Abstract
Olfactory receptor (OR) genes are of vital importance for animals to find food, identify mates, and avoid dangers. In mammals, the number of OR genes is large and varies extensively among different orders, whereas, in insects, the extent of interspecific variation appears to be small, although only a few species have been studied. To understand the evolutionary changes of OR genes, we identified all OR genes from 12 Drosophila species, of which the evolutionary time is roughly equivalent to that of eutherian mammals. The results showed that all species examined have similar numbers (≈60) of functional OR genes. Phylogenetic analysis indicated that the ancestral species also had similar numbers of genes, but there were frequent gains and losses of genes that occurred in each evolutionary lineage. It appears that tandem duplication and random inactivation of duplicate genes are the major factors of gene number change. However, chromosomal rearrangements have contributed to the establishment of genome-wide distribution of OR genes. These results suggest that the repertoire of OR genes in Drosophila has been quite stable compared with the mammalian genes. The difference in evolutionary pattern between Drosophila and mammals can be explained partly by the differences of gene expression mechanisms and partly by the environmental and behavioral differences.
Keywords: birth-and-death evolution, insect evolution, multigene family
Olfactory receptor (OR) genes form one of the largest multigene families in animals, and the number of genes varies extensively among different mammalian orders (≈400–1,200 genes) (1, 2). Insects also have many OR genes, but these genes are remotely related to vertebrate OR genes, and there is virtually no sequence similarity between them (3). In addition to the extensive sequence divergence, there is a structural difference between insect and vertebrate OR genes. Both of the genes belong to the G protein-coupled receptor gene superfamily, but insect OR genes contain introns (4) whereas vertebrate OR genes have no introns in the protein-coding region (5). OR genes have been studied in a few insect species, and it has been reported that fruit flies (4), mosquitoes (6), and honey bees (7) have ≈60, ≈80, and ≈160 genes, respectively. This suggests that variation in the number of OR genes is smaller in insects than in mammals. However, to understand the evolutionary dynamics of OR genes in insects, we need more information about the gene repertoire of closely related species.
Fortunately, draft genome sequences of 12 Drosophila species have been released from the Assembly/Alignment/Annotation (AAA) database. The 12 species are D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. virilis, D. mojavensis, and D. grimshawi (the genus name will be omitted in the following). Molecular data have suggested that these species evolved and diverged during the last 63 million years (MY) (8), which is somewhat lower than but similar to the divergence times of eutherian mammals (<100 MY) (9). This allows us to compare the evolutionary dynamics of OR genes of Drosophila and mammals. We have therefore conducted an evolutionary study of OR genes from these 12 Drosophila species. The results obtained are presented in this article.
Results
Numbers of OR Genes in Drosophila Species.
Our homology search (see Materials and Methods) detected 711 functional, 67 nonfunctional, and 34 partial OR genes in the genomes of 12 Drosophila species. All species examined have similar numbers of functional OR genes and much smaller numbers of pseudogenes (Table 1). Although simulans, sechellia, persimilis, and virilis show somewhat smaller numbers of functional genes, they have larger numbers of pseudogenes and partial genes. Here a partial gene refers to a gene with an open reading frame (ORF) truncated at the end of a genomic contig studied. This gene may therefore become a functional gene when the entire genomic sequence is assembled. Some pseudogenes might also become functional genes later because the nonsense or frameshift mutations identified could be caused by sequencing errors. For these reasons, our estimates of functional genes are likely to be minimums except in melanogaster, where the genome sequence is well established. In particular, the Hawaiian fruit fly grimshawi, which has many pseudogenes and partial genes, may turn out to have the largest number of functional genes. In this connection, it should be mentioned that the gene OR83b (OR49 in our notation) is known to be coexpressed with another OR gene in most olfactory receptor neurons (ORNs) (10, 11) and is highly conserved even among different orders of insects (12). Yet, this gene in simulans was judged as a pseudogene because it contained a stop codon. However, if we consider the functional importance of this gene, the stop codon is likely to have occurred because of sequencing errors. We have therefore decided to regard it as a functional gene in this paper. Furthermore, the previous study showed that melanogaster has 62 functional OR genes (4), but we regarded one of them (OR85e or OR55) as a pseudogene because of large deletions.
Table 1.
Numbers of OR genes in the 12 Drosophila species
| Subgenus | Species | Funct | Pseudo | Partial | Total |
|---|---|---|---|---|---|
| Sophophora | D. melanogaster | 61 | 1 | 0 | 62 |
| D. simulans | 52 | 4 | 10 | 66 | |
| D. sechellia | 54 | 6 | 2 | 62 | |
| D. yakuba | 63 | 0 | 1 | 64 | |
| D. erecta | 61 | 1 | 1 | 63 | |
| D. ananassae | 66 | 4 | 3 | 73 | |
| D. pseudoobscura | 64 | 8 | 0 | 72 | |
| D. persimilis | 52 | 12 | 3 | 67 | |
| D. willistoni | 65 | 7 | 3 | 75 | |
| Drosophila | D. virilis | 53 | 8 | 1 | 62 |
| D. mojavensis | 59 | 4 | 0 | 63 | |
| D. grimshawi | 61 | 12 | 10 | 83 |
Funct, functional genes; Pseudo, pseudogenes; Partial, partial genes.
Chromosomal Locations of OR Genes and Their Phylogenetic Relationships.
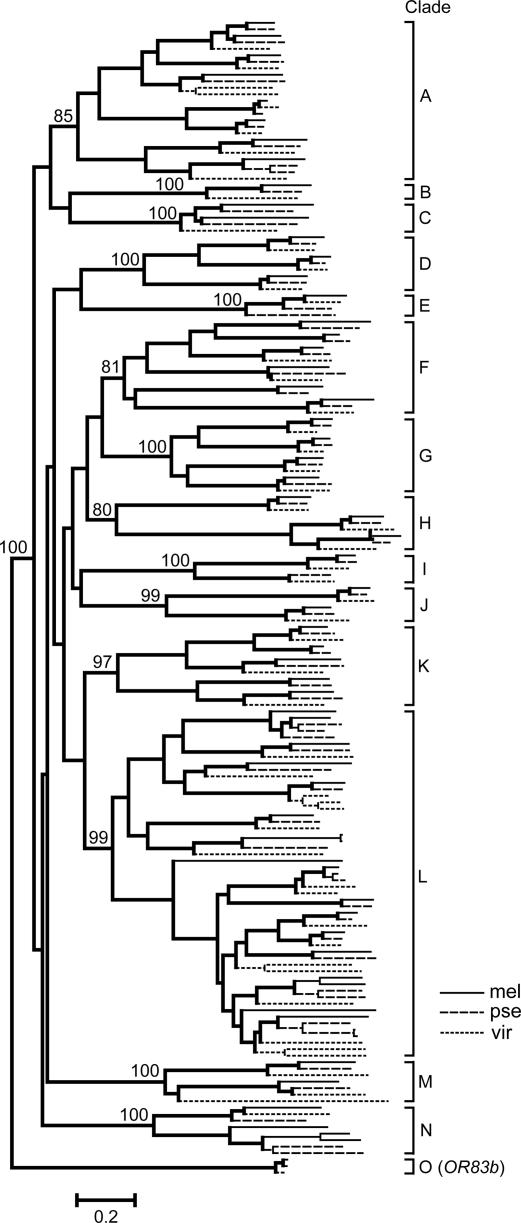
Fig. 1 shows the chromosomal locations of OR genes in melanogaster, yakuba, and pseudoobscura, whose genome sequences are better assembled than others. OR genes are numbered from the left-hand side of the genome to the right-hand side in each species. In all these species, OR genes are widely distributed in the genome except for the dot chromosome (chromosome 4 in melanogaster and yakuba, and chromosome 5 in pseudoobscura), where no OR gene was observed. Phylogenetic analysis of these OR genes revealed their orthologous and paralogous relationships among the three species (see Fig. 2 for a segment of the phylogenetic tree). The orthologous genes between melanogaster and yakuba are arranged in the genome in essentially the same order. The genomic arrangement of OR genes in pseudoobscura was quite different from that in the other two species apparently because of the gene rearrangements that occurred in the past. However, Fig. 2 indicates that pseudoobscura almost always has a gene orthologous to the OR genes from the other two species.
Fig. 1.
Chromosomal locations of OR genes and their orthologous and paralogous relationships in melanogaster (mel), yakuba (yak), and pseudoobscura (pse). Long rods show functional OR genes, whereas short rods indicate either pseudogenes or partial genes. Rods above and below the lines indicate that the OR genes are located on opposite strands. Broken and solid lines show orthologous and paralogous relationships of OR genes, respectively. Open triangles and rectangles indicate the genes whose orthologous genes were lost after the divergence of melanogaster and yakuba and after the divergence of yakuba and pseudoobscura, respectively. Stars indicate the position of OR83b (OR49) orthologs. Filled circles represent the centromeres. L and R indicate the left and right arms of chromosomes, respectively. The subscript r indicates that the chromosomal location of the sequences remains undetermined. In melanogaster and yakuba, the genes are numbered from chromosomes X to 3R, and the numbers in melanogaster are shown above the figure. In pseudoobscura, the genes are arranged from chromosomes XL to 2, because melanogaster chromosomes X, 2L, 2R, 3L, and 3R correspond to pseudoobscura chromosomes XL, 4, 3, XR, and 2, respectively (13). Species relationship is shown in the left-hand side. Three OR pseudogenes whose chromosomal locations are unidentified in pseudoobscura are not shown.
Fig. 2.
Partial segment of the neighbor-joining (NJ) tree (14) of functional OR genes from melanogaster (m), yakuba (y), and pseudoobscura (p) (236 aa used). In each species, the genes are named as mentioned in Fig. 1. Filled circles indicate that the interior branches are supported by ≥80% bootstrap value. The NJ tree was constructed by using the complete deletion option of MEGA3.1 (15) after sequence alignment with CLUSTAL W (16).
Comparison of the melanogaster and yakuba genomic sequences shows that there are two lost genes in melanogaster (corresponding to the open triangles in the yakuba sequence) and one lost gene in yakuba. One melanogaster pseudogene OR55 is apparently the ortholog of yakuba gene OR55 and is likely to be one of the two lost genes because its adjacent OR genes showed 1:1 orthologous relationships with the same gene order. In addition, the melanogaster genome contains one set of duplicate genes (solid lines) that apparently occurred after divergence of the two species. Similarly, the yakuba genome has two sets of duplicate genes. In other words, it appears that melanogaster acquired one gene and lost two genes whereas yakuba acquired two genes and lost one gene after their divergence. Comparison of the yakuba and pseudoobscura sequences suggests that yakuba acquired four genes and lost two genes (open rectangles) whereas pseudoobscura acquired seven genes and lost four genes after their divergence (Fig. 1). However, these estimates may not be reliable, because pseudoobscura and yakuba (or melanogaster) diverged ≈55 million years ago (MYA) (8). Therefore, we used different methods to estimate the numbers of gene gains and losses as shown in the next section.
The genomic maps presented in Fig. 1 are also useful for identifying chromosomal inversion and translocation events. Genes OR61 and OR62 in melanogaster are inverted relative to their orthologs of yakuba, and they are located on the DNA strand opposite to that of yakuba. In addition, some genes (e.g., OR24 to OR27) moved from one chromosome (2R) arm to another (2L) or vice versa. Multiple events of inversion and translocation apparently occurred between yakuba and pseudoobscura, so that it is difficult to infer every evolutionary event.
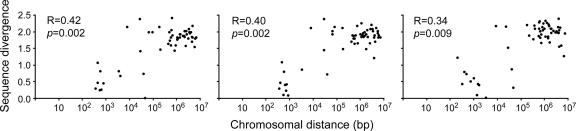
We noticed that 7 of the 12 duplication events observed in the three species occurred by tandem duplication (<10 kb apart on the same strand). This can also be seen in Fig. 2, which shows that the genes contiguously located on the chromosomes are phylogenetically very close. Fig. 3 shows the relationships between the genomic distance between two consecutive OR genes and their phylogenetic distance (Poisson-correction distances of amino acid sequences) within chromosomes. It is clear that there is a high correlation between the two quantities, suggesting that duplicate genes eventually spread through the entire chromosome by repeated chromosomal rearrangements.
Fig. 3.
Relationships between sequence divergence (Poisson-correction distances) and chromosomal distance of two consecutive functional OR genes for melanogaster (Left), yakuba (Center), and pseudoobscura (Right). Chromosomal distances are shown in logarithm. R and p indicate the correlation coefficient and its significance level, respectively.
Fig. 4 shows the NJ tree of functional OR genes from melanogaster, pseudoobscura (subgenus Sophophora), and virilis (subgenus Drosophila). The melanogaster gene OR83b (OR49) and its orthologs from different species were used as outgroups, because the gene is known to have diverged from other OR genes a long time ago (4) and have a function different from other OR genes (10, 11). OR genes of each species do not form a species-specific clade but are scattered throughout the tree. We classified OR genes into 15 phylogenetic clades (A–O), each of which was defined as the largest cluster of similar genes supported by a bootstrap value of ≥80%. These clades remained unchanged even in the tree constructed for all functional OR genes from the 12 Drosophila species [supporting information (SI) Fig. 7]. Because all these clades contained OR genes from the Sophophora and Drosophila species, they must have existed in the most recent common ancestor (MRCA) of these subgenera.
Fig. 4.
NJ tree of functional OR genes from melanogaster (mel), pseudoobscura (pse), and virilis (vir) (264 aa used). Bootstrap values are shown only for the clades, which were supported by ≥80% bootstrap values. Note that most of the interior branches in each clade are supported by ≥80% bootstrap values.
The numbers of genes for the 15 phylogenetic clades in each of the 12 Drosophila species are presented in Table 2. The number of OR genes varies considerably among these clades, clade L having the largest number of genes. In all species, clade O has only one gene, which is orthologous to OR83b (OR49) in melanogaster. The genes belonging to this clade are distantly related to other OR genes (Fig. 4). There are some other clades, which contain only one or two genes, but it is uncertain whether they have any distinct function or not. In this study, OR gene groups are defined by phylogenetic relationships only, and therefore the classification may not be related to gene function. Yet, it is interesting to note that the number of genes in each clade is more or less the same for all species.
Table 2.
Numbers of functional OR genes for 15 clades in each Drosophila species
| Species* | Clade |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O† | |
| mel | 8 | 1 | 2 | 3 | 1 | 6 | 4 | 3 | 1 | 2 | 5 | 18 | 2 | 4 | 1 |
| sim | 8 | 1 | 2 | 3 | 1 | 4 | 3 | 1 | 1 | 2 | 4 | 15 | 2 | 4 | 1‡ |
| sec | 8 | 1 | 2 | 3 | 0 | 6 | 4 | 1 | 2 | 2 | 5 | 15 | 2 | 2 | 1 |
| yak | 9 | 1 | 2 | 3 | 1 | 6 | 4 | 3 | 2 | 2 | 5 | 17 | 2 | 5 | 1 |
| ere | 8 | 1 | 3 | 3 | 1 | 6 | 4 | 3 | 2 | 1 | 5 | 17 | 2 | 4 | 1 |
| ana | 8 | 0 | 1 | 3 | 1 | 5 | 4 | 3 | 2 | 2 | 5 | 24 | 3 | 4 | 1 |
| pse | 9 | 1 | 2 | 3 | 2 | 6 | 4 | 3 | 2 | 2 | 5 | 19 | 2 | 3 | 1 |
| per | 7 | 1 | 0 | 1 | 0 | 6 | 4 | 2 | 2 | 2 | 4 | 18 | 2 | 2 | 1 |
| wil | 12 | 1 | 1 | 3 | 1 | 7 | 3 | 2 | 1 | 2 | 5 | 19 | 4 | 3 | 1 |
| vir | 8 | 1 | 1 | 3 | 1 | 3 | 4 | 3 | 2 | 2 | 3 | 17 | 3 | 1 | 1 |
| moj | 8 | 1 | 1 | 3 | 1 | 3 | 4 | 2 | 2 | 2 | 5 | 21 | 3 | 2 | 1 |
| gri | 9 | 1 | 1 | 2 | 1 | 2 | 4 | 4 | 2 | 2 | 7 | 20 | 4 | 1 | 1 |
The phylogenetic clades in this table are defined in Fig. 4.
*The first three characters are used as the abbreviation for each species.
†Orthologous to the melanogaster gene OR83b (OR49).
‡Contains one internal stop codon.
Numbers of OR Genes in Ancestral Species.
We estimated the numbers of OR genes in ancestral species and gains and losses of genes during Drosophila evolution, using two different methods. One is the modified reconciled-tree (MR) method (17). This method is based on the comparison of a bootstrap condensed gene tree (18) with the species tree, and the numbers are estimated under the parsimony principle. In this study, we used a 50% bootstrap condensed tree of OR genes. The other is the gene order (GO) method developed in this study. In this method, the adjacent genes of each OR gene are considered, and when at least one of the two adjacent genes for a pair of OR genes gives the same best hit gene in the Blast search (19), the OR genes are regarded as orthologs or paralogs. By using this information, the number of gene gains and losses in the evolutionary process can be estimated (see Materials and Methods). For this analysis, we used representative five species including both subgenera, because the application of the GO method for a larger number of species was complicated.
Fig. 5 shows estimates of the numbers of OR genes in ancestral species and gene gains and losses when the five Drosophila species were used. The estimates obtained by the MR and GO methods are generally similar to each other. Both methods showed that substantial numbers of gene gains and losses have occurred for each branch of the tree. Nevertheless, both extant and ancestral species contain similar numbers of OR genes despite these gains and losses. Similar results were obtained when we used different bootstrap threshold values (30–95%) in the MR method. These observations suggest that the OR repertoire of Drosophila species has been quite stable in the evolutionary process.
Fig. 5.
Estimates of the numbers of genes in ancestral species and gains and losses of genes during Drosophila evolution. (a) Estimates by the MR method using a 50% condensed tree. (b) Estimates by the GO method. Numbers in circles indicate the numbers of OR genes in the extant species, whereas those in squares show the numbers in ancestral species. Numbers above and below each branch indicate the numbers of gains (+) and losses (−) of genes, respectively. The time scale shown below the tree is from ref. 8.
Comparing the genomic maps of OR genes for melanogaster and yakuba, we previously concluded that melanogaster probably gained one gene and lost two genes after their divergence, whereas yakuba gained two genes and lost one gene. This conclusion is the same as that obtained by the MR method and similar to that obtained by the GO method. However, comparison of the genomic maps of OR genes for yakuba and pseudoobscura gave much smaller number of gene gains and losses compared with those obtained by the MR and GO methods. These results suggest that when distantly related species are compared, the MR and GO methods give more accurate estimates than the comparison of genomic sequences.
Discussion
We have seen that the repertoire of OR genes in Drosophila species has been nearly the same for the entire period of Drosophila evolution. Yet, we identified frequent gains and losses of OR genes in the evolutionary process, indicating that the birth-and-death process (20, 21) has operated during the evolution of Drosophila OR genes. Unequal crossing over appears to be the major mechanism for increasing gene number, because closely related genes in the phylogenetic tree were observed in tandem arrays in the genome. Although gene retroposition can increase the number of genes in a genome, this factor appears to have played negligible roles in the present case, because only two of 711 functional OR genes were intronless. Intronless genes could occur by retroposition, but they can also be generated by deletion of introns. Chromosomal rearrangements have contributed to the establishment of genome-wide distribution of OR genes, because the gene order of the orthologous OR genes is substantially different between distantly related species. These aspects of evolutionary changes of OR genes are essentially the same for both Drosophila and mammalian species (22).
However, there are two major differences in the olfactory system between mammals and Drosophila. One is the number of glomeruli in the olfactory bulb of mammals and in the antennal lobe of Drosophila relative to the number of OR genes. In Drosophila, each ORN expresses only one or two OR genes (in addition to the co-expression of gene OR83b in most neurons), and the olfactory information from ORNs, which express the same OR genes, is transmitted to a single glomerulus (23). The number of glomeruli is therefore similar to the number of different ORs. For example, melanogaster has ≈60 ORs (4) and ≈50 glomeruli (24), and the honey bee has ≈160 ORs (7) and ≈160 glomeruli (25). In mammals, each ORN also expresses only one OR gene, but the number of glomeruli is considerably greater than the number of ORs. For example, the mouse has ≈1,000 ORs (22) and 1,800 glomeruli (26), and the human has ≈400 ORs (27) and ≈8,000 glomeruli (28). These observations suggest that the olfactory information received by ORs can be transmitted to glomeruli in a more flexible way in mammals than in Drosophila.
The other difference is the expression pattern of OR genes. In Drosophila, a specific OR gene tends to be expressed deterministically in a specific ORN, which produces precise expression pattern of a given OR gene (29). Therefore, if an OR gene is duplicated or lost from the genome, the gene expression pattern may be disturbed. In mammals, however, one of the clustered OR genes in the genome is stochastically chosen to be expressed in each ORN (30). Therefore, the expression pattern of OR genes appears to be considerably different among different individuals, and consequently the number of OR genes may change relatively easily in the evolutionary process. Actually, at least 26 loci of OR genes are known to be polymorphic between functional and nonfunctional alleles among human individuals (31).
Of course, the most important factor for determining the number of OR genes in mammals would be environmental conditions. Mammalian species inhabit a wide range of environments, including various temperature zones, diverse habitats (terrestrial, aquatic, and aerial living), etc. It is therefore possible that a varying degree of olfaction required for living in different environments has caused the evolutionary change of OR genes and this change has occurred because of the flexible system of olfaction in mammals. Drosophila species also inhabit highly variable environments (32), but the variation in the number of OR genes may not have changed greatly because of the rigid expression and the signal pathway mentioned above.
It should be noted that mammalian species generally have a large number of OR pseudogenes. It is also known that removal of a substantial proportion of the olfactory bulb in laboratory rats does not seriously affect their survival (33). Therefore, a considerable portion of the change in the number of OR genes could be more or less neutral.
Materials and Methods
Identification of OR Genes in the Draft Genome Sequences.
The genome sequences of melanogaster (release 4.3) and pseudoobscura (release 2.0) were downloaded from the FlyBase (34). Other genome sequences were obtained from the AAA database (http://rana.lbl.gov/drosophila/): simulans (dsim_washu_2jun05_mosaic.tar.gz), sechellia (dsec_broad_28oct05.tar.gz), yakuba (dyak_washu_13dec05.tar.gz), erecta (dere_freeze1.tar.gz), ananassae (dana_freeze1.tar.gz), persimilis (dper_broad_28oct05.tar.gz), willistoni (dwil_caf1.tar.gz), virilis (dvir_freeze1.tar.gz), mojavensis (dmoj_freeze1.tar.gz), and grimshawi (dgri_freeze1.tar.gz).
To identify the OR genes, we performed a two-round TBlastN (19) search with E-value ≤10−10 against each genome sequence. In the first round, amino acid sequences of 61 OR genes annotated in the melanogaster genome were used as queries. After collecting hit sequences, we manually annotated each hit sequence, because Drosophila OR genes have introns (4) and it was not easy to determine the ORF by using computer programs. In the second round, the procedures were repeated by using the functional OR genes identified in the first round to find additional OR genes. To extract only unique genes, we compared the nucleotide sequences of all functional OR genes with one another and eliminated any overlapping genes. The flowchart for the detailed procedure is shown in SI Fig. 8. Note that the alternative transcripts from the same locus were considered as different genes. There were two such loci in the melanogaster genome. Coding sequences of functional OR genes and the genomic locations of all OR genes are available from SI Data Set 1 and SI Table 3, respectively.
Estimation of the Numbers of OR Genes in Ancestral Species.
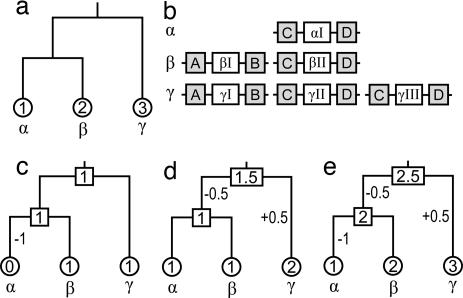
The numbers of genes in ancestral species and gains and losses of OR genes in evolution were estimated by the MR (17) and the GO methods. The latter method is expected to give more reliable results when the bootstrap support of gene trees is low, although the adjacent genes may not always be identifiable because of gene rearrangements in the past. The detailed procedure of this method is described in Fig. 6. The flowchart for the identification of adjacent genes is shown in SI Fig. 9.
Fig. 6.
Example illustrating the GO method for estimating the numbers of genes in ancestral species and gains and losses of genes. (a) Phylogenetic tree for three species α, β, and γ. We assume that α, β, and γ have one, two, and three OR genes, respectively, as shown in circles. (b) Linkages of the OR genes (αI-γIII) and their adjacent genes (A–D). The pattern of linkages of the OR genes with adjacent genes A and B suggests that βI and γI are orthologs, whereas there is no such OR gene in species α. Similarly, αI, βII, and γII and αI, βII, and γIII are each orthologs, whereas γII and γIII are paralogs. (c) From the information of adjacent genes A and B, we can infer that ancestral species had one gene (shown in squares) and one gene loss occurred in species α using parsimony principle. (d) From the information of adjacent genes C and D, two equally parsimonious pathways are found. If the MRCA of these three species had one gene, one gene gain occurred in species γ, whereas if the MRCA had two genes, one gene loss occurred in the ancestral species of α and β. In this study, we use the average numbers of genes in ancestral species and gene gains (+) and losses (−) for these two pathways. (e) The numbers of genes in ancestral species and gains and losses of genes obtained by summing up the estimates in c and d. When a large number of genes is used, the fractional numbers often disappear. Theoretically, there are more complicated cases. For example, species α may have the linkage A–αI–D instead of C–αI–D. In this case, we assume that all OR genes are either orthologous or paralogous even though some pairs of OR genes do not share the same adjacent genes. In the present analysis, however, such cases rarely occurred.
Supplementary Material
Acknowledgments
We thank Saby Das, Zhenguo Lin, Jongmin Nam, Nikos Nikolaidis, Alex Rooney, Shigeru Saito, Claire T. Saito, Shozo Yokoyama, and Jianzhi Zhang for valuable comments on earlier versions of the manuscript. Steve Schaeffer provided us with information on the chromosome assembly of pseudoobscura. Jongmin Nam and Yoshihito Niimura gave us the scripts for the MR method. This work was supported by National Institutes of Health Grant GM020293 (to M. Nei).
Abbreviations
- GO
gene order
- MR
modified reconciled-tree
- OR
olfactory receptor
- ORN
olfactory receptor neuron.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702133104/DC1.
References
- 1.Niimura Y, Nei M. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aloni R, Olender T, Lancet D. Genome Biol. 2006;7:R88. doi: 10.1186/gb-2006-7-10-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann CI. Nature. 2006;444:295–301. doi: 10.1038/nature05402. [DOI] [PubMed] [Google Scholar]
- 4.Robertson HM, Warr CG, Carlson JR. Proc Natl Acad Sci USA. 2003;100:14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niimura Y, Nei M. Proc Natl Acad Sci USA. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 7.Robertson HM, Wanner KW. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K, Subramanian S, Kumar S. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Hedges SB. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 10.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Benton R, Sachse S, Michnick SW, Vosshall LB. Plos Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krieger J, Klink O, Mohl C, Raming K, Breer H. J Comp Physiol A. 2003;189:519–526. doi: 10.1007/s00359-003-0427-x. [DOI] [PubMed] [Google Scholar]
- 13.Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, et al. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 16.Thompson J, Higgins DG, Gibson TI. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam J, Nei M. Mol Biol Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. pp. 165–186. [Google Scholar]
- 19.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zheng Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ota T, Nei M. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- 21.Nei M, Rooney AP. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niimura Y, Nei M. Gene. 2005;346:13–21. doi: 10.1016/j.gene.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Ache BW, Young JM. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Fishilevich E, Vosshall LB. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- 25.Galizia CG, Menzel R. J Insect Physiol. 2001;47:115–130. doi: 10.1016/s0022-1910(00)00106-2. [DOI] [PubMed] [Google Scholar]
- 26.Royet JP, Souchier C, Jourdan F, Ploye H. J Comp Neurol. 1988;270:559–568. doi: 10.1002/cne.902700409. [DOI] [PubMed] [Google Scholar]
- 27.Niimura Y, Nei M. Proc Natl Acad Sci USA. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meisami E, Mikhail L, Baim D, Bhatnagar KP. Ann NY Acad Sci. 1998;855:708–715. doi: 10.1111/j.1749-6632.1998.tb10649.x. [DOI] [PubMed] [Google Scholar]
- 29.Ray A, van der Goes van Naters W, Shiraiwa T, Carlson JR. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 31.Menashe I, Man O, Lancet D, Gilad Y. Nat Genet. 2003;34:143–144. doi: 10.1038/ng1160. [DOI] [PubMed] [Google Scholar]
- 32.Powell JR. Progress and Prospects in Evolutionary Biology. New York: Oxford Univ Press; 1997. pp. 143–184. [Google Scholar]
- 33.Bisulco S, Slotnick B. Chem Senses. 2003;28:361–370. doi: 10.1093/chemse/28.5.361. [DOI] [PubMed] [Google Scholar]
- 34.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM. The FlyBase Consortium. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.