 was identified as the gene responsible for vitamin K epoxide reductase. VKORC1 is essential in maintaining sufficient vitamin K levels when dietary vitamin K is limited. Figure 1B ▶ depicts the variability in stable dose for patients grouped by both the three VKORC1 variants (GG, CG and CC) and CYP2C9 genotype. The *1/*1 subjects with variants GG, CG and CC show median doses of 42.5, 32.5 and 20.5, respectively, while *1/*3 subjects with the same variants show median doses of 30.0, 22.5 and 14.75, respectively.
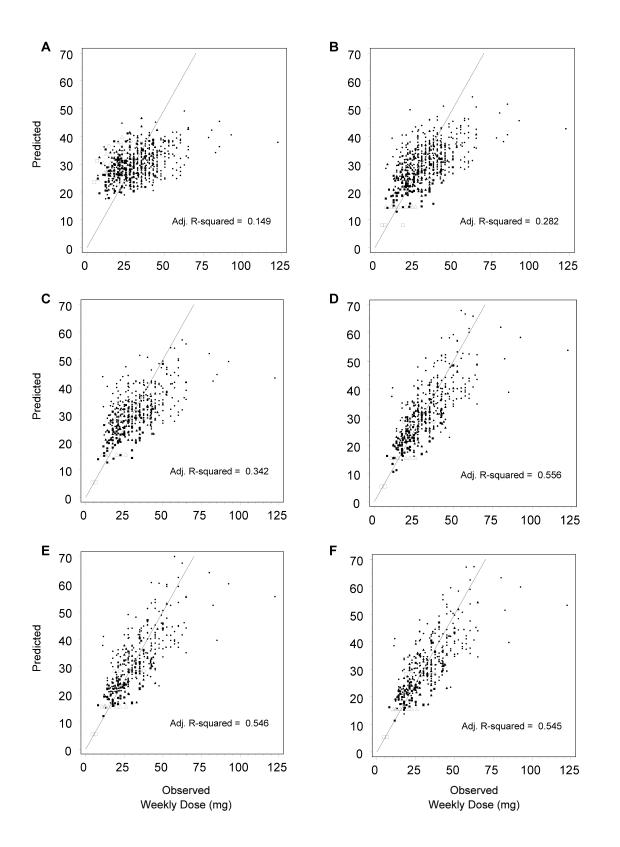
We depict the impact of CYP2C9, VKORC1 and the additional genetic variants in figure 2 ▶. The paired plots in this figure show changes in the relationship between observed stable dose and dose predicted by the model as we serially add each genetic factor. CYP2C9 contributed substantially and significantly (P<0.001) to a predictive model for stable warfarin dose which included only clinical factors (figure 2 ▶, panels A–B), increasing the R2adj from 15% to 28%. VKORC1 contributed substantially and significantly (P<0.001) to a predictive model for stable warfarin dose which included both CYP2C9 and clinical factors (figure 2 ▶, panels C–D), increasing the R2adj from 34% (in the subset with valid VKORC1 results) to 56%. Our results confirm that both VKORC1 and CYP2C9 are important factors associated with stable warfarin dose.
Figure 2.
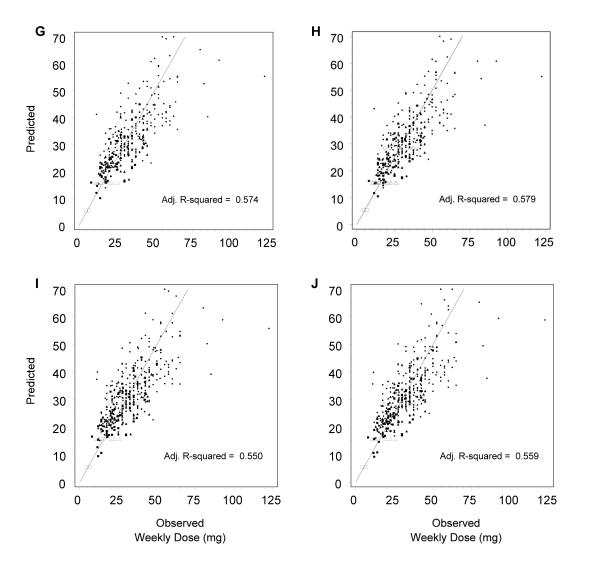
Paired scatter plots of predicted versus observed weekly stable dose. Models involving different genetic variables were compared for the study cohort with complete data. Each pair incorporates another gene: CYP2C9 (A, B), VKORC1 (C, D), Apo E (E, F), gamma carboxylase (G, H) and factor VII (I, J). Note: panels A–F appear on page 12.
In contrast, inspection of the plots and corresponding R2adj statistics show little additional benefit to adding any of the other genetic variants (figure 2 ▶, panels E–J). Therefore, our recommended model includes CYP2C9, VKORC1 and clinical factors (i.e., age, gender, body surface area, and adjustments for patients with diabetes and/or artificial heart valves), and explains about 56% of the variability in therapeutic warfarin dose.
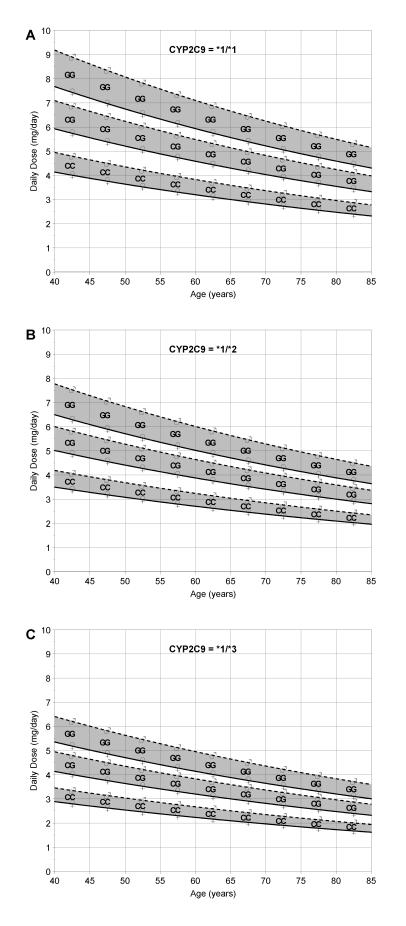
The results from our model could be used to construct visual aids, such as nomograms, for personalized warfarin dosing. Figure 3 ▶ illustrates a series of nomograms for predicting the warfarin dose based on a simplified version of our model which includes the patient’s CYP2C9 and VKORC1 genotype, age and gender. Each nomogram represents a different CYP2C9 genotype with separate results presented for each VKORC1 genotype, specified as homozygous GG, heterozygous CG or homozygous CC. Patient age is referenced on the horizontal scale in the nomograms.
Figure 3.
Nomograms based on CYP2C9 and VKORC1 genotype, age and gender. These nomograms indicate daily dose when the patient’s CYP2C9 and VKORC1 genotype, age and gender are known. Nomograms are provided for (A) CYP2C9 *1/*1, (B) CYP2C9 *1/*2, and (C) CYP2C9 *1/*3. Patient age is specified on the horizontal axis and predicted dose is specified on the vertical axis. Dose is calculated by selecting the correct nomogram based on CYP2C9 genotype, locating the patient age across the bottom, identifying the correct predictive line based on VKORC1 genotype and gender, and reading the predicted dose from the left axis.
Discussion
We have confirmed that individual variation in stable warfarin dose is related to CYP2C9 and VKORC1 genotypes in addition to clinical factors. In contrast, our results do not support a substantial role (which we would define as explaining at least 5% of the remaining variability) for the polymorphisms in factor VII, gamma carboxylase or Apo E as determinants of stable warfarin dose. Polymorphisms in these genes have been previously suggested as contributors to stable dose variation.18,19,30–35 However, in each of the previous publications, the individual genes accounted for a very small amount (<5%) of the predictive value for warfarin dose. Given the low explanatory value of these genes and differences between our patient population and the Japanese, Italian and Swedish populations used in the previous studies, it is not surprising that these variants do not contribute to our model. Consistent with our results, the possible effect of Apo E was not reproduced in an Italian population36 suggesting that if there is an effect of Apo E on stable warfarin dose, it may be population specific.
Our model explains about 56% of the variation in stable dose. We acknowledge that this model requires validation in other populations. We expect some reduction in predictive ability, even in our own population, for cases not used in the model development. We also note a relatively small number of our current patients for whom dose is not well predicted by the model. It is possible that these patients carry additional genetic mutations in genes involved in the coagulation pathway. We fully expect that the model will continue to evolve as we discover new factors that contribute to variability in dosing and acquire additional data from cases with rare genotypes.
We have illustrated dosing nomograms that are simple to use but allow for a more accurate and personalized approach to warfarin dosing. These nomograms graphically display and highlight the wide variation in warfarin stable dose, and allow physicians the ability to visually determine a more precise dose when genetic, gender and age parameters are known. Our ultimate goal is to develop a computer-based application that will be both easy to use and easy to update as the model is improved.
As methods for personalized dosing of warfarin are improved, it is likely that physician comfort with this drug will increase. Although the use of genetic testing and dosing algorithms is currently feasible in the United States, these tools are not currently available to clinical practice. Two studies have demonstrated the feasibility of genetic testing prior to warfarin dosing.17,37 Both studies concluded that genetic testing is technically feasible, acceptable to the patient and physician, and likely to improve the clinical management of anticoagulation.
Acknowledgments
We are grateful to the subjects who participated in this research. We thank Jane Carl for assistance with subject recruitment and follow-up and Carla Rottscheit for assistance with database maintenance and quality assurance. The assistance of Alice Stargardt and Linda Weis in preparation of the manuscript is also highly appreciated.
Grant Support: This study was supported by Disease Specific Restricted Funds donated to Marshfield Clinic Research Foundation.
References
- 1.Hardwick ME, Colwell CW Jr. Advances in DVT prophylaxis and management in major orthopaedic surgery. Surg Technol Int 2004;12:265–268. [PubMed] [Google Scholar]
- 2.Wittkowsky AK. Effective anticoagulation therapy: defining the gap between clinical studies and clinical practice. Am J Manag Care 2004;10(10 suppl):S297–306. [PubMed] [Google Scholar]
- 3.Gutt CN, Oniu T, Wolkener F, Mehrabi A, Mistry S, Buchler MW. Prophylaxis and treatment of deep vein thrombosis in general surgery. Am J Surg 2005;189:14–22. [DOI] [PubMed] [Google Scholar]
- 4.Evans A, Davis S, Kilpatrick C, Gerraty R, Campbell D, Greenberg P. The morbidity related to atrial fibrillation at a tertiary centre in one year: 9.0% of all strokes are potentially preventable. J Clin Neurosci 2002;9:268–272. [DOI] [PubMed] [Google Scholar]
- 5.Caro JJ. An economic model of stroke in atrial fibrillation: the cost of suboptimal oral anticoagulation. Am J Manag Care 2004;10(14 suppl):S451–458. [PubMed] [Google Scholar]
- 6.Miller PS, Andersson FL, Kalra L. Are cost benefits of anticoagulation for stroke prevention in atrial fibrillation underestimated? Stroke 2005;36:360–366. [DOI] [PubMed] [Google Scholar]
- 7.Redman AR. Implications of cytochrome P450 2C9 polymorphism on warfarin metabolism and dosing. Pharmacotherapy 2001;21:235–242. [DOI] [PubMed] [Google Scholar]
- 8.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz HJ, Lappegard K, Seifried E, Scharrer I, Tuddenham EG, Muller CR, Strom TM, Oldenburg J. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004;427:537–541. [DOI] [PubMed] [Google Scholar]
- 9.Bodin L, Verstuyft C, Tregouet DA, Robert A, Dubert L, Funck-Brentano C, Jaillon P, Beaune P, Laurent-Puig P, Becquemont L, Loriot MA. Cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood 2005;106:135–140. [DOI] [PubMed] [Google Scholar]
- 10.D’Andrea G, D’Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V, Grandone E, Margaglione M. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood 2005;105:645–649. [DOI] [PubMed] [Google Scholar]
- 11.Harrington DJ, Underwood S, Morse C, Shearer MJ, Tuddenham EG, Mumford AD. Pharmacodynamic resistance to warfarin associated with a Val66Met substitution in vitamin K epoxide reductase complex subunit 1. Thromb Haemost 2005;93:23–26. [DOI] [PubMed] [Google Scholar]
- 12.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, Blough DK, Thummel KE, Veenstra DL, Rettie AE. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 2005;352:2285–2293. [DOI] [PubMed] [Google Scholar]
- 13.Yuan HY, Chen JJ, Lee MT, Wung JC, Chen YF, Charng MJ, Lu MJ, Hung CR, Wei CY, Chen CH, Wu JY, Chen YT. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet 2005;14:1745–1751. [DOI] [PubMed] [Google Scholar]
- 14.Wajih N, Sane DC, Hutson SM, Wallin R. The inhibitory effect of calumenin on the vitamin K-dependent gamma-carboxylation system. Characterization of the system in normal and warfarin-resistant rats. J Biol Chem 2004;279:25276–25283. [DOI] [PubMed] [Google Scholar]
- 15.Hillman MA, Wilke RA, Caldwell MD, Berg RL, Glurich I., Burmester JK. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics 2004;14:539–547. [DOI] [PubMed] [Google Scholar]
- 16.McCarty CA, Wilke RA, Giampietro PF, Wesbrook SD, Caldwell MD. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Medicine 2005;2:49–79. [DOI] [PubMed] [Google Scholar]
- 17.Hillman MA, Wilke RA, Yale SH, Vidaillet HJ, Caldwell MD, Glurich I, Berg RL, Schmelzer J, Burmester JK. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clin Med Res 2005;3:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Ambrosio RL, D’Andrea G, Cappucci F, Chetta M, Di Perna P, Brancaccio V, Grandone E, Margaglione M. Polymorphisms in factor II and factor VII genes modulate oral anticoagulation with warfarin. Haematologica 2004;89:1510–1516. [PubMed] [Google Scholar]
- 19.Shikata E, Ieiri I, Ishiguro S, Aono H, Inoue K, Koide T, Ohgi S, Otsubo K. Association of pharmacokinetic (CYP2C9) and pharmacodynamic (factors II, VII, IX, and X; proteins S and C; and gamma-glutamyl carboxylase) gene variants with warfarin sensitivity. Blood 2004;103:2630–2635. [DOI] [PubMed] [Google Scholar]
- 20.SeattleSNPs. Output from PHASE v2.0. NHLBI Program for Genomic Applications, SeattleSNPs, Seattle, WA; 2006 (http://pga.gs.washington.edu/data/f7/fct07.phase.out) Accessed April 2006.
- 21.Keene ON. The log transformation is special. Stat Med 1995;14:811–819. [DOI] [PubMed] [Google Scholar]
- 22.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics 1994;4:39–42. [DOI] [PubMed] [Google Scholar]
- 23.Steward DJ, Haining RL, Henne KR, Davis G, Rushmore TH, Trager WF, Rettie AE. Genetic association between sensitivity to warfarin and expression of CYP2C9*3. Pharmacogenetics 1997;7:361–367. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki H, Inoue K, Shimada T. Roles of two allelic variants (Arg144Cys and Ile359Leu) of cytochrome P4502C9 in the oxidation of tolbutamide and warfarin by human liver microsomes. Xenobiotica 1998;28:103–115. [DOI] [PubMed] [Google Scholar]
- 25.Daly AK, Day CP, Aithal GP. CYP2C9 polymorphism and warfarin dose requirements. Br J Clin Pharmacol 2002;53:408–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanuh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002;287:1690–1698. [DOI] [PubMed] [Google Scholar]
- 27.Peyvandi F, Spreafico M, Siboni SM, Moia M, Mannucci PM. CYP2C9 genotypes and dose requirements during the induction phase of oral anticoagulant therapy. Clin Pharmacol Ther 2004;75:198–203. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med 2005;7:97–104. [DOI] [PubMed] [Google Scholar]
- 29.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, Wood P, Kesteven P, Daly AK, Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood 2005;106:2329–2333. [DOI] [PubMed] [Google Scholar]
- 30.Kohnke H, Sorlin K, Granath G, Wadelius M. Warfarin dose related to apolipoprotein E (APOE) genotype. Eur J Clin Pharmacol 2005;61:381–388. [DOI] [PubMed] [Google Scholar]
- 31.Wadelius M, Chen LY, Downes K, Ghori J, Hunt S, Eriksson N, Wallerman O, Melhus H, Wadelius C, Bentley D, Deloukas P. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J 2005;5:262–270. [DOI] [PubMed] [Google Scholar]
- 32.Herman D, Peternel P, Stegnar M, Breskvar K, Dolzan V. The influence of sequence variations in factor VII, gamma-glutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Thromb Haemost 2006;95:782–787. [PubMed] [Google Scholar]
- 33.Chen LY, Eriksson N, Gwilliam R, Bentley D, Deloukas P, Wadelius M. Gamma-glutamyl carboxylase (GGCX) microsatellite and warfarin dosing. Blood 2005;106:3673–3674. [DOI] [PubMed] [Google Scholar]
- 34.Sconce EA, Daly AK, Khan TI, Wynne HA, Kamali F. APOE genotype makes a small contribution to warfarin dose requirements. Pharmacogenet Genomics 2006;16:609–611. [DOI] [PubMed] [Google Scholar]
- 35.Herman D, Peternel P, Stegnar M, Breskvar K, Dolzan V. The influence of sequence variations in factor VII, gamma-glutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Thromb Haemost 2006;95:782–787. [PubMed] [Google Scholar]
- 36.Kohnke H, Scordo MG, Pengo V, Padrini R, Wadelius M. Apolipoprotein E (APOE) and warfarin dosing in an Italian population. Eur J Clin Pharmacol 2005;61:781–783. [DOI] [PubMed] [Google Scholar]
- 37.Voora D, Eby C, Linder MW, Milligan PE, Bukaveckas BL, McLeod HL, Maloney W, Clohisy J, Burnett RS, Grosso L, Gatchel SK, Gage BF. Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost 2005;93:700–705. [DOI] [PubMed] [Google Scholar]