Abstract
Bacteriophage N4 minivirion RNA polymerase (mini-vRNAP), the RNA polymerase (RNAP) domain of vRNAP, is a member of the T7-like RNAP family. Mini-vRNAP recognizes promoters that comprise conserved sequences and a 3-base loop–5-base pair (bp) stem DNA hairpin structure on single-stranded templates. Here, we defined the DNA structural and sequence requirements for mini-vRNAP promoter recognition. Mini-vRNAP binds a 20-nucleotide (nt) N4 P2 promoter deoxyoligonucleotide with high affinity (Kd = 2 nM) to form a salt-resistant complex. We show that mini-vRNAP interacts specifically with the central base of the hairpin loop (−11G) and a base at the stem (−8G) and that the guanine 6-keto and 7-imino groups at both positions are essential for binding and complex salt resistance. The major determinant (−11G), which must be presented to mini-vRNAP in the context of a hairpin loop, appears to interact with mini-vRNAP Trp-129. This interaction requires template single-strandedness at positions −2 and −1. Contacts with the promoter are disrupted when the RNA product becomes 11–12 nt long. This detailed description of vRNAP interaction with its promoter hairpin provides insights into RNAP–promoter interactions and explains how the injected vRNAP, which is present in one or two copies, recognizes its promoters on a single copy of the injected genome.
Keywords: T7-like RNA polymerase
Bacteriophage N4 virion RNA polymerase (vRNAP), which is responsible for the transcription of the phage early genes, is present in virions in one or two copies and is injected into the host together with the phage genome at the onset of infection (1, 2). vRNAP is a 3,500-aa polypeptide that lacks extensive sequence similarity to either of the two known families of DNA-dependent RNAPs (3). Using controlled trypsin proteolysis and catalytic autolabeling, we defined a stable and transcriptionally active 1,106-aa domain (mini-vRNAP) located at the center of the vRNAP polypeptide (3). Mini-vRNAP possesses the same initiation, elongation, termination, and product displacement properties as full-length vRNAP (3, 4).
Mutational, biochemical, and phylogenetic analyses indicated that mini-vRNAP is a highly diverged member of the T7 RNAP family (3). However, although T7 RNAP recognizes its promoters in double-stranded templates (5), vRNAP transcribes promoter-containing, single-stranded templates with in vivo specificity (6), recognizing a 3-base loop–5-bp stem hairpin structure and specific sequences at the promoter (7). In vivo, vRNAP promoter recognition requires template supercoiling (8) to drive extrusion of the promoter hairpins from the double-stranded N4 genome (9, 10) and Escherichia coli single-stranded DNA-binding protein to stabilize single-stranded regions surrounding the site of transcription initiation (11). We have used single-stranded templates to identify unambiguously the site of transcription initiation as well as the sequence and structural DNA determinants of vRNAP–promoter interaction. The results reveal two major determinants of vRNAP hairpin recognition: a purine at the center (−11G preferred) of the hairpin loop that must be presented to vRNAP in the context of a loop, and a specific interaction in the major groove (−8G) of the hairpin stem. Both interactions (−11G, −8G) are required for the characteristic salt resistance of the mini-vRNAP–promoter complex (12). RNAP UV cross-linking to a −11 5-iododeoxyuracil (5-IdU)-substituted, 5′ end-labeled promoter oligonucleotide and mapping of the cross-link suggest that −11G interacts with Trp-129, most likely through stacking, yielding a salt-resistant RNAP–promoter complex. The results presented provide a detailed description of the determinants of recognition of the promoter DNA hairpin and explain the unusual characteristics of the vRNAP–promoter complex and how the injected vRNAP recognizes its promoters on a single copy of the injected genome.
Results
Mini-vRNAP Initiates Transcription 11 Nucleotides Downstream from the Center of the Hairpin Triloop.
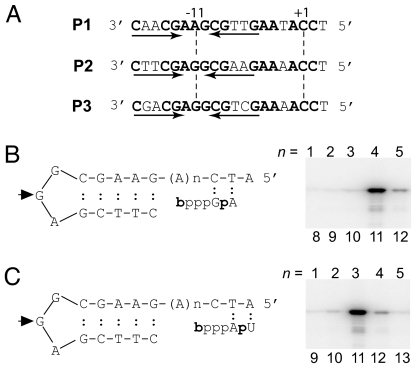
Three N4 vRNAP promoters (P1, P2, and P3) are present in the N4 genome (6) (Fig. 1A). The bottom stem base pair, 3′-C:G-5′, of the vRNAP promoter hairpin is connected to downstream cytosines by four adenines in promoters P2 and P3. We used a series of P2 templates containing increasing numbers of adenines followed by the sequence CTA to identify the determinants of mini-vRNAP initiation site selection. We measured mini-vRNAP catalytic autolabeling by cross-linking the hydroxybenzaldehyde esters of GTP (bGTP) or ATP (bATP) to the enzyme and then adding [α-32P]ATP (Fig. 1B) or [α-32P]UTP, respectively (Fig. 1C), in a template-directed manner (3). The RNAP polypeptide is labeled as a result of phosphodiester bond formation. Transcription initiation at promoter P2 was most efficient when either bGTP or bATP could pair with the base present 11 nt downstream from the center of the hairpin triloop. Therefore, the first cytosine at the vRNAP promoters serves as the template for transcription initiation.
Fig. 1.
Mini-vRNAP initiates transcription 11 nucleotides downstream from the center of the hairpin triloop. (A) Sequences of the bacteriophage N4 early promoters (P2 corresponds to the P2–3 oligonucleotide). Arrows, inverted repeats; +1, site of transcription initiation; −11, center of hairpin loop. (B) Schematic representation of first phosphodiester bond formation by catalytic autolabeling by using DNA templates with increasing number (n) of As between the promoter hairpin and the sequence CTA. Derivatized GTP (bGTP) and [α-32P]ATP are shown. (C) Derivatized ATP (bATP) and [α-32P]UTP. Arrowhead, hairpin loop central position. The number of nt (n) from the central loop position of the promoter hairpin to the transcription start site is shown below each lane. PhosphorImages of 10% SDS/polyacrylamide gels are shown.
The Center of the Loop and a DNA Sequence at the Hairpin Stem Are the Major Determinants of vRNAP Promoter Recognition.
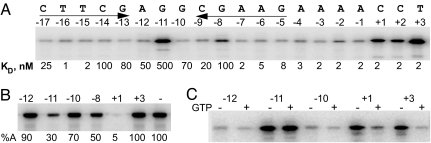
To identify the sequence determinants of vRNAP promoter recognition, we used a series of 5′ end-labeled and singly 5-IdU-substituted oligonucleotides containing the promoter hairpin and downstream sequences up to position +3 (Fig. 2A). Note that the 5-IdU-pairing base in the stem was changed to dA in each substituted DNA to maintain the integrity of the stem. The ability of 5-IdU at a specific position to cross-link to mini-vRNAP was determined after 312-nm light irradiation followed by SDS/PAGE and autoradiography (4). The cross-link to 5-IdU determines direct (0 Å) interactions between the iodine in 5-IdU, in either single-stranded or double-stranded (major groove) nucleic acids and a protein, preferentially at aromatic residues (13). Equilibrium dissociation binding constants were determined by solution competition (SC) using surface plasmon resonance (SPR). Mini-vRNAP activity was assessed by catalytic autolabeling using unlabeled oligonucleotides.
Fig. 2.
A purine at the center of the hairpin triloop (−11) is the major determinant of promoter recognition. (A) UV cross-linking of mini-vRNAP to 5-IdU-substituted and 32P end-labeled 20-mer P2–3 DNA. Above each lane, base and position substituted with 5-IdU are shown. Below each lane, Kd values obtained by SC using SPR for each singly 5-IdU-substituted DNA oligonucleotide are shown. For unsubstituted DNA, Kd = 2 nM. (B) Mini-vRNAP catalytic autolabeling with bGTP and [α-32P]ATP and unsubstituted or 5-IdU-substituted DNA at positions −12, −11, −10, −8, +1, +3. %A, percent activity relative to unsubstituted DNA. (C) GTP effect on mini-vRNAP UV-cross-linking to promoter-containing oligonucleotides substituted with 5-IdU at positions −12, −11, −10, +1, or + 3.
Oligonucleotides containing 5-IdU at positions −11, −8, +1, +2, and +3 cross-linked efficiently to mini-vRNAP (Fig. 2A). Substitution with 5-IdU at the center of the hairpin loop (−11G in P2) resulted in a 250-fold decrease in affinity (Fig. 2A) and a 3-fold decrease in first phosphodiester bond formation at 1 μM promoter concentration (Fig. 2iB). Therefore, position −11 must provide a critical vRNAP contact.
5-IdU substitutions at position −8 or −14 resulted in a 50-fold decrease in affinity (Fig. 2A) and a 2-fold decrease in catalytic autolabeling for substitution at position −8 (Fig. 2B). We surmise that position −8 represents an additional mini-vRNAP-specific contact, whereas the effect of substitution at position −14 on affinity is due to the presence of A at the base-pairing position (−8) in the −14 5-IdU-substituted promoter.
vRNAP interactions with bases at positions +1, +2, and +3 were not sequence-specific because 5-IdU substitutions at positions +1, +2, and +3 had no effect on mini-vRNAP binding. However, the efficiency of cross-linking to positions +1 and +3 decreased in the presence of GTP (Fig. 2C), suggesting that changes in the environment of promoter DNA at the site of transcription initiation occur upon binding of one of the initiating nucleotides. Note that the decrease in catalytic autolabeling observed for 5-IdU at +1 is the result of imposing a shift to initiate transcription at position +2 with bGTP (Fig. 1).
Single-Stranded Template at Positions −2, −1, and +1 Is Required for Efficient Mini-vRNAP Binding.
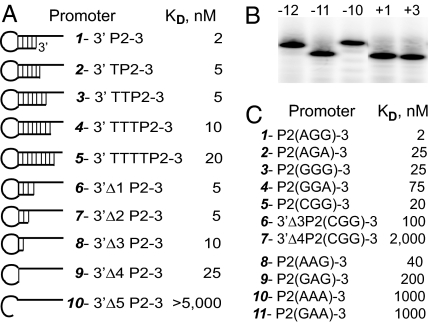
To test the relevance of the hairpin stem length in mini-vRNAP–promoter interactions, the affinity of mini-vRNAP for synthetic oligonucleotides with increasing stem lengths (lengthening at the 3′ end of P2–3) was determined (Fig. 3A). Increasing the length of the stem by the addition of 1 (6-bp stem) or 2 (7-bp stem) nt to the 3′ end of the template did not affect binding (Fig. 3A, 2 and 3) or catalytic autolabeling (data not shown) significantly. However, further lengthening of the stem (Fig. 3A, 5) led to a 10-fold reduction in binding, indicating that single-strandedness around the site of transcription initiation (−2 and −1) is required for vRNAP promoter binding and that vRNAP cannot unwind a double-stranded template.
Fig. 3.
Role of hairpin stem length and hairpin loop sequence on mini-vRNAP promoter binding, determined by SC using SPR. (A) Effect of promoter hairpin stem length on mini-vRNAP promoter binding. 3′ P2–3 is P2(AGG)-3. (B) Mobility of 5′ end-labeled 5-IdU-substituted DNA at −12, −11, −10, +1, or + 3 in 6 M urea/8% polyacrylamide gel. Mobilities of the +1- and +3-substituted DNA oligonucleotides correspond to that of unsubstituted P2–3 DNA. (C) Effect of promoter hairpin loop sequence, shown in parentheses, on mini-vRNAP binding. Kd values were obtained by SC using SPR.
A Hairpin Promoter Loop Is Required for Mini-vRNAP Binding.
vRNAP promoter hairpins are unusually stable because of specific sequences of the loop (−12A and −10G) and the loop-closing base pair (3′-G:C-5′) (9, 14–16). This sequence-dependent stability is reflected in the increased electrophoretic mobility in the presence of 6 M urea of oligonucleotides containing these unusually stable DNA hairpins (Fig. 3B, −11, +1, +3 vs. −12, −10, 5-IdU-substituted oligonucleotides). Decreasing the length of the promoter hairpin stem to 2 bp (Tm, 76°C; ref. 15) or 1 bp reduced the affinity by only 5- to 10-fold (Fig. 3A, 1 vs. 8 and 9), whereas abolishing the closing base pair (Fig. 3A, 10) led to loss of binding. To investigate the role of hairpin stability in vRNAP promoter binding, we tested the effect of substitutions at positions −12 and −10, which destabilize the hairpin (9). A 12- to 30-fold decrease in binding affinity was observed, whether promoters had G or A at the center of the hairpin loop (Fig. 3C, 1 vs. 2, 3, 4, 5; 8 vs. 9, 10, 11). Additionally, although the affinities for −12-substituted promoters containing 5-bp (Fig. 3C, 5) or 2-bp (Fig. 3C, 6) hairpin stems decreased by 10-fold with respect to their wild-type counterparts (Fig. 3A, 1 and 8), the affinity for the 1-bp stem −12-subtituted promoter decreased 80-fold (Fig. 3C, 7 vs. Fig. 3A, 9), reflecting destabilization of the loop-closing base pair and indicating that position −11 recognition must occur in the context of a loop structure.
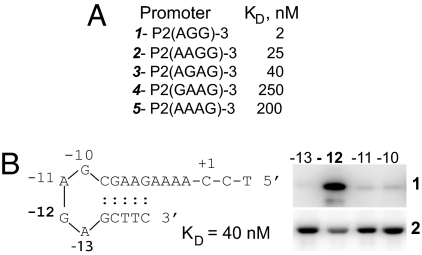
To determine the effect of the hairpin loop size on mini-vRNAP binding, we used promoter oligonucleotides with purine tetraloop hairpins. The interaction with tri- and tetraloop hairpins was sequence-dependent (Figs. 3C and Fig. 4A). Comparison of the relative binding affinities of triloop and tetraloop hairpin promoters (Fig. 3C, 2 vs. Fig. 4A, 3; Fig. 3C, 8 vs. Fig. 4A, 2) indicated that an increase in the size of the loop by one nucleotide affects binding by 10-fold compared with the optimal P2 triloop. In some cases, the enzyme had higher affinity for a tetraloop than for a triloop hairpin promoter (Fig. 3C, 9 vs. Fig. 4A, 3).
Fig. 4.
vRNAP interaction with promoters containing tetraloop hairpins. (A) Effect of promoter hairpin loop sequence and length on mini-vRNAP promoter binding, measured by SPR. (B) (Left) Sequence of a promoter with a tetraloop hairpin; numbers indicate positions at which IdU substitutions were introduced with respect to + 1C. (Right) 1, mini-vRNAP DNA UV cross-linking; 2, mini-vRNAP catalytic autolabeling, in the presence of 10 nM oligonucleotide replaced with 5-IdU at the indicated positions.
To identify the tetraloop hairpin base contacted by vRNAP, 5′ end-labeled oligonucleotides with single 5-IdU substitutions at each loop position were analyzed in UV cross-linking experiments (Fig. 4B). The contact shifted from position −11 in a triloop hairpin to position −12 in a tetraloop hairpin (Fig. 4B, 1), with a corresponding decrease in the ability of mini-vRNAP to support catalytic autolabeling when 5-IdU was present at −12 (Fig. 4B, 2).
The Salt Resistance of the vRNAP–Promoter Complex Is Promoter Sequence-Dependent.
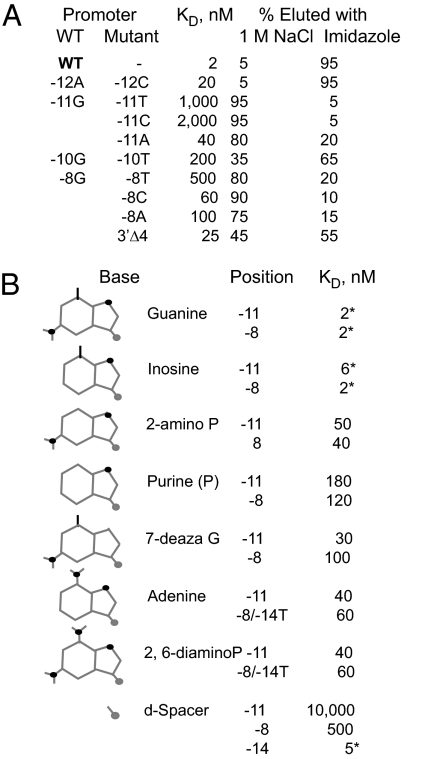
We have shown previously that the vRNAP–promoter P2 complex is resistant to 2 M NaCl, although the binding process is salt-sensitive (12). To define the basis for complex salt resistance, we investigated the salt sensitivity of mini-vRNAP complexes with promoter oligonucleotides containing substitutions at positions required for vRNAP recognition or promoter hairpin stability. We incubated 5′ end-labeled oligonucleotides with C-terminally His6-tagged mini-vRNAP prebound to Talon resin. After binding, the resin was washed with 1 M NaCl buffer to dissociate salt-sensitive complexes and then washed with the same buffer containing 100 mM imidazole to elute salt-resistant complexes (Fig. 5A). Complexes formed with promoter P2 were salt-resistant. The resistance of the complex to salt decreased upon substitution of −11G with A, whereas substitution with a pyrimidine (−11T or C) yielded low-affinity, salt-sensitive complexes. Similar results were obtained with promoters containing substitutions at −8 (−8T, C, or A), a position that is also critical for vRNAP–promoter interaction. In contrast, substitutions at positions essential for hairpin stability (−12A → C, −10G → T) did not greatly affect the salt resistance of the complex, although a reduction in promoter affinity by 5- to 100-fold was observed, as when the hairpin stem length is reduced to 1 bp. We conclude that complex salt resistance is determined by the identity of the hairpin triloop central base and not by the hairpin stability or the affinity of vRNAP for the promoter. Promoters with tetraloop hairpins (AAGG, AGAG) yielded complexes that were partially salt-resistant.
Fig. 5.
Hairpin-sequence recognition by vRNAP. (A) Effect of promoter sequence on the affinity and salt resistance of vRNAP–promoter complexes. Binding affinity was determined by SC using SPR. (B) Role of H bond donors and acceptors at positions −11 and −8 on vRNAP–promoter interaction. Kd and salt resistance of the complex were determined as described. *, salt-resistant complex.
Identification of −11G and −8G Hydrogen Bond Donors and Acceptors Essential for vRNAP–Promoter Interaction.
The contributions of −11G and −8G hydrogen bond donors and acceptors to vRNAP–promoter interaction were determined by using oligonucleotides with substitutions at each of these positions (Fig. 5B). Removal of the G 2-amino group at −8 (inosine) had no effect on binding or salt resistance. In contrast, removal of the 6-keto (2-amino P) or 7-imino (7-deaza G) groups resulted in salt-sensitive complexes and 20- and 50-fold loss in binding affinity, respectively. Substitution with A yielded a salt-sensitive complex with a 30-fold decrease in binding affinity. These results indicate that mini-vRNAP interacts with −8G in the major groove of the hairpin stem. Substitution at −8 with an abasic spacer led to a 250-fold decrease in binding affinity. In contrast, the absence of a base at the −8 base-pairing partner (−14) had little effect on the affinity and no effect on the salt resistance of the complex, indicating that position −14 does not interact with mini-vRNAP.
Removal of the −11G 2-amino group had a small decrease in binding affinity with no effect on the salt resistance of the complex. The absence of the 6-keto or 7-imino groups resulted in salt-sensitive complexes and 90- and 15-fold decreases in binding affinity, respectively. The affinity reached the micromolar range when an abasic spacer was present at −11, highlighting the importance of the purine heterocycle at this position. We conclude that −11G represents the major recognition determinant for N4 vRNAP.
The Base at the Center of the Hairpin Loop, −11G, Interacts with vRNAP Trp-129.
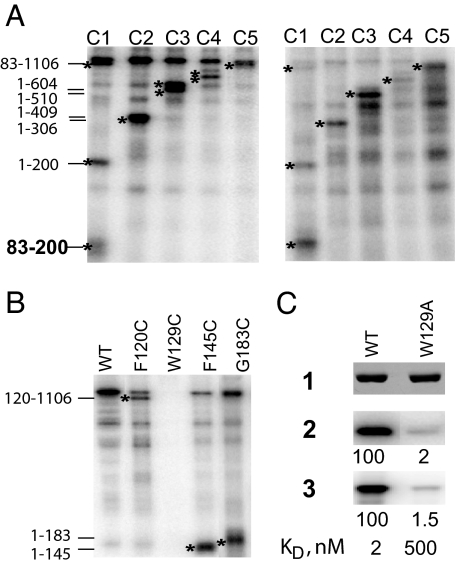
The mini-vRNAP polypeptide lacks cysteines (3). To identify the mini-vRNAP residue that interacts with −11G, we used five active mini-vRNAPs, each containing a pair of cysteines (di-Cys) substituting residues separated by ≈100 aa (E.K.D. and K. Kazmierczak, unpublished results). Each di-Cys-containing mini-vRNAP was UV cross-linked to a 5′ end-labeled −11 5-IdU-substituted promoter oligonucleotide. Cross-linked complexes were then treated with 2-nitro-5-thiocyanobenzoic acid (NTCB) and analyzed by SDS/PAGE (Fig. 6A Left). Analysis of the pattern of labeled polypeptides arising from cleavage of all five di-Cys-containing mini-vRNAPs indicates that a residue in the 83–200-aa interval was cross-linked to the −11 5-IdU-substituted promoter DNA. The cross-link was not probe-dependent because similar results were obtained when a −11 4-thio-T-substituted oligonucleotide was used (Fig. 6A Right).
Fig. 6.
Mini-vRNAP Trp-129 cross-links to −11 position of promoter DNA. (A) NTCB cleavage of C1–C5 RNAPs after UV cross-linking to −11 5-IdU- (Left) and −11 4-thio-T- (Right) substituted, 5′ 32P-labeled P2–3 DNA oligonucleotides. Stars indicate bands of interest. (B) NTCB cleavage of F120C, W129C, F145C, and G183C enzymes cross-linked to −11 5-IdU-labeled DNA. PhosphorImages of 10% SDS/PAGE are shown in A and B. (C) Effects of the W129A substitution on UV cross-linking to −11 5-IdU-labeled DNA (2), catalytic autolabeling (3), and binding affinity. 1, silver-stained gel; 2 and 3, PhosphorImages.
Three mini-vRNAPs containing Cys substitutions in the 83- 200-aa mini-vRNAP interval were used to identify the cross-linked residue. The NTCB cleavage pattern of F120C-, F145C-, and G183C-labeled polypeptides mapped the site of cross-linking to the 120–145-aa interval (Fig. 6B), which contains a single aromatic residue, Trp-129. The W129C substitution completely abolished cross-linking. To confirm that Trp-129 is indeed the target of −11 5-IdU cross-linking, we generated and tested the W129A enzyme. The W129A substitution decreased promoter-binding affinity 250-fold and abolished cross-linking to −11 5-IdU, catalytic autolabeling (Fig. 6C, 2 and 3), and salt resistance (data not shown), indicating that Trp-129 interacts with the center of the promoter hairpin loop. We propose that a stacking interaction between −11G and Trp-129 leads to the unusual salt resistance of the complex.
Mini-vRNAP Clears the Promoter When the RNA Product is 11–12 Nucleotides Long.
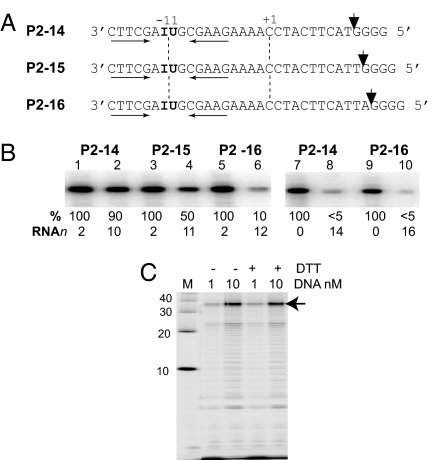
To define the step in transcription at which vRNAP undergoes promoter clearance, 5′ end-labeled templates containing 5-IdU at position −11 and 10 (P2–14), 11 (P2–15), or 12 (P2–16) G-less nucleotides in the transcribed region, followed by four G residues were used (Fig. 7A). Transcription in the presence of GTP, ATP, and UTP halted mini-vRNAP before the terminal G tract (Fig. 7B, 2, 4, and 6). Contacts with the promoter, measured as the ability of mini-vRNAP to cross-link to 5-IdU at −11, were present in the RNAP promoter (Fig. 7B, 7 and 9) and initiation (Fig. 7B, 1, 3, and 5) complexes. Contacts were weakened when the transcript was 11 nt long (Fig. 7B, 4) and lost upon the addition of the next nucleotide (Fig. 7B, 6).
Fig. 7.
Mini-vRNAP promoter clearance. (A) P2–14, P2–15, and P2–16 DNA. Arrowheads show positions where the mini-vRNAP stalls transcription in the absence of CTP. (B) Mini-vRNAP was incubated with 5′ 32P end-labeled −11 IdU-substituted oligonucleotides P2–14, P2–15, or P2–16 in the presence of 1 μM GTP (lanes 1, 3, and 5), 1 μM each GTP, ATP, and UTP (lanes 2, 4, and 6), no NTPs (lanes 7 and 9), all four NTPs (lanes 8 and 10) followed by UV cross-linking. The relative cross-linking efficiency (%) and transcript length (RNAn) are shown below each lane. (C) Mini-vRNAP runoff transcription on a −16/−7 cross-linked stem (−DTT) or uncross-linked (+DTT) P2–32 template. Arrow, runoff product.
Because mini-vRNAP–promoter complexes are resistant to 2 M NaCl and can be disrupted by raising the temperature in the absence of salt (12), we hypothesized that promoter clearance might be elicited by vRNAP-dependent melting of the hairpin. To test this possibility, we used templates containing an intramolecular cross-link (17) between two derivatized (N6-thioethyl) dA residues located on opposite strands of the hairpin stem at nonconserved positions (−7 and −16), whereas the corresponding base-pairing positions (−6 and −15) were substituted with dT. As a control, the same oligonucleotide containing the reduced form of the cross-link was used. Results of runoff transcription experiments showed that cross-linked stem and uncross-linked templates were equally active (Fig. 7C), indicating that melting of the promoter hairpin is not required for promoter clearance. In contrast, the results presented suggest that the length of the RNA transcript must signal mini-vRNAP to disrupt promoter contacts.
Discussion
The three N4 vRNAP promoters are characterized by a conserved sequence and a 3-nt loop and a 5- to 7-bp stem hairpin (Fig. 1). We have shown previously that specific conserved elements of the promoter [the base identities of the loop-closing base pair (3′-G:C-5′) and the 3′ (−12A) and 5′ (−10G) positions of the triloop, which determine the unusual stability of this hairpin] are required for its extrusion from double-stranded DNA in a process that depends on template supercoiling (9, 18).
Here, we identified conserved sequences that are essential for vRNAP–promoter interaction. The main determinant of promoter recognition is a purine at the center of the hairpin triloop (−11), with G being preferred. The presence of G at −11 has two consequences: it increases the affinity 20-fold (relative to −11A), and yields a vRNAP–promoter complex that is resistant to high salt concentrations (Fig. 5). Replacement of −11G by pyrimidines reduces the affinity 500- to 1,000-fold, indicating that the interaction with the purine heterocycle is essential for vRNAP binding (Fig. 5). Furthermore, −11G must be presented to vRNAP in the context of a hairpin loop (Fig. 3). Based on the structure of the promoter hairpin, −11G is the only unpaired base with one surface exposed for interaction with the protein because the flanking bases (3′A and 5′G) form a sheared base pair (15).
The identification of mini-vRNAP Trp-129 as the likely residue that contacts −11G provides an explanation for the mini-vRNAP complex salt resistance through a proposed stacking interaction. In contrast, it is not obvious why the replacement of −11 and −8G with purine analogs leads to salt sensitivity of the vRNAP–promoter complex (Fig. 5). We propose that the RNAP interaction with −8G in the major groove of the hairpin stem places the hairpin on the enzyme such that −11 G is positioned close to Trp-129. Furthermore, we propose that the −11G 6-keto and 7-imino groups are required for interactions with another residue, probably a bidentate interaction with arginine, which would place the purine heterocycle at the correct position to stack with Trp-129.
We have proposed that the N4 vRNAP active site must resemble that of T7 RNAP, although N4 vRNAP is the most evolutionarily diverged member of the T7 RNAP family (3). T7 RNAP recognizes two sets of sequences at its promoters on double-stranded templates: (i) base pairs −17 to −13, recognized in the minor groove by a flexible surface loop at the T7 RNAP N-terminal domain; and (ii) base pairs −10 to −7, recognized through interactions with the template strand in the major groove by the “specificity loop” of the fingers domain. Two T7 RNAP–promoter interactions are worth noting: Arg-756 and Arg-746 make bidentate interactions with the 7-imino and 6-keto groups of −9G and −7G, respectively (19). We propose that vRNAP recognition of −8G from the hairpin stem major groove is equivalent to recognition of ′9G or ′7G at the T7 RNAP promoters, whereas recognition of −11G by Trp-129 is unique to the existence of the vRNAP promoter as a DNA hairpin. This proposition is entirely plausible based on the structure of mini-vRNAP (K. Murakami, E.D., and L.B.R.-D., unpublished results).
Surprisingly, a hairpin triloop is not essential because a promoter with a hairpin tetraloop was recognized efficiently; indeed, 3′-AGAG-5′ and 3′-AAG-5′ loop promoters are recognized with similar affinity (Fig. 5). The results of cross-linking experiments using 5-IdU-substituted promoters with a tetraloop hairpin indicate that vRNAP contacts the hairpin loop 2 bases 5′ from the 3′ base of the loop-closing base pair (Fig. 5). Although the structures of hairpins with tetraloops are unknown, we expect that the base present 2 nt 5′ from the loop-closing base pair will be positioned on the same face of the hairpin as −8G, the other sequence determinant of vRNAP–hairpin interaction. This configuration is observed in hairpins with triloops of the same sequence as in the N4 promoter hairpins (15).
These findings bear on the process that specifies the transcriptional start site. We showed that vRNAP initiates transcription 11 nt downstream from the center of the loop (Fig. 1B). The importance of the base identity at position −11 suggests that vRNAP selects the site of transcription initiation by measuring the distance from the center of the hairpin loop. However, analysis of the interaction of vRNAP with tetraloop hairpins indicates that the site of transcription initiation is selected by measuring the distance from the loop closing base pair or −8G to the active center of the enzyme.
In some cases, multisubunit RNAPs initiate transcription specifically on single-stranded templates. E. coli RNAP initiates RNA synthesis at secondary structures on single-stranded templates at the minus strand origin of M13 DNA replication (20) and at the leading region of the conjugative ColIIb-P9 plasmid (21). Yeast RNAP III initiates transcription on the nontranscribed strand of the U6 promoter in a transcription initiation factor IIIB-dependent manner (22). However, within the single-subunit T7 RNAP family, initiation of specific transcription on single-stranded templates is so far limited to N4 vRNAP. N4 vRNAP is present in virions in one or two copies and must recognize the three early promoters on the single copy of the injected N4 genome, whereas T7 RNAP is synthesized abundantly early in phage infection (23). We propose that the ability of N4 vRNAP to discriminate against other single-stranded templates (8) and to recognize its promoter hairpin with high affinity (2 nM) provides a combinatorial strategy for restricted template specificity in the infected cell.
Materials and Methods
Expression and Purification of C-Terminally His6-Tagged Mini-vRNAP and Mini-vRNAP Mutant Enzymes.
Growth of E. coli BL21 cells bearing pEKD27 and induction and purification of C-terminally His6-tagged mini-vRNAP were performed as described previously (3). pEKD27 is a derivative of pKMK25 (3) where the His6 tag is directly attached to the mini-vRNAP C terminus, resulting in the C-terminal sequence KVAKA-His6. The di-Cys-containing RNAPs (C1–C5) have the following substitutions: C1, G83C/G200C; C2, G306C/S408C; C3, S510C/S604C; C4, A709C/S812C; C5, A918C/A1020C (E.K.D. and K. Kazmierczak, unpublished results). The F120C, W129C, F145C, G183C, and W129A mutant RNAPs were constructed by using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA), and the sequences were confirmed. All enzymes were stable.
Catalytic Autolabeling Assay.
Catalytic autolabeling was performed as described previously (3).
UV Cross-Linking of 5-IdU-Substituted Promoter Oligonucleotides to Mini-vRNAP.
Twenty deoxyoligonucleotides (Integrated DNA Technologies, Coralville, IA) containing single 5-IdU substitutions at each position of the 20-mer P2–3 DNA (TCCAAAAGAAGCGGAGCTTC) were 5′ end-labeled by using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and [γ-32P]ATP (3,000 Ci/mmol; Amersham Pharmacia, Piscataway, NJ). When substitutions were made in the stem (underlined positions), the pairing base was changed to dA. Mini-vRNAP (1 μM) and a 5′ 32P-labeled 5-IdU-substituted deoxyoligonucleotide (100 nM) were mixed in 5 μl of buffer A (10 mM Tris·HCl, pH 8/10 mM MgCl2/50 mM NaCl/1 mM DTT) in the wells of a polystyrene 72-well minitray (MicroWell; Nunc, Rochester, NY). Samples were irradiated and processed as described previously (4). When present during the cross-linking reaction, GTP was added at 1 mM. Halted RNAP elongation complexes were prepared, cross-linked, and analyzed as described (4).
Mapping of the −11 5-IdU contact to the mini-vRNAP polypeptide. To map the −11 5-IdU contact to mini-vRNAP, di-Cys-containing mini-vRNAP mutants were radiolabeled as described above and processed as described previously (24) with modifications. Radiolabeled protein (1–10 pmol) was incubated for 15 min at 37°C in 10 μl of 8 M urea/0.2M Tris·HCl, pH 8.3/5 mM DTT. Cysteines were modified by addition of 1 μl of freshly prepared 10% NTCB in methanol and incubation for 15 min at 37°C. The pH was adjusted to 9.3–9.5 by the addition of 1 μl of 1 N NaOH. Further incubation at 37°C for 30 min led to specific cleavage of the peptide bond at cysteine residues. The cleavage pattern was analyzed by SDS/PAGE and PhosphorImaging.
Determination of Equilibrium Dissociation Binding Constants.
Equilibrium dissociation binding constants of mini-vRNAP–promoter complexes were measured by SC using SPR (Biacore 3000). The Sensor Chip SA (Biacore, Piscataway, NJ) containing immobilized streptavidin was prepared according to the manufacturer's recommendations. 5′-biotinylated P2–15 DNA oligonucleotide 5′-GGCATTACTTCATCCAAAAGAAGCGGAGCTTC-3′ (Integrated DNA Technologies) at 100 nM was injected in one channel (DNA channel) for 1 min at 20 μl/min at 25°C, resulting in binding of 80 resonance units of DNA. All further experiments were performed at 4°C in HBS-EP buffer [0.01 M Hepes, pH 7.4/0.15 M NaCl/0.005% Surfactant P20 (Biacore)]. Regeneration after each cycle was achieved by two 10-s injections of 0.25 M NaCl/1.5 mM NaOH at 30 μl/min. For SC analysis, each DNA analyte was serially diluted 2-fold in 1 nM mini-vRNAP in HBS-EP buffer and kept at 4°C for at least 15 min to reach equilibrium binding. Samples were injected for 1 min at 20 μl/min into two channels: the DNA channel and the control channel (no DNA), whose signal was subtracted from the DNA channel signal. Kd was estimated as the concentration of the DNA analyte at which the binding level was 50% compared with the binding of 1 nM mini-vRNAP with no DNA added. At least four analyte concentrations within the 20–80% binding interval were analyzed in each experiment. In all experiments, 1 nM mini-vRNAP (100% binding, 80–120 resonance units) and promoters P2–3 at 8, 4, 2, and 1 nM were used as controls. The difference in two to five independently determined Kd values for each promoter tested was <30%.
Salt Sensitivity of vRNAP–Promoter Complexes.
C-terminally His6-tagged mini-vRNAP was added to 10 μM to a 0.5-ml slurry of Talon resin in buffer A (20 mM Tris·HCl, pH 8/50 mM NaCl) in a spin column (BD Biosciences, San Jose, CA). The column was washed with 2 ml of 1 M NaCl in buffer A followed by 2 ml of buffer A. The resin was made a 50% slurry in buffer A, slurry aliquots (50 μl) were transferred into new MicroSpin columns (Amersham Biosciences), and 5′ 32P-labeled DNA oligonucleotides were added to 10 nM. After a 10-min incubation on ice, the columns were washed three times with 100 μl of buffer A, three times with 100 μl of 1 M NaCl in buffer A, and they were eluted with 100 μl of 100 mM imidazole in buffer A. All procedures were performed at 4°C. The radioactivity in each of the fractions was determined by the Cherenkov method.
Cross-Linked Stem Hairpin Template for Promoter Clearance.
An intrastrand cross-linked hairpin derivative of the P2 promoter deoxyoligonucleotide (TTTCATACAGGATTGGAT G C A T T ACTTCATCCAAAAGT〈pI〉GCGGAGCT〈pI〉C), containing 32 nt in the transcribed region, was synthesized with the standard phosphoramidites of O6-phenyl-2′-deoxyinosine (O6-phenyl-dI, pI) (Glen Research, Sterling, VA) at positions −7 and −16 (Oligonucleotide Synthesis Facility, Cornell University, Ithaca, NY) and T at positions −6 and −15. The templates were processed as described previously (17) to yield a cross-link between N6-thioethyl-dAs at −7 and −16, and they were purified in a prewarmed (55°C) 8 M urea/20% polyacrylamide denaturing gel at 55°C. The 100% cross-linked material was excised and eluted from the gel. Runoff transcription reactions were performed for 5 min at 37°C in the presence of 10 μΜ E. coli SSB and analyzed as described previously (4).
Acknowledgments
We thank Dr. A. Mustaev (Public Health Research Institute, Newark, NJ) for providing the bGTP and bATP and for advice on the NTCB cleavage procedure, Dr. M. Yousef for advice on SPR experiments, Dr. K. M. Kazmierczak for the construction of cysteine-containing mini-vRNAP variants, E. A. Davydova for technical assistance, Dr. E. P. Geiduschek for enlightening discussions, and Dr. E. P. Geiduschek and Irene Kaganman for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Grant R01 AI12575 (to L.B.R.-D.). T.J.S. was supported by NIH Grant R01 GM21941 (to Jeffrey W. Roberts).
Abbreviations
- bATP
hydroxybenzaldehyde ester of ATP
- bGTP
hydroxybenzaldehyde ester of GTP
- 5-IdU
5-iododeoxyuracil
- mini-vRNAP
mini-virion RNA polymerase
- NTCB
2-nitro-5-thiocyanobenzoic acid
- RNAP
RNA polymerase
- SC
solution competition
- SPR
surface plasmon resonance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Falco SC, VanderLaan K, Rothman-Denes LB. Proc Natl Acad Sci USA. 1977;74:520–523. doi: 10.1073/pnas.74.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falco SC, Zehring W, Rothman-Denes LB. J Biol Chem. 1980;255:4339–4347. [PubMed] [Google Scholar]
- 3.Kazmierczak KM, Davydova EK, Mustaev AA, Rothman-Denes LB. EMBO J. 2002;21:5815–5823. doi: 10.1093/emboj/cdf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davydova EK, Rothman-Denes LB. Proc Natl Acad Sci USA. 2003;100:9250–9255. doi: 10.1073/pnas.1133325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diaz G, Raskin C, McAllister W. J Mol Biol. 1993;229:805–811. doi: 10.1006/jmbi.1993.1086. [DOI] [PubMed] [Google Scholar]
- 6.Haynes LL, Rothman-Denes LB. Cell. 1985;41:597–605. doi: 10.1016/s0092-8674(85)80032-5. [DOI] [PubMed] [Google Scholar]
- 7.Glucksmann MA, Markiewicz P, Malone C, Rothman-Denes LB. Cell. 1992;70:491–500. doi: 10.1016/0092-8674(92)90173-a. [DOI] [PubMed] [Google Scholar]
- 8.Falco SC, Zivin R, Rothman-Denes LB. Proc Natl Acad Sci USA. 1978;75:3220–3224. doi: 10.1073/pnas.75.7.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai X, Greizerstein M, Nadas-Chinni K, Rothman-Denes LB. Proc Natl Acad Sci USA. 1997;94:2174–2179. doi: 10.1073/pnas.94.6.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai X, Rothman-Denes LB. Genes Dev. 1998;12:2782–3172. doi: 10.1101/gad.12.17.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glucksmann-Kuis MA, Dai X, Markiewicz P, Rothman-Denes LB. Cell. 1996;84:147–154. doi: 10.1016/s0092-8674(00)81001-6. [DOI] [PubMed] [Google Scholar]
- 12.Davydova EK, Kazmierczak KM, Rothman-Denes LB. Methods Enzymol. 2003;370:83–94. doi: 10.1016/S0076-6879(03)70008-1. [DOI] [PubMed] [Google Scholar]
- 13.Willis MC, Hicke BJ, Uhlenkeck OC, Cech TR, Koch TH. Science. 1993;262:1255–1257. doi: 10.1126/science.7694369. [DOI] [PubMed] [Google Scholar]
- 14.Hirao I, Nishimura Y, Tagawa Y-I, Watanabe K, Miura K-I. Nucleic Acids Res. 1992;20:3891–3896. doi: 10.1093/nar/20.15.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirao I, Kawai G, Yoshizawa S, Nishimura Y, Ishido Y, Watanabe K, Miura K. Nucleic Acids Res. 1994;22:576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshizawa S, Kawai G, Watanabe K, Miura K, Hirao I. Biochemistry. 1997;36:4761–4767. doi: 10.1021/bi961738p. [DOI] [PubMed] [Google Scholar]
- 17.Santangelo TJ, Roberts JW. Methods Enzymol. 2003;371:120–132. doi: 10.1016/S0076-6879(03)71008-8. [DOI] [PubMed] [Google Scholar]
- 18.Dai X, Kloster M, Rothman-Denes LB. J Mol Biol. 1998;283:43–58. doi: 10.1006/jmbi.1998.2096. [DOI] [PubMed] [Google Scholar]
- 19.Cheetham GMT, Jeruzalmi D, Steitz TA. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 20.Zenkin N, Severinov K. Proc Natl Acad Sci USA. 2004;101:4396–4400. doi: 10.1073/pnas.0400886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasim MT, Eperon IC, Wilkins BM, Brammar WJ. Mol Microbiol. 2004;53:405–417. doi: 10.1111/j.1365-2958.2004.04114.x. [DOI] [PubMed] [Google Scholar]
- 22.Schröder O, Geiduschek EP, Kassavetis GA. Proc Natl Acad Sci USA. 2003;100:934–939. doi: 10.1073/pnas.242735699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studier FW. Science. 1972;176:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- 24.Markotsov V, Mustaev A, Goldfarb A. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]