Abstract
Recent studies indicate that the interaction between rabies virus (RV) phosphoprotein and the dynein light chain 8 (LC8) is essential for RV pathogenesis. Through its association with the dynein motor complex, LC8 has been suggested as a molecular factor that links the viral ribonucleoprotein to the host cell transport system. Recent structural investigations, however, dispute this model. To understand the role of LC8 in RV pathogenesis, we generated recombinant RVs with or without the LC8 binding domain (LC8-BD) deleted from the RV phosphoprotein. Peripheral infection of adult mice showed that removal of the LC8-BD did not inhibit entry into the CNS, although it prevented onset of RV-induced CNS disease. However, deletion of the LC8-BD significantly attenuated viral transcription and replication in the CNS. Studies in RAG2 knockout (KO) mice infected with the same recombinant RVs confirmed this finding and indicated that the adaptive immune system is not a factor in the attenuation of viral replication early in the infection. In cell culture, the deletion of the LC8-BD greatly attenuated growth on neuronal cells whereas the growth pattern on nonneuronal cells remained unchanged. However, deletion of the LC8-BD did not affect production of RV virions. We provide evidence that removal of the LC8-BD decreases primary transcription. In this study, we propose that LC8 does not play a role in the retrograde axonal transport of RV and that the deletion of the LC8-BD impairs the infectivity of the virions by reducing early transcription and replication in neurons.
Keywords: pathogenesis, negative-strand RNA viruses, rhabdovirus, viral transport
Rabies virus (RV) is a member of the family Rhabdoviridae that infects the CNS. RV has a relatively simple genome that encodes the five structural proteins: nucleoprotein (N), phosphoprotein (P), matrix (M), glycoprotein (G), and RNA polymerase (L). Pathogenic RV infection without immediate postexposure prophylaxis frequently causes fatal encephalomyelitis in both humans and animals and has the distinction of having the highest case-fatality ratio among infectious pathogens (1). Past experiments in mice have suggested that street strains of RV initiate infection at the nerve terminals of the peripheral nervous system (PNS) and then travel along the axons located in the spinal cord (2, 3). Consequently, this axonal transport leads to RV infection of the brain (4). Yet, the molecular basis for the transport of the RV particle remains unclear and controversial. Most transneuronal studies of the CNS in nonhuman primates (5, 6) and mice (7, 8) reveal that fixed (laboratory-adapted) strains of RV spread exclusively in the retrograde direction; however, past in vitro and in vivo studies suggest that anterograde transport might also be involved (7, 9–11). The discovery that the dynein light chain, LC8 (DNLC1), interacts with the RV phosphoprotein (RVP) provides a putative molecular link between the virus and host cell transport system and, thus, a potential mechanism for the neuroinvasive property of RV.
Originally identified as a subunit of cytoplasmic dynein motor complex, LC8 is a small (10.3 kDa) homodimer that binds a diverse set of proteins (12). This ability to bind to a large number of targets has led to the hypothesis that LC8 and the other dynein light chains might mediate cargo binding to the dynein motor (13, 14). Within the ≈1.2-MDa dynein motor complex, LC8 specifically interacts with an N-terminal region of the dynein intermediate chain (15). The intermediate chain (74 kDa) functions as a scaffold that links the different components of the dynein motor complex, which includes the heavy chains (530 kDa), the light intermediate chains (52–61 kDa), the p150glued subunit of dynactin, and the light chains (16). Structural studies indicate that most of these targets contain the same sequence as the dynein intermediate chain, KETQT (residues 132–136 of human intermediate chain 2C). Additional LC8 targets [e.g., neuronal nitric oxide synthase (nNOS)] encode a different sequence, TGIQV (residues 236–240 in nNOS). Structural data show that peptides bearing this sequence bind in an identical manner as the intermediate chain peptides (J.C.W., unpublished data). Because LC8 is a homodimer with two equivalent binding sites, it is possible to envision a mechanism where one site binds the intermediate chain and the other site binds to the cargo. However, recent thermodynamic measurements preclude this model (J.C.W., unpublished data). Specifically, the intermediate chain is dimeric under physiological conditions and, thus, acts as a bivalent target for the bivalent LC8. As such, substantial gains in binding affinity were observed for the dimeric target over the monomeric target. Consequently, LC8 does not directly mediate binding of cargo for retrograde transport. Consistent with these findings, a structural homolog of LC8, TcTex1, which binds VP26 protein of herpes simplex virus 1 and also has been implicated as a dynein adaptor, was found to be dispensable for the transport of herpes simplex virus 1 in vitro (17).
In light of these results, the role of LC8 in the RVP remains unclear. Previous studies indicate that the interaction of LC8 and the transcription factor tricho-rhino-phalangeal syndrome 1 releases its suppressor activity to promote the transcription of GATA consensus sequences (18). Likewise, the interaction of LC8 with P53 binding protein 1 regulates the nuclear distribution of p53-binding protein 1 and p53 (19). In fact, LC8 binds to several transcription factors, including nuclear respiratory factor 1 (20) and Drosophila swallow (21).
Herein, we present data indicating that the RVP–LC8 interaction is not directly involved in the retrograde axonal transport of RV from the periphery to the CNS. Rather, the RVP–LC8 has a significant role in the transcriptional activity of viral polymerase at the onset of infection in vivo and in vitro. This role was demonstrated in the infection of immunocompetent adult mice with recombinant RVs expressing RVP with and without the LC8 target sequence. Removal of the LC8 binding domain (LC8-BD) prevented mice from succumbing to RV-induced CNS disease but did not inhibit viral RNA dissemination into the CNS. Analysis of viral genomic and messenger transcripts showed viral replication attenuation in both the brain and the spinal cord in mice infected with the recombinant RV without the LC8-BD. Infection of immunocompromised RAG2 knockout (KO) mice showed a similar effect in mice infected with the mutant RV, which indicated that the adaptive immune system was not a factor in the attenuation. Detailed analysis using different cell lines reveals that deletion of the LC8-BD has a significant effect in de novo viral transcription. Thus, the LC8-BD in the RVP is important for efficient viral RNA polymerase activity, consistent with its role as a cofactor of the viral polymerase L.
Results
Construction of Recombinant RVs.
A number of studies suggest that microtubules and motor proteins play an important role in the intracellular replication cycle of viruses (22, 23). The only motor protein subunit found to associate with RV, thus far, is LC8 (24, 25). In fact, LC8 is incorporated into mature RV virions (26), providing additional evidence that LC8 might be involved in the attachment of the viral particle to the dynein motor complex on entry into the host cell. However, subsequent experiments in 7-day-old mice demonstrated that removing the LC8-BD simply delayed onset of disease, still allowing the transport of RV into the CNS (27, 28).
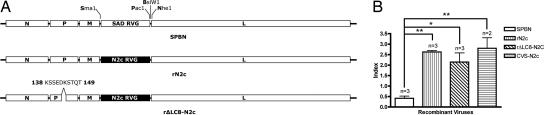
To further elucidate the role of the LC8-BD on the RVP in the axonal transport of RV and on CNS disease progression, we constructed several recombinant RVs that allowed us to analyze the function of LC8-BD in an adult mouse model. Because vaccine strains do not cause disease in immunocompetent adult mice when infected peripherally, we replaced the vaccine strain RV glycoprotein (RVG) of our vector SPBN with an RVG from a highly neurotropic and pathogenic strain, CVS-N2c (29) (rN2c) (Fig. 1A). To investigate the role of LC8, we deleted 11 aa from the RVP containing the LC8-BD as previously described by Mebatsion (27) (rΔLC8-N2c) (Fig. 1A).
Fig. 1.
Construction of recombinant RVs. (A) Recombinant RV SPBN (first letter of each restriction site) is a recombinant clone derived from the wildlife vaccine strain SAD-B19. The SPBN RVG was replaced with a glycoprotein from a highly neurotropic and pathogenic strain, CVS-N2c (rN2c). An additional recombinant clone was designed to examine the role of the LC8 binding site by removing 11 aa containing the LC8 binding site from RVP (rΔLC8-N2c). (B) Relative neuronal specificity index. Viral stocks of CVS-N2c, SPBN, rN2c, and rΔLC8-N2c were titered on both hamster BSR and mouse NA cells in parallel. One-way ANOVA followed by Tukey's test was used to determine the significance of differences between means (∗, P < 0.01; ∗∗, P < 0.001; error bars, standard deviations).
In Vitro Characterization of RVG Exchange Recombinant RVs.
The RVG is the sole viral protein on the surface of the RV envelope and is responsible for both receptor-mediated attachment to the cellular receptor and endosomal membrane fusion (30–32). We hypothesized that the substitution of the rN2c RVG on a vaccine backbone would confer cell tropism similar to that of its neurotropic parental strain, CVS-N2c (29). Thus, as shown in the relative neuronal specificity index of the recombinant RVs in Fig. 1B, substitution of the N2c RVG showed a significant increase in the tropism of rN2c and rΔLC8-N2c (Tukey's test; P < 0.001 and P < 0.01, respectively) for mouse neuroblastoma (NA) cell lines compared with hamster kidney (BSR) cell lines; similar results were obtained with CVS-N2c (Tukey's test; P < 0.001).
Deletion of the LC8-BD Prevents Disease.
The mouse-adapted strain, CVS-N2c, is pathogenic in adult mice infected in the periphery (29). Next, we determined whether the deletion of the LC8-BD would be sufficient for preventing disease and/or viral dissemination into the CNS of adult mice. As a control, we examined whether a recombinant RV based on a vaccine backbone that expressed the N2c RVG would have a phenotype similar to that of CVS-N2c. Therefore, ten 6- to 8-week-old Swiss–Webster mice were infected intramuscularly with 3.5 × 105 focus-forming units of SPBN, CVS-N2c, rN2c, or rΔLC8-N2c in the gastrocnemius muscle of the hind leg and were monitored daily for weight loss and CNS disease. We have previously shown that infection of adult mice with a recombinant vaccine strain does not cause disease (32), and SPBN-infected mice did not show symptoms of RV-induced encephalitis or weight loss as late as 24 days postinfection (dpi). Mice infected with CVS-N2c showed signs of disease as early as 5 dpi (data not shown) and all mice either succumbed to or were killed because of disease by 7 dpi. Mice infected with rN2c had a 70% mortality rate [supporting information (SI) Fig. 5], which demonstrated that the expression of an RVG from a more neurotropic and pathogenic strain of RV could partially confer the disease phenotype of the parental strain. Mice infected with rΔLC8-N2c did not show any signs of disease or weight loss, which indicates that the deletion of the LC8 is sufficient to prevent onset of disease in immunocompetent adult mice.
Deletion of the LC8-BD Does Not Prevent Viral Entry into the CNS.
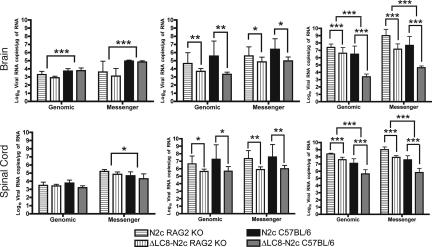
Next, we determined whether the absence of disease in rΔLC8-N2c-infected mice is the result of the failure of the virus to enter the CNS. A previous study in the peripheral infection of 7-day-old suckling mice revealed that deletion of the LC8-BD did not prevent viral invasion of the CNS or development of disease (28). Therefore, to correlate the presence of RV to disease, we used Northern blot analysis (data not shown) and real-time PCR to detect viral RNA from CNS samples. Fifteen 6- to 8-week-old C57BL/6 mice were infected with 3.5 × 105 focus-forming units of rN2c or rΔLC8-N2c in the gastrocnemius muscle of the hind legs. Mice were monitored daily for weight loss and individually scored for disease progression (SI Fig. 6). At 3, 6, and 8/9 dpi, five mice per group were randomly killed, their brains and spinal cords were harvested, and total RNA was extracted from the CNS homogenates. As shown in Fig. 2, deletion of the LC8-BD did not prevent viral entry of the CNS, because both viral genomic RNA and messenger RNA in rΔLC8-N2c-infected mice CNS were present at all time points.
Fig. 2.
Detection of viral RNA in the CNS. Fifteen 6- to 8-week-old RAG2 KO and C57BL/6 mice were infected with rN2c or rΔLC8-N2c in the gastrocnemius muscle of the hind legs. Five mice were randomly killed at 3 (Left), 6 (Center), or 8/9 (Right) dpi. Brain and spinal cord were harvested and total RNA was extracted from the CNS homogenates. RT-PCR and quantitative real-time PCR were used to amplify and quantify viral RNA from individual samples. Each bar represents the average of five mice and the error bars are the standard deviation. Two-way ANOVA was used to determine the significance of differences between means (∗, P < 0.01; ∗∗, P < 0.001; and ∗∗∗, P < 0.0001).
LC8-BD Deleted Recombinant RV Shows the Same Reduction in Replication Rate in Immunodeficient Mice.
Although deletion of the LC8-BD prevented disease, it did not inhibit viral entry into the CNS. To further investigate whether the absence of disease in rΔLC8-N2c-infected mice was due to either a reduction of replication of the rΔLC8-N2c in vivo or the presence of an adaptive immune system, we infected RAG2 KO mice with either rN2c or rΔLC8-N2c. Mice were analyzed daily for weight loss and disease progression and at 3, 6, and 9 dpi, five mice were randomly killed. Real-time quantitative PCR data indicated that deletion of the LC8-BD did not affect viral entry, nor did it significantly reduce viral replication at 3 dpi (Fig. 2 and SI Fig. 6). Of note is the fact that both viruses were significantly slower in reaching the brain in RAG2 KO mice at 3 dpi. However, at 6 dpi, removal of the LC8-BD significantly reduced the viral genomic and messenger RNA load in the CNS similarly in RAG2 KO and C57BL/6 mice (Fig. 2). By 9 dpi, however, rΔLC8-N2c- and rN2c-infected C57BL/6 mice had a significantly lower amount of viral RNA than rΔLC8-N2c- or rN2c-infected RAG2 KO mice, indicating an immune system role in viral clearance. Our in vivo data indicate that the presence of B and T cells is not a factor in the attenuation of viral transcription/replication in the CNS at the onset of infection but does have a role at a later time (Fig. 2). In the late stage of the disease, survival of immunocompetent mice infected with rΔLC8-N2c is due to slower viral replication and the mounting of an adaptive immune response, which is too late to be beneficial in mice infected with the faster-replicating rN2c.
Deletion of the LC8-BD Attenuates Growth on Neuronal Cell Lines.
In vivo data clearly indicated that deletion of the LC8-BD attenuates viral replication. In contrast, Mebatsion (27) had previously demonstrated that deletion of the dynein LC8-BD did not affect viral replication in nonneuronal BHK-1 cells. However, when murine neuroblastoma (NA) cells were infected synchronously at a multiplicity of infection (MOI) of 10, rΔLC8-N2c grew to 5- to 10-fold lower titers than rN2c (SI Fig. 7), whereas rN2c grew to titers identical to those of the parental strain, CVS-N2c. To confirm the decrease in viral titer, we infected several cell lines with rN2c or rΔLC8-N2c. We observed that the two viruses grew similarly on nonneuronal cell lines, such as BSR hamster kidney and 293T human kidney cells, whereas on neuronal cell lines such as NA or N2a, rΔLC8-N2c grew to a significantly lower titer than rN2c (SI Fig. 8).
Viral Protein Production Is Not Affected.
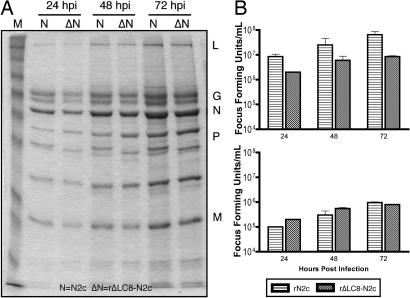
To determine whether the titer difference on neuronal cell lines was due to a defect in viral protein production, we infected NA cells with rN2c or rΔLC8-N2c at an MOI of 5. Supernatants were harvested at 24, 48, and 72 h postinfection (hpi), and 95% of the virions from the supernatant were purified over a 20% sucrose gradient. A small aliquot from the supernatant was used to determine the viral titers on neuronal and nonneuronal cells. Purified viral proteins were resolved by using SDS/12% PAGE and visually quantified by using Coomassie blue staining. In Fig. 3A, virion production over time was similar for the two viral strains and thus not affected by the deletion of the LC8 binding site. Of note, the infectious titers were identical when supernatants were titered on BSR cells, in contrast to the 5-fold different titers observed on NA cells (Fig. 3B). Furthermore, overall viral protein expression was also not affected by the deletion the LC8-BD (SI Fig. 9).
Fig. 3.
Analysis of viral protein production and infectious RV titers. (A) Viral protein production is not attenuated. NA cells were infected with rN2c or rΔLC8-N2c at an MOI of 5. At 24, 48, and 72 hpi, supernatants were harvested and RV particles were purified by centrifugation through 20% sucrose (see Materials and Methods). Viral structural proteins were visually quantified by Coomassie blue staining. (B) Viral attenuation in neuronal cell line supernatants harvested from NA cells from above were titered on NA (Upper) and BSR (Lower) in parallel.
Deletion of the LC8-BD Affects Infectious Titers.
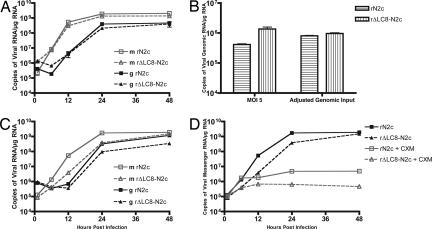
The disparity between neuronal and nonneuronal infectious titer units led us to focus on the amount of genome that enters the cell lines. Therefore, NA cells were infected with rN2c or rΔLC8-N2c at an MOI of 5, by using titers determined on the same cells. Infected cells were lysed at 0.5, 6, 12, 24, and 48 hpi and total RNA was then extracted. Quantification of viral genomic and messenger RNA by real-time PCR at 12, 24, and 48 hpi showed small differences between rN2c- and rΔLC8-N2c-infected NA cells (Fig. 4A). However, viral genomic input determined at 0.5 hpi indicated that there were more RV genomes in rΔLC8-N2c-infected cells at the onset of infection (Fig. 4B, MOI of 5). We therefore adjusted the infectious titers so that the amounts of viral genomes at 0.5 hpi were equal for the two recombinant RVs were equal (Fig. 4B, adjusted genomic input). In this setting, NA cells infected with rΔLC8-N2c had fewer copies of genome and messenger transcripts compared with rN2c-infected NA cells at 24 and 48 hpi (Fig. 4C). Thus, with a similar level of genome entering the cell, the ability of rΔLC8-N2c genomes to establish an infectious cycle was reduced.
Fig. 4.
Real-time PCR quantification of genomic input RNA and viral transcription of rN2c and rΔLC8-N2c. (A) RVN quantification in NA. NA cells were infected with rN2c or rΔLC8-N2c at an MOI of 5 and genomic RNA input was determined at 0.5 hpi followed by viral genomic (g) and messenger (m) RNA quantification at 6, 12, 24, and 48 hpi. (B) Viral genomic input. Titer from the “NA infectious titer” was adjusted to equal genomic input. (C) RVN quantification in NA. With viral genome input adjusted by the results in B, viral genomic and messenger RNA expression were quantified as in A at 6, 12, 24, and 48 hpi. (D) Primary transcription. Primary viral mRNA transcription was determined in the presence of cycloheximide (CXM), and viral messenger RNA expression was quantified at 6, 12, 24, and 48 hpi. Viral messenger RNA was amplified by RT-PCR and quantified by quantitative real-time PCR. All data are representative of one of three experiments.
Primary Transcription Is Attenuated.
An earlier study reported that deletion of the LC8-BD did not significantly affect viral polymerase activity in a minigenome assay (26). However, our in vivo and in vitro data suggest that viral polymerase replication is hampered in neuronal cell lines. To determine whether primary transcription is affected after the deletion of the LC8-BD from the RVP, we inhibited host and viral de novo protein synthesis with cycloheximide. Equal genomic input for both viruses was used, and primary transcription was determined at 0.5, 6, 12, and 48 hpi. Real-time PCR revealed that the transcriptional activity for the rΔLC8-N2c was significantly lower than rN2c (Fig. 4D), suggesting that the LC8-BD is necessary for efficient primary transcription.
Discussion
Previous reports indicated that viral RNA burden is a major determinant in the survival of an infected individual. For instance, patient records from the largest recorded outbreak of Ebola hemorrhagic fever revealed that survivors of the infection had, on average, two orders of magnitude lower amount of viral RNA copies in their blood than patients that succumbed to the disease (33). Additionally, increased amounts of viral RNA burden in the CNS are directly correlated with increased mortality in transgenic C57BL/6 mice infected intracranially with measles viruses (34). Thus, it is not surprising to observe that rΔLC8-N2c-infected mice with a low amount of viral RNA in the CNS did not progress to RV-induced CNS disease.
We initially thought that the absence of disease in mice infected with rΔLC8-N2c could be due to the failure of RV to enter the CNS. However, on closer examination of the CNS of mice infected with rΔLC8-N2c by using Northern blot analysis (not shown) and real-time PCR, we detected viral RNA in the CNS and thus concluded that LC8 or the LC8-BD of the RVP is not directly involved in the dynein-mediated transport of RV along the axons. In fact, we detected similar amounts of viral genomic and messenger RNA in mice infected with either recombinant RVs as early as 3 dpi. At 6 dpi, however, mice infected with rΔLC8-N2c had a significantly lower viral RNA burden in both the brain and spinal cord compared with rN2c in both immunocompetent and immunocompromised mice. This finding demonstrated two important observations: first, that deletion of the LC8-BD significantly lowered viral replication in both mouse strains in a similar fashion; and second, that the adaptive immune system did not play a role in the attenuation of viral replication at the onset of RV infection. Only with the mounting of the adaptive immune response combined with the slower replication of rΔLC8-N2c at 9 dpi did immunocompetent mice infected with rΔLC8-N2c have a significantly lower amount of viral RNA than RAG2 KO mice infected with the same recombinant RV. Our present mouse experiments suggest that LC8 or the LC8-BD of the RVP has a more crucial role in viral polymerase activity than in the intracellular transport of RV.
Given that the LC8 molecule has a diverse set of binding partners with different known functions, which include nNOS (35), two isomers of the Bcl-2 interacting molecule (BimXL/BIML) (36), Drosophila swallow gene (21), guanylate kinase domain-associated protein (37), the 3′-untranslated region of the parathyroid hormone mRNA (38), transforming growth factor β (TGF-β) (39), GTPase Rab4A (40), and other miscellaneous proteins (12), it is difficult to assess the exact role LC8 or the LC8-BD plays in RV pathogenesis.
Interestingly, we had also detected that recombinant RVs with an intact LC8-BD up-regulated intracellular LC8 mRNA (not shown) and protein levels (SI Fig. 9), whereas cells infected with rΔLC8-N2c did not. Thus, the up-regulation of LC8 could be an important factor in viral replication. As one of many innate immune effectors, nitric oxide synthase activity has been previously reported to be up-regulated during RV infection (41, 42) and is involved in the inhibition of RV RNA synthesis (43). Conversely, LC8 was originally reported to be a protein inhibitor of nNOS (35). Thus, we initially correlated the lower titers from rΔLC8-N2c-infected cells with its inability to induce the up-regulation of LC8 and inhibit nNOS activity. However, the presence of competitive nNOS inhibitors such as 7-nitroindazole or l-Nω-nitroarginine-2,4-l-diaminobutyric amide did not increase viral titers of rΔLC8-N2c as with the levels of rN2c-infected cells (data not shown). Consistent with recent reports, our data also indicate that LC8 does not inhibit nNOS activity (44, 45).
However, what is evident in the mouse studies is that the removal of the LC8-BD severely attenuated production of viral RNA transcripts in the CNS, contrary to what was found earlier in a minigenome assay (26). LC8s have additional binding targets that may provide clues to its regulatory role in viral polymerase activity. Interestingly, LC8 binds to several known factors that regulate transcriptional activities of genes. Interaction with the tricho-rhino-phalangeal syndrome 1 protein inhibits the suppressor activity of tricho-rhino-phalangeal syndrome 1 and increases the transcriptional activity of GATA-consensus sequences. LC8 was also shown to interact with nuclear respiratory factor 1, which has been shown to be involved in the regulation of cytochrome c expression and mitochondrial DNA transcription (20).
In the same way, we propose that LC8 or the LC8-BD is critical in the optimal activity of the viral RNA-dependent RNA polymerase and not involved in dynein-mediated viral transport. Data from cell culture experiments demonstrated that deletion of the LC8-BD decreased viral infectious titers in neuronal cell lines by 5- to 10-fold compared with nonneuronal cell lines, yet viral protein production was not affected. Quantification of viral RNA transcripts by real-time PCR at the onset of infection indicated that recombinant RVs lacking the LC8-BD were not as efficient.
Ultimately, we identified that primary viral mRNA transcription was severely decreased in neuronal cells infected with rΔLC8-N2c by inhibiting host and viral de novo protein synthesis. The following finding may perhaps explain the attenuation seen in vivo and the subsequent survival of mice infected with rΔLC8-N2c. Yet, the exact mechanism by which LC8 or the LC8-BD affects efficiency or processivity of the viral polymerase complex remains unanswered.
We know from other studies that there are cases in which a host molecule can modulate the transcriptional activity of a viral polymerase. For example, heat shock protein 72, an inducible member of the 70-kDa family of heat shock proteins, was previously shown to interact with the canine distemper virus ribonucleoprotein (46) and to increase polymerase activity (47). More recently, heat shock protein 72 was shown to interact with the nucleoprotein of measles viruses (48), enhance viral polymerase processivity, and subsequently increase mortality in mice, as well (34). Likewise, it is conceivable that LC8 may also regulate the processivity of the RV viral polymerase complex in a mechanism that has yet to be defined.
Materials and Methods
Construction of Recombinant RVs.
To construct a recombinant RV without the LC8 binding motif, two fragments were amplified by using Vent polymerase (New England Biolabs, Ipswich, MA) upstream and downstream of the 11-aa (138–149) stretch that contained the LC8 binding site of the RVP in SPBN. Primer pair RP 64 and RP 168 was used to amplify a 2.0-kb fragment upstream whereas primer pair RP 167 and RP 169 was used to amplify a 1.4-kb fragment downstream of the LC8 binding site. The two fragments were digested with XhoI and then ligated together, resulting in a 3.4-kb fragment containing an XhoI restriction site at the position of the dynein LC8-BD. The 3.4-kb fragment was digested with PstI and XmaI and cloned into SPBN (rΔLC8).
To replace the vaccine glycoprotein with one from CVS-N2c, the N2c RVG was amplified by using primer pair RP 13 and RP 185 from a plasmid encoding the N2c RVG. The 1.6-kb fragment was digested with XmaI and PacI and cloned into SPBN (rN2c) or rΔLC8 (rΔLC8-N2c). Infectious RVs were recovered as described in ref. 49.
Relative Neuronal Specificity Index.
Viral stocks of CVS-N2c, SPBN, rN2c, and rΔLC8-N2c were titered on both BSR (a derivative of BHK-21) and on NA cells, in parallel. Briefly, 10 μl of viral stock was added to 90 μl of either RPMI medium 10 (NA) or DMEM (BSR) and serially diluted (1:10) in a 96-well flat-bottom plate. Foci were detected by direct immunofluorescence with an anti-RV nucleoprotein (RVN) conjugate (Centecor, Jacksonville Beach, FL). The index was calculated by taking the log10 titer difference between the two cell lines (log10[NA] − log10[BSR]).
Mice.
Fifteen 6- to 8-week-old C57BL/6 and RAG2 KO mice (Taconic Farms, Germantown, NY) were infected intramuscularly with 3.5 × 105 focus-forming units of rN2c and rΔLC8-N2c in the gastrocnemius muscle. Mice were monitored and scored daily for weight changes and signs of rabies (0, none; 1, hunched back; 2, motor impairment; 3, one hind leg paralysis; 4, two hind leg paralysis; 5, moribund). At 3, 6, and 8/9 dpi (C57BL/6 infected with rN2c were killed at 8 dpi for humane reasons), five mice from each group were randomly killed. Serum, brain, and spinal cord were harvested for further analysis.
CNS Tissue Samples.
At 3, 6, and 8/9 dpi, brain and spinal cord tissue samples were removed from individual mice. CNS tissues were stored in either a 15-ml conical or a 1.5-ml Eppendorf tube (Eppendorf-5 Prime, Boulder, CO) and immediately flash frozen in liquid nitrogen. Brain and spinal cord samples were homogenized in TRIzol Reagent (Invitrogen, Carlsbad, CA) by using a 7-ml Ten Broeck tissue homogenizer (Corning, Acton, MA).
RNA Purification.
Total RNA from CNS tissue homogenates was isolated by using a combination of a phenol/chloroform protocol and the Qiagen RNeasy kit (Qiagen, Valencia, CA). In brief, choloroform was added to an aliquot of CNS tissue homogenate and shaken vigorously for 15 s. Samples were then incubated at room temperature for 10 min and spun at 12,000 × g for 15 min at 4°C. After centrifugation, the aqueous phase was harvested. Buffer RLT (Qiagen) was added to the harvested aqueous phase to make a volume of 700 μl. The rest of the RNA purification follows the Qiagen protocol.
Primer and TaqMan Probe Design.
All primers and probes for the quantitative real-time PCR were designed by the program Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) using as input the leader and RVN nucleotide sequence of the RV strain SAD B19 (GenBank accession no. M31046), which is identical to the nucleotide sequence of SPBN. The TaqMan probes were labeled at the 5′ end with FAM (6-carboxyfluorescein) as a reporter dye and the quencher TAMRA (6-carboxytetramethylrhodamine) at the 3′ end. The primers for generating a PCR amplicon containing both the target sequence for the RVN genomic and the messenger RNA primers were chosen manually.
cDNA Generation.
One microgram of total RNA of all brain and spinal cord samples was reverse-transcribed by using Omniscript reverse transcriptase (QIAGEN) according to the manufacturer's protocol and a gene-specific primer either for genomic SPBN RVN RNA (RP381) or for SPBN RVN messenger RNA (RP389). The reactions were incubated for 1 h at 37°C and the enzyme was subsequently inactivated by a 5-min step at 95°C. cDNAs were stored at −20°C.
Quantitative Real-Time PCR.
Virus loads in brains and spinal cords were determined by a TaqMan probe-based real-time PCR assay. Quantitative real-time PCRs were set up in LightCycler capillaries (Roche Applied Science, Indianapolis, IN). Each 20-μl reaction contained 1× DyNAmo Probe qPCR Master Mix (New England Biolabs), 500 nM forward primer, 500 nM reverse primer, 100 nM TaqMan probe, and 100 ng of template cDNA. Reactions were performed as triplicates in a LightCycler 1.5 instrument (Roche Applied Science) along with a positive control triplicate and a no-template control by using the following cycling program: 15 min at 95°C; 45 cycles (15 s at 95°C, 1 min at 60°C); 30 s at 40°C. The fluorescence of the hydrolyzed probes was measured in a single step at the end of each amplification cycle, and threshold cycles were obtained by the second derivative method through the LightCycler software ver. 3.5.3 (Roche Applied Science).
For absolute quantification, a standard curve was generated for SPBN RVN genomic RNA and SPBN RVN messenger RNA from serial dilutions of cDNA of a known copy number (SPBN genomic RVN, y = −3.1968x + 38.373, R2 = 0.9991; SPBN RVN mRNA, y = −3.0271x + 36.319, R2 = 0.9994) and the copy numbers were normalized to 1 μg/μl total RNA.
Virion Purification.
Murine NA cells were seeded in a T75 flask and infected with rN2c or rΔLC8-N2c at an MOI of 5. At 24, 48, and 72 hpi, total supernatants were harvested and the RV particles were purified by centrifugation through 20% sucrose (buffered in 0.1 M Tris·Cl, pH 8.0/0.01 M EDTA/1 M NaCl). Samples were then spun at 20,000 rpm (SW-28 rotor; Beckman Coulter, Fullerton, CA) for 1 h at 4°C and the supernatants were decanted. Viral particle pellets were suspended in 100 μl of 1× PBS, and 10 μl of each sample was resolved by SDS/10% PAGE. The gel was then incubated in Coomassie blue at room temperature for 1 h and washed overnight with destaining buffer to visualize the viral proteins. A 100-μl aliquot was removed from each sample before purification for titering on BSR and NA cells.
Primary Transcription.
NA cells were seeded onto six-well plates at a concentration of 3.0 × 105 cells per well and infected at an MOI of 5. Cells grew with or without the presence of cycloheximide (150 μg/ml) (50) to inhibit protein translation. At 0.5, 6, 12, 24, and 48 hpi, cells were lysed with RLT buffer and total RNA was isolated by using an RNeasy kit (Qiagen). Viral genomic and messenger RNAs were amplified and quantified by reverse transcription and quantitative PCR, respectively. Viral input was determined by the amount of viral genomic RNA at 0.5 hpi, and the infectious titer was adjusted accordingly to infect NA cells with equal viral inputs.
Statistics.
GraphPad Prism 4.0 software (GraphPad Software, San Diego, CA) was used to analyze data in this work. One-way ANOVAs followed by Tukey's tests were used to determine the significance of the relative neuronal specificity index. Real-time PCR data were analyzed by two-way ANOVAs to determine whether the deletion of the LC8-BD had a significant effect on viral RNA burden in the CNS within a mouse strain or had a significant effect on the difference between mouse strains.
Supplementary Material
Acknowledgments
This work was supported by a Center for Neuroanatomy and Neurotropic Viruses grant to Peter Strick (Center for Neuroscience, University of Pittsburgh) (USPHS-NCRR P40RR018604, subcontract to M.J.S.). G.S.T. was supported in part by National Institutes of Health Training Grant T32AI007492.
Abbreviations
- dpi
days postinfection
- G
glycoprotein
- hpi
hours postinfection
- KO
knockout
- L
RNA polymerase
- LC8
dynein light chain 8
- LC8-BD
LC8 binding domain
- M
matrix protein
- MOI
multiplicity of infection
- N
nucleoprotein
- NA
neuroblastoma
- nNOS
neuronal nitric oxide synthase
- P
phosphoprotein
- RNP
ribonucleoprotein
- RV
rabies virus
- RVG
RV glycoprotein
- RVN
RV nucleoprotein
- RVP
RV phosphoprotein.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701397104/DC1.
References
- 1.Hemachudha T, Laothamatas J, Rupprecht CE. Lancet Neurol. 2002;1:101–109. doi: 10.1016/s1474-4422(02)00041-8. [DOI] [PubMed] [Google Scholar]
- 2.Bulenga G, Heaney T. J Gen Virol. 1978;39:381–385. doi: 10.1099/0022-1317-39-2-381. [DOI] [PubMed] [Google Scholar]
- 3.Tsiang H. J Neuropathol Exp Neurol. 1979;38:286–299. doi: 10.1097/00005072-197905000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Jackson AC, Wunner WH, editors. Rabies. San Diego: Academic; 2002. [Google Scholar]
- 5.Kelly RM, Strick PL. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- 6.Kelly RM, Strick PL. J Neurosci. 2003;23:8432–8444. doi: 10.1523/JNEUROSCI.23-23-08432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y, Rampin O, Giuliano F, Ugolini G. J Comp Neurol. 1999;414:167–192. [PubMed] [Google Scholar]
- 8.Ugolini G. J Comp Neurol. 1995;356:457–480. doi: 10.1002/cne.903560312. [DOI] [PubMed] [Google Scholar]
- 9.Gillet JP, Derer P, Tsiang H. J Neuropathol Exp Neurol. 1986;45:619–634. doi: 10.1097/00005072-198611000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Jackson AC, Reimer DL. Acta Neuropathol (Berl) 1989;78:159–165. doi: 10.1007/BF00688204. [DOI] [PubMed] [Google Scholar]
- 11.Tsiang H, Lycke E, Ceccaldi PE, Ermine A, Hirardot X. J Gen Virol. 1989;70:2075–2085. doi: 10.1099/0022-1317-70-8-2075. [DOI] [PubMed] [Google Scholar]
- 12.Lo KW, Naisbitt S, Fan JS, Sheng M, Zhang M. J Biol Chem. 2001;276:14059–14066. doi: 10.1074/jbc.M010320200. [DOI] [PubMed] [Google Scholar]
- 13.Pfister KK. Structure (London) 2005;13:172–173. doi: 10.1016/j.str.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Tai AW, Chuang J-Z, Bode C, Wolfrum U, Sung C-H. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 15.Makokha M, Hare M, Li M, Hays T, Barbar E. Biochemistry. 2002;41:4302–4311. doi: 10.1021/bi011970h. [DOI] [PubMed] [Google Scholar]
- 16.Vale RD. Cell. 2003;112:467–468. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 17.Dohner K, Radtke K, Schmidt S, Sodeik B. J Virol. 2006;80:8211–8224. doi: 10.1128/JVI.02528-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser FJ, Tavassoli K, Van den Bemd GJ, Chang GT, Horsthemke B, Moroy T, Ludecke HJ. Hum Mol Genet. 2003;12:1349–1358. doi: 10.1093/hmg/ddg145. [DOI] [PubMed] [Google Scholar]
- 19.Lo KW, Kan HM, Chan LN, Xu WG, Wang KP, Wu Z, Sheng M, Zhang M. J Biol Chem. 2005;280:8172–8179. doi: 10.1074/jbc.M411408200. [DOI] [PubMed] [Google Scholar]
- 20.Herzig RP, Andersson U, Scarpulla RC. J Cell Sci 113 Pt. 2000;23:4263–4273. doi: 10.1242/jcs.113.23.4263. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Hare M, Hays TS, Barbar E. Biochemistry. 2004;43:4611–4620. doi: 10.1021/bi036328x. [DOI] [PubMed] [Google Scholar]
- 22.Radtke K, Dohner K, Sodeik B. Cell Microbiol. 2006;8:387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 23.Greber UF, Way M. Cell. 2006;124:741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Jacob Y, Badrane H, Ceccaldi PE, Tordo N. J Virol. 2000;74:10217–10222. doi: 10.1128/jvi.74.21.10217-10222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raux H, Flamand A, Blondel D. J Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poisson N, Real E, Gaudin Y, Vaney MC, King S, Jacob Y, Tordo N, Blondel D. J Gen Virol. 2001;82:2691–2696. doi: 10.1099/0022-1317-82-11-2691. [DOI] [PubMed] [Google Scholar]
- 27.Mebatsion T. J Virol. 2001;75:11496–11502. doi: 10.1128/JVI.75.23.11496-11502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasalingam P, Rossiter JP, Mebatsion T, Jackson AC. Virus Res. 2005;111:55–60. doi: 10.1016/j.virusres.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Morimoto K, Hooper DC, Carbaugh H, Fu ZF, Koprowski H, Dietzschold B. Proc Natl Acad Sci USA. 1998;95:3152–3156. doi: 10.1073/pnas.95.6.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wunner WH, Reagan KJ, Koprowski H. J Virol. 1984;50:691–697. doi: 10.1128/jvi.50.3.691-697.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitt MA, Buonocore L, Prehaud C, Rose JK. Virology. 1991;185:681–688. doi: 10.1016/0042-6822(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 32.Morimoto K, Foley HD, McGettigan JP, Schnell MJ, Dietzschold B. J Neurovirol. 2000;6:373–381. doi: 10.3109/13550280009018301. [DOI] [PubMed] [Google Scholar]
- 33.Towner JS, Rollin PE, Bausch DG, Sanchez A, Crary SM, Vincent M, Lee WF, Spiropoulou CF, Ksiazek TG, Lukwiya M, et al. J Virol. 2004;78:4330–4341. doi: 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carsillo T, Traylor Z, Choi C, Niewiesk S, Oglesbee M. J Virol. 2006;80:11031–11039. doi: 10.1128/JVI.01438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffrey SR, Snyder SH. Science. 1996;274:774–777. doi: 10.1126/science.274.5288.774. [DOI] [PubMed] [Google Scholar]
- 36.Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. Mol Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 37.Naisbitt S, Valtschanoff J, Allison DW, Sala C, Kim E, Craig AM, Weinberg RJ, Sheng M. J Neurosci. 2000;20:4524–4534. doi: 10.1523/JNEUROSCI.20-12-04524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Epstein E, Sela-Brown A, Ringel I, Kilav R, King SM, Benashski SE, Yisraeli JK, Silver J, Naveh-Many T. J Clin Invest. 2000;105:505–512. doi: 10.1172/JCI8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Q, Staub CM, Gao G, Jin Q, Wang Z, Ding W, Aurigemma RE, Mulder KM. Mol Biol Cell. 2002;13:4484–4496. doi: 10.1091/mbc.E02-05-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bielli A, Thornqvist PO, Hendrick AG, Finn R, Fitzgerald K, McCaffrey MW. Biochem Biophys Res Commun. 2001;281:1141–1153. doi: 10.1006/bbrc.2001.4468. [DOI] [PubMed] [Google Scholar]
- 41.Koprowski H, Zheng YM, Heber-Katz E, Fraser N, Rorke L, Fu ZF, Hanlon C, Dietzschold B. Proc Natl Acad Sci USA. 1993;90:3024–3027. doi: 10.1073/pnas.90.7.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hooper DC, Ohnishi ST, Kean R, Numagami Y, Dietzschold B, Koprowski H. Proc Natl Acad Sci USA. 1995;92:5312–5316. doi: 10.1073/pnas.92.12.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ubol S, Hiriote W, Anuntagool N, Utaisincharoen P. Virus Res. 2001;81:125–132. doi: 10.1016/s0168-1702(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 44.Hemmens B, Woschitz S, Pitters E, Klosch B, Volker C, Schmidt K, Mayer B. FEBS Lett. 1998;430:397–400. doi: 10.1016/s0014-5793(98)00704-2. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Crespo I, Straub W, Gavilanes F, Ortiz de Montellano PR. Arch Biochem Biophys. 1998;359:297–304. doi: 10.1006/abbi.1998.0928. [DOI] [PubMed] [Google Scholar]
- 46.Oglesbee M, Ringler S, Krakowka S. J Gen Virol. 1990;71:1585–1590. doi: 10.1099/0022-1317-71-7-1585. [DOI] [PubMed] [Google Scholar]
- 47.Oglesbee M, Liu Z, Kenney H, Brooks CL. J Gen Virol. 1996;77:2125–2135. doi: 10.1099/0022-1317-77-9-2125. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Bourhis JM, Longhi S, Carsillo T, Buccellato M, Morin B, Canard B, Oglesbee M. J Virol. 2005;337:162–174. doi: 10.1016/j.virol.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 49.Schnell MJ, Mebatsion T, Conzelmann KK. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, Gong X, Foley HD, Schnell MJ, Fu ZF. J Virol. 2002;76:4153–4161. doi: 10.1128/JVI.76.9.4153-4161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.