Abstract
In the cilia of the nematode Caenorhabditis elegans, anterograde intraflagellar transport (IFT) is mediated by two kinesin-2 complexes, kinesin II and OSM-3 kinesin. These complexes function together in the cilia middle segments, whereas OSM-3 alone mediates transport in the distal segments. Not much is known about the mechanisms that compartmentalize the kinesin-2 complexes or how transport by both kinesins is coordinated. Here, we identify DYF-5, a conserved MAP kinase that plays a role in these processes. Fluorescence microscopy and EM revealed that the cilia of dyf-5 loss-of-function (lf) animals are elongated and are not properly aligned into the amphid channel. Some cilia do enter the amphid channel, but the distal ends of these cilia show accumulation of proteins. Consistent with these observations, we found that six IFT proteins accumulate in the cilia of dyf-5(lf) mutants. In addition, using genetic analyses and live imaging to measure the motility of IFT proteins, we show that dyf-5 is required to restrict kinesin II to the cilia middle segments. Finally, we show that, in dyf-5(lf) mutants, OSM-3 moves at a reduced speed and is not attached to IFT particles. We propose that DYF-5 plays a role in the undocking of kinesin II from IFT particles and in the docking of OSM-3 onto IFT particles.
Keywords: cilia length, dyf-5, intraflagellar transport
Cilia are present on almost every vertebrate cell and have important functions in motility or sensation. Within cilia, structural components and signaling molecules are transported by a specialized system, called intraflagellar transport (IFT) (1–5). Transport from the base of the cilia to the tip (anterograde) is mediated by kinesin-2 motor complexes, whereas dynein motor complexes mediate transport back to the base (retrograde). The nematode Caenorhabditis elegans has 60 ciliated neurons, including eight pairs of amphid neurons exposed to the environment (6). The cilia of these neurons can be divided into a middle segment with nine doublet microtubules and a distal segment with nine singlet microtubules (7). In the middle segments, two distinct kinesin-2 motor complexes mediate anterograde transport, heterotrimeric kinesin II, encoded by klp-11, klp-20, and kap-1 and homodimeric OSM-3 kinesin (8). In the distal segments, transport is mediated by only OSM-3 (8). Live imaging of the movement of these kinesins suggests that kinesin II alone moves at 0.5 μm/s, and OSM-3 alone moves at 1.3 μm/s, whereas the two motor complexes together move at 0.7 μm/s (8). Recently, Pan et al. (9) have shown that these in vivo transport rates can be reconstituted in vitro by using purified kinesin II and OSM-3 motors. Thus far, two proteins have been identified that are required to stabilize IFT complexes transported by both kinesin II and OSM-3, BBS-7 and BBS-8 (10). However, it remains unclear how kinesin II is restricted to the cilia middle segments, whereas OSM-3 is allowed to enter the distal segments and what the functional significance is of this compartmentalization.
Recently, Evans et al. (11) have suggested that differences in the activities of the two kinesins contribute to morphological and functional differences between the cilia of individual neurons. Not much is known about the molecular mechanisms that control length and morphology of cilia, although it has been proposed that cilia length is controlled by a balance between cilia assembly and disassembly regulated by IFT (12–14). Genetic approaches have thus far identified three proteins involved in the regulation of cilia length: two Chlamydomonas reinhardtii proteins, LF1 and LF2, and the Chlamydomonas MAP kinase LF4 and its Leishmania mexicana homologue LmxMPK9 (15–18).
The identification of genes involved in IFT in C. elegans has profited significantly from the fact that a subset of C. elegans' amphid neurons exposed to the environment can take up fluorescent dyes via their cilia, a process called dye-filling (19). This phenomenon has allowed the identification of mutants with cilia defects, often caused by mutations in components of the IFT machinery (1, 2, 7, 20). Here, we describe the identification of the gene mutated in dyf-5 animals, a dye filling-defective (Dyf) mutant isolated previously in a forward genetic screen (20). dyf-5 encodes a predicted serine–threonine kinase homologous to three human MAP kinases with unknown functions, Chlamydomonas LF4 and Leishmania LmxMPK9. We found that mutations in dyf-5 affect cilia length and morphology. We also show that dyf-5(lf) affects the constitution and coordination of IFT particles. First, kinesin II can enter the distal segments in dyf-5(lf) mutants. Second, OSM-3 is separated from the IFT particles and moves at a reduced speed in dyf-5(lf) mutants. Third, dyf-5(lf) animals show accumulation of six different IFT proteins in the cilia.
Results
dyf-5 Encodes a Conserved MAP Kinase.
dyf-5 has been mapped to a small region of chromosome I (20). We identified dyf-5 as M04C9.5 using transgenic rescue and candidate gene sequencing (Fig. 1A). Independently, others have also identified the dyf-5 gene (21).
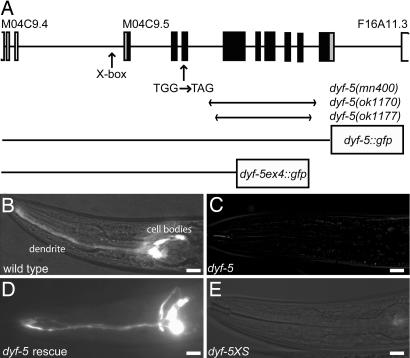
Fig. 1.
dyf-5 encodes a putative protein kinase. (A) Schematic representation of the dyf-5 gene structure (M04C9.5). Coding exons are indicated as black boxes; 5′ and 3′ UTR are depicted as gray boxes. Exons of the flanking genes are indicated as open boxes. The predicted X-box is indicated. Arrows indicate the position of the G-to-A substitution in dyf-5(mn400) animals, and the regions deleted in the ok1170 and ok1177 alleles. The two dyf-5::gfp fusion constructs have been indicated. (B–E) DiI dye-filling of wild-type (B), dyf-5(mn400) (C), dyf-5(ok1170) gjEx824(dyf-5) rescue strain (D), and dyf-5XS(gjEx817) (E) animals. Both dyf-5(lf) and dyf-5(XS) animals are dye-filling-defective. (Scale bars, 20 μm.) Anterior is to the left.
dyf-5(mn400) contains a G-to-A transition in exon 3 (Fig. 1A), which introduces a stop codon at amino acid position 49. In addition, we characterized two deletion alleles of dyf-5, ok1170 and ok1177, generated by the C. elegans gene knockout consortium. Both deletion alleles remove most of the dyf-5 coding region and are very likely null alleles (Fig. 1A). All three dyf-5(lf) alleles show Dyf phenotypes (Fig. 1 B and C and results not shown). Dye-filling could be restored by introducing low concentrations (5 ng/μl) of a 7.5-kb genomic fragment containing the M04C9.5 gene or a full-length dyf-5::gfp fusion construct in dyf-5(ok1170) and dyf-5(mn400) animals (Fig. 1D and results not shown), confirming that this gene indeed encodes dyf-5. Transgenic animals carrying high copy numbers of the dyf-5 gene (dyf-5XS, injected with 75 ng/μl) or the full-length dyf-5::gfp construct showed dye-filling defects (Fig. 1E and results not shown), suggesting that the levels of DYF-5 are important for its function.
Characterization of the dyf-5 gene structure using RT-PCR identified an additional upstream exon as well as some errors in the predicted M04C9.5 gene structure. We have communicated the confirmed dyf-5 gene structure to Wormbase (www.wormbase.org). dyf-5 encodes a protein of 471 aa, which shows extensive homology to three mammalian proteins that form a small subfamily of MAP kinases. This subfamily consists of MAK (male germ cell-associated kinase) (22), ICK or MRK (intestinal cell kinase and MAK-related kinase, respectively) (23, 24), and MOK (MAPK/MAK/MRK overlapping kinase) (25), with highly conserved N-terminal catalytic domains and more divergent C-terminal noncatalytic domains. Although the functions of these kinases in mammals are not known, studies in other organisms suggest a function in the cilia. Mutations in a DYF-5 homologue in Leishmania mexicana, LmxMPK9 (17), and in one of the two homologues in Chlamydomonas reinhardtii, LF4 (18), affect the length of the flagella of these organisms. In addition, several genome-wide approaches have identified DYF-5 and its homologues as likely ciliary proteins (21, 26–30).
dyf-5 Functions in Ciliated Neurons.
We examined the expression pattern of dyf-5 using two dyf-5::gfp fusion constructs: a full-length, functional dyf-5::gfp fusion and a construct in which GFP is fused in frame to the fourth exon of dyf-5 (dyf-5ex4::gfp) (Fig. 1A). The full-length dyf-5::gfp construct showed weak GFP expression in many neurons in the head, including amphid and labial sensory neurons (Fig. 2A) and three pairs of neurons in the tail, including the phasmid sensory neurons. In addition, we observed expression in many cells in the male tail (Fig. 2B). This DYF-5::GFP fusion could be detected uniformly in axons, cell bodies, and dendrites. In addition, we observed strong fluorescence at the transition zones, which connect the cilia with the dendrites, and weak fluorescence uniformly in the cilia (Fig. 2C). The dyf-5ex4::gfp fusion construct essentially showed the same dyf-5 expression pattern, albeit stronger and more restricted to the cell bodies. In addition, DYF-5ex4::GFP could be detected in the CAN cells, neurons associated with the excretory canal (6) and in a pair of neurons in the posterior lateral ganglion (results not shown).
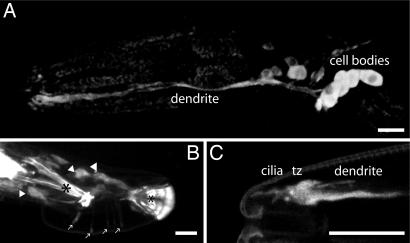
Fig. 2.
dyf-5::gfp expression pattern. (A) Expression of dyf-5::gfp in the dendrites and cell bodies of many neurons, including amphid sensory neurons in the head. (B) Strong dyf-5::gfp expression in many neurons in the male tail. Cell bodies are indicated with arrowheads, expression in the sensory rays is indicated with arrows, and autofluorescence of the spicule and the posterior end of the fan is indicated with asterisks. (C) Strong dyf-5::gfp expression in the tip of the head in dendrites and transition zone (tz) and weakly in cilia. [Scale bars, 20 μm (A and B) or 10 μm (C).] Anterior is to the left.
Expression of many ciliary genes is regulated by the DAF-19 transcription factor (31). This regulation occurs through an X-box promoter element. The dyf-5 promoter region contains a predicted X-box promoter element, 193 bp upstream of the SL1 trans splice site and 271 bp upstream of the translational start (Fig. 1A). Previous genomic analyses have identified many genes that contain X-box elements, including M04C9.5 (21, 26, 28). Chen et al. (21) have confirmed that expression of dyf-5 is indeed regulated by daf-19.
Mutation of dyf-5 Affects Cilia Morphology and Length.
We wondered whether mutations in dyf-5 affect cilia morphology and length, as in Chlamydomonas and Leishmania (17, 18). Visualization of the microtubular axonemes of the cilia using anti-tubulin antibodies revealed that the cilia of dyf-5(lf) animals were not properly aligned into the amphid pore. The cilia were more dispersed, misdirected, and sometimes turned back toward the transition zone (Fig. 3B). In contrast, in the two dyf-5XS strains, gjIs828 and gjIs831, we observed very weak anti-tubulin staining and only very short or no cilia structures (results not shown).
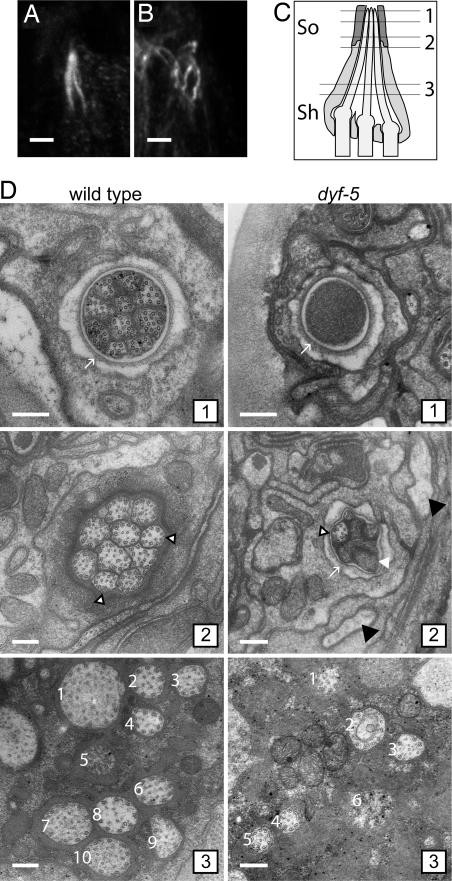
Fig. 3.
Mutation of dyf-5 affects cilia length and morphology. (A and B) Anti-tubulin immunostaining of wild-type (A) and dyf-5(ok1177) (B) animals. (Scale bar, 2 μm.) Anterior is up. (C) Schematic representation of three of the amphid channel cilia embedded in the sheath cell (sh) and the socket cell (so). The approximate positions of the EM cross-sections in D have been indicated. (D) EM cross-section of a wild-type (Left 1–3) and dyf-5(lf) (Right 1–3) animal (1). Sections through the socket cell (arrow), showing the distal segments of the 10 channel cilia that contain singlet microtubules in the wild-type animal and the channel filled with electron-dense material in the dyf-5(lf) animal (2). Ten channel cilia embedded in the sheath cell are present in the wild-type animal. Most cilia contain doublet microtubules; some contain singlets (open arrowheads). Section of a dyf-5(lf) animal through the socket cell (arrow), showing some cilia with singlet microtubules (open arrowhead) and some cilia filled with electron-dense material (white arrowhead). Black arrowheads indicate oblique sections through cilia (3). Sections through the middle segments close to the base of the cilia. In the wild-type animal, 10 channel cilia (numbered 1–10) embedded in the sheath cell are present, containing doublet microtubules. In the dyf-5(lf) animal, six channel cilia (1–6) embedded in the sheath cell could be identified, containing doublet microtubules. The cilia are more dispersed than in wild-type animals. (Scale bars, 200 nm.)
To study the morphological defects in more detail, we examined cross-sections of wild-type and dyf-5(lf) animals with EM. In the wild-type animal, the 10 cilia were enveloped by the sheath cell and projected to the tip of the animal, where the cilia ended in a channel lined by the socket cell (Fig. 3 C and D; refs. 7, 32, and 33). The microtubular axoneme in the middle segment consisted of a ring of nine doublet microtubules and up to six singlet microtubules (Fig. 3D) (7, 32, 33). In the tip of the cilium the nine microtubules were all singlets (Fig. 3D) (7, 32, 33). In the dyf-5(lf) mutant, the morphology of the cilia was severely affected. In the most proximal section, the cilia were more dispersed, consistent with the anti-tubulin data (Fig. 3D). We could not identify all 10 cilia, perhaps because of their dispersion and their altered morphology. In the cilia, the microtubular axoneme consisted of an array of singlet and doublet microtubules (Fig. 3D). More distally, we observed some cilia entering into the channel, indicating that the socket cell is present and open. At this level, the microtubules were singlets. Not all cilia entered the channel. Next to the socket cell, we observed two structures resembling oblique sections through cilia, which could be misdirected channel cilia (Fig. 3D). In the most distal sections, the channel was completely filled with electron-dense matrix material, probably accumulated proteins (Fig. 3D).
To study the effect of dyf-5(lf) on the morphology of individual amphid cilia, we examined transgenic animals expressing gpa-4::gfp or gpa-15::gfp. The Gα subunits gpa-4 and gpa-15 are expressed in only the ASI amphid neurons or in the ADL, ASH, and ASK neurons, respectively (34) and localized along the length of the cilia in wild-type animals (Fig. 4 A and J). Again, we observed that the cilia were more dispersed and misdirected. In addition, the cilia of dyf-5(lf) animals were significantly longer than cilia of wild-type animals (Fig. 4 B and K and Table 1). In contrast, in dyf-5XS animals, we observed very short cilia or, in 27% of the animals, no discernible cilia at all (Fig. 4 C and L and Table 1).
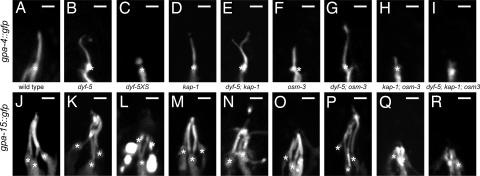
Fig. 4.
Mutation of dyf-5 affects cilia length. Visualization of ASI cilia structures using gpa-4::gfp (A–I) and ADL, ASH, and ASK cilia morphology using gpa-15::gfp (J–R) in wild-type (A and J), dyf-5(ok1177) (B and K), dyf-5XS(gjIs828) (C and L), kap-1(ok676) (D and M), dyf-5; kap-1 (E and N), osm-3(p802) (F and O), dyf-5; osm-3 (G and P), kap-1; osm-3 (H and Q), and dyf-5; kap-1; osm-3 (I and R) animals. In dyf-5, dyf-5; kap-1, and dyf-5; osm-3 animals, cilia were elongated and misdirected. In dyf-5XS animals, cilia were very short or could not be detected. We observed branching of the cilia in 30% of dyf-5; kap-1 animals. Transition zones are indicated with asterisks. (Scale bars, 2 μm.) Anterior is up.
Table 1.
Cilia lengths of dyf-5 mutant animals
| Genotype |
gpa-4::gfp |
gpa-15::gfp |
||
|---|---|---|---|---|
| Length, μm | n | Length, μm | n | |
| Wild type | 4.94 ± 0.57 | 60 | 6.83 ± 0.40 | 40 |
| dyf-5 | 5.66 ± 0.97* | 83 | 8.62 ± 0.86* | 30 |
| dyf-5XS(gjIs828) | 1.89 ± 0.49* | 18 | ND | |
| dyf-5XS(gjIs831) | 2.13 ± 0.66* | 15 | 4.56 ± 0.72* | 28 |
| kap-1 | 4.78 ± 0.24 | 23 | 6.79 ± 0.40 | 31 |
| dyf-5; kap-1 | 5.48 ± 1.11†‡ | 54 | 8.32 ± 1.23*‡ | 50 |
| osm-3 | 3.77 ± 0.32* | 24 | 4.89 ± 0.47* | 41 |
| dyf-5; osm-3 | 6.48 ± 0.86*§¶ | 43 | 8.70 ± 1.35*¶ | 47 |
| kap-1; osm-3 | 1.84 ± 0.31*‡¶ | 26 | ND | |
| dyf-5; kap-1; osm-3 | 1.86 ± 0.39*‡§¶ | 23 | ND | |
| dyf-5; gjEx312 | 4.96 ± 0.97‖ | 24 | ND | |
| dyf-5; gjEx324 | 4.83 ± 0.83§ | 22 | ND | |
Indicated are the average lengths of GPA-4::GFP and GPA-15::GFP staining cilia ±SD, the significance tested by using a two-tailed paired t test compared with wild-type animals (*, P < 0.001; †, P < 0.005); kap-1(‡, P < 0.001); dyf-5 (§, P < 0.001; ‖, P < 0.005); and osm-3(¶, P < 0.001), and the number of animals (n). The dyf-5XS cilia lengths indicated represent the lengths of the cilia where cilia could be seen; in an additional 27%, no cilia could be detected. gjEx312 and gjEx324 are two independent gpa-4::dyf-5::gfp strains that express dyf-5 only in the ASI neurons. ND, not determined.
The above-described approaches all showed similar effects of dyf-5(lf) on the length and morphology of the cilia. To determine whether dyf-5 regulates cilia length in a cell-autonomous fashion, we expressed the full-length dyf-5::gfp construct specifically in the ASI neurons of dyf-5(lf) animals and determined the lengths of the ASI cilia of these animals by using gpa-4::gfp. In two of the five transgenic lines that expressed dyf-5::gfp specifically in the ASI neurons, the cilia were significantly shortened, to approximately wild-type length (Table 1). These results indicate that dyf-5 functions cell-autonomously to regulate cilia length.
Kinesin II Localizes to the Distal Segments in dyf-5(lf) Animals.
Next, we wondered whether elongation of the cilia in dyf-5(lf) animals requires the function of either of the two anterograde motors, kinesin II or OSM-3. To this end, we generated dyf-5(lf); kap-1 and dyf-5(lf); osm-3 double mutants. osm-3 animals lack the cilia distal segments, because OSM-3 is essential for transport in these segments (8) (Fig. 4 F and O). kap-1 animals have lost an essential component of the heterotrimeric kinesin II complex but have full-length cilia, because OSM-3 can mediate IFT by itself (8) (Fig. 4 D and M).
Both dyf-5(lf); osm-3 and dyf-5(lf); kap-1 double-mutant animals were dye filling-defective (results not shown). Anti-tubulin antibody staining and gpa-4::gfp and gpa-15::gfp fluorescence showed that the cilia of both double mutants were as long as or even longer than the dyf-5(lf) cilia (Fig. 4 E, G, N, and P and Table 1). This suggests that both motors can transport cargo along the full length of the cilia and, thus, that DYF-5 is required to restrict kinesin II to the middle segments. In addition, these results suggest that the elongation of the cilia in dyf-5(lf) does not depend solely on either of the two kinesin-2 motors.
Interestingly, we observed an additional morphological defect of the cilia of dyf-5(lf); kap-1 animals: in 30% of the animals, the cilia were branched (Fig. 4E), a defect that was observed in only 7% of dyf-5(lf) animals and never in wild-type or kap-1 animals. In dyf-5(lf); osm-3 animals, branching was observed in only 1 animal of 33 analyzed. These results suggest that DYF-5 and kinesin II both play a role in cilium morphogenesis.
To determine whether transport in the cilia of dyf-5(lf) animals can be mediated by a kinesin II- and OSM-3-independent mechanism, we generated dyf-5(lf); kap-1; osm-3 triple-mutant animals. Visualization of the cilia using anti-tubulin antibodies and gpa-4::gfp and gpa-15::gfp showed that these triple mutants have no or only very short cilia, similar to kap-1; osm-3 animals (Fig. 4 H, I, Q, and R and Table 1), indicating that either kinesin II or OSM-3 is required for ciliogenesis.
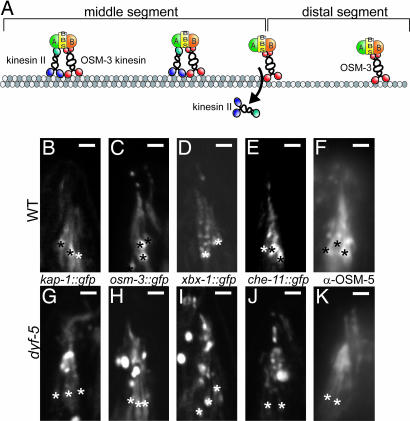
Because both kinesin-2 motors are individually dispensable for cilia elongation in dyf-5(lf) animals, we analyzed the localization of OSM-3::GFP and KAP-1::GFP fusion constructs in dyf-5(lf) animals (8). In wild-type animals, OSM-3::GFP could be detected along the length of the cilia, whereas KAP-1::GFP could be detected only in the cilia middle segment (Fig. 5 A–C), as reported (8). In dyf-5(lf) animals, OSM-3::GFP could be detected along the full length of the cilia. Additionally, we observed strong accumulation of OSM-3::GFP starting ≈2–3 μm from the transition zone and in the distal region of the cilia (Fig. 5H). Also KAP-1::GFP fluorescence could be seen along the full length of the cilia of dyf-5(lf) animals (Fig. 5G), confirming that DYF-5 is required to restrict KAP-1::GFP to the middle segments. We also observed accumulation of KAP-1::GFP halfway along the cilia. dyf-5XS animals showed both OSM-3::GFP and KAP-1::GFP fluorescence at the transition zones, but not at all or only very weakly in the cilia of these animals (results not shown), confirming that dyf-5 overexpression blocks cilia assembly.
Fig. 5.
DYF-5 is required to restrict kinesin II to cilia middle segments. (A) Model of anterograde IFT in C. elegans (based on model in ref. 10). In the cilia middle segments, cargo is transported by IFT particles that contain OSM-3 kinesin-2, kinesin II, complex A (A) and B (B) proteins and the BBS-7 and BBS-8 proteins (BBS). At the end of the middle segments, kinesin II is probably released from the IFT particle. Transport in the distal segment is mediated by OSM-3. (B–K) Visualization of KAP-1::GFP (B and G), OSM-3::GFP (C and H), XBX-1::GFP (D and I), CHE-11::GFP (E and J), and OSM-5 (F and K) in wild-type (B–F) and dyf-5(ok1170) (G–K) animals. All IFT proteins, including KAP-1::GFP, could be detected along the length of the cilia of dyf-5 animals. We observed accumulation of all IFT proteins, approximately halfway along the cilia and in the distal segment. Because these GFP constructs are expressed in all channel cilia, it was impossible to determine the length of a specific cilium. Transition zones are indicated with asterisks. (Scale bars, 2 μm.) Anterior is up.
To determine whether the localization of other IFT proteins was affected, we used a che-11::gfp fusion construct to visualize this complex-A IFT protein (35), a che-13::gfp fusion construct and anti-OSM-5 antibodies to visualize these two complex-B proteins (35–37), and xbx-1::gfp to visualize the dynein complex (38). All these IFT proteins could be detected along the length of the cilia of dyf-5(lf) animals, and they all showed accumulation starting approximately halfway along the cilia and into the distal segments (Fig. 5 D–F and I–K and results not shown). Also dyf-5(lf); kap-1 and dyf-5(lf); osm-3 double-mutant animals showed accumulation of CHE-11 and OSM-5 in the cilia (results not shown), suggesting that these effects of dyf-5(lf) do not depend on either kinesin II or OSM-3.
dyf-5(lf) Separates the OSM-3 Motor from IFT Particles.
Because DYF-5 is required to restrict kinesin II to the cilia middle segments, we wondered whether dyf-5(lf) affects the transport rates of the two kinesin-2 motors. In the middle segments of wild-type animals, kinesin II and OSM-3 traveled jointly at 0.7 μm/s (Table 2). In the absence of kinesin II OSM-3 moved faster (1.15 μm/s), and in the absence of OSM-3, kinesin II moved slower (0.5 μm/s; Table 2). In the distal segments, only OSM-3 is found, moving at a rate of 1.14 μm/s. These speeds are in agreement with previous reports (8). Examples of live imaging of CHE-11::GFP in wild-type and dyf-5(lf) animals and corresponding kymographs are given in supporting information (SI) and Movies 1 and 2.
Table 2.
dyf-5 separates the OSM-3 motor from IFT particles
| IFT particle | Genotype | Middle segment |
Distal segment |
||
|---|---|---|---|---|---|
| Speed, μm/s | n | Speed, μm/s | n | ||
| KAP-1::GFP | Wild type | 0.70 ± 0.14 | 189 | ||
| osm-3 | 0.49 ± 0.10* | 91 | |||
| dyf-5 | 0.50 ± 0.12* | 110 | 0.51 ± 0.16 | 56 | |
| OSM-3::GFP | Wild type | 0.70 ± 0.19 | 117 | 1.14 ± 0.23 | 43 |
| kap-1 | 1.17 ± 0.24* | 124 | 1.23 ± 0.28 | 42 | |
| dyf-5 | 0.59 ± 0.15*† | 210 | 0.59 ± 0.17*‡ | 84 | |
| dyf-5; kap-1 | 0.58 ± 0.15* | 91 | |||
| CHE-11::GFP | Wild type | 0.69 ± 0.20 | 118 | 1.20 ± 0.27 | 49 |
| osm-3 | 0.50 ± 0.09* | 90 | |||
| dyf-5 | 0.49 ± 0.11* | 170 | 0.49 ± 0.11* | 45 | |
| dyf-5; kap-1 | 0.60 ± 0.16*§ | 118 | 0.61 ± 0.12*§ | 38 | |
| dyf-5; osm-3 | 0.49 ± 0.12* | 109 | 0.49 ± 0.11* | 31 | |
| OSM-5::GFP | Wild type | 0.70 ± 0.17 | 136 | 1.13 ± 0.29 | 66 |
| osm-3 | 0.50 ± 0.11* | 88 | |||
| dyf-5 | 0.51 ± 0.11* | 151 | 0.50 ± 0.11* | 42 | |
| dyf-5; kap-1 | 0.59 ± 0.19*§ | 117 | 0.58 ± 0.12*§ | 45 | |
| dyf-5; osm-3 | 0.51 ± 0.13* | 103 | |||
| XBX-1::GFP | Wild type | 0.70 ± 0.17 | 141 | 1.11 ± 0.19 | 55 |
| dyf-5 | 0.50 ± 0.11* | 139 | 0.47 ± 0.08* | 42 | |
Indicated are average speeds ±SD and the number of tracks (n). Footnotes indicate significant differences when comparing speeds of: *, IFT protein in a mutant background with the same IFT protein in wild type (P < 0.001); † and ‡, OSM-3::GFP with the other IFT proteins in dyf-5(lf) animals (‡, P < 0.001; ‡, P < 0.01); and §, CHE-11 and OSM-5 in dyf-5; kap-1 with their speeds in dyf-5 and dyf-5; osm-3 animals. The speeds of all IFT proteins in dyf-5 and dyf-5; osm-3 animals, except OSM-3::GFP, were not significantly different from the speeds in osm-3 animals. Speeds in dyf-5; kap-1 animals were not significantly different from the speed of OSM-3::GFP in dyf-5 animals.
We found that, in the middle segments of dyf-5(lf) animals, kinesin II traveled at a rate of 0.5 μm/s, and OSM-3 traveled at 0.59 μm/s (Table 2). We found very similar speeds in the distal segments, although imaging was very difficult in these segments because of the twisted appearance of the cilia, often only partly in focus, and accumulation of the GFP fusion proteins. These results suggest that kinesin II and OSM-3 move at least partly separately in the cilia. Ou et al. (10) have recently described two mutants that cause separation of kinesin II and OSM-3-mediated transport: osm-12/bbs-7 and bbs-8. In these animals, kinesin II transports the complex-A proteins, and OSM-3 transports the complex-B proteins. To test whether a similar separation occurs in dyf-5(lf) animals, we determined the anterograde speeds of the complex-A protein CHE-11, the complex-B protein OSM-5, and the dynein subunit XBX-1. Interestingly, all these proteins moved at 0.5 μm/s in the middle and distal segments of the cilia of dyf-5(lf) animals (Table 2), a rate very similar to kinesin II.
The difference in transport rates of kinesin II, CHE-11, OSM-5, and XBX-1 on the one hand and OSM-3 on the other hand suggests that OSM-3 is no longer part of the joint IFT complex in dyf-5(lf) animals. However, the speed of OSM-3::GFP observed in dyf-5(lf) animals is lower than that of free traveling OSM-3 (Table 1) (8, 10). This effect can be explained by a reduction of the motility of OSM-3 itself. Alternatively, OSM-3 containing particles may be transported predominantly by the slower moving kinesin II, thereby reducing their average speed. To test the latter possibility, we measured transport rates of OSM-3::GFP, CHE-11::GFP, and OSM-5::GFP in dyf-5(lf); kap-1 and dyf-5(lf); osm-3 double-mutant animals. We found that, in dyf-5(lf); kap-1 animals, OSM-3, CHE-11, and OSM-5 moved at 0.59–0.60 μm/s (Table 2), whereas, in dyf-5(lf); osm-3 animals, CHE-11 and OSM-5 moved at 0.5 μm/s. These findings suggest that the speeds of OSM-3 and kinesin II in dyf-5(lf) animals are independent of each other and that these motors indeed move separately. Therefore, we favor the hypothesis that dyf-5(lf) reduces the speed of OSM-3 itself. In addition, these results confirm that, in the absence of kinesin II, IFT still can be mediated by OSM-3.
Discussion
In this article, we identify the gene mutated in dyf-5 animals. DYF-5 encodes a putative MAP kinase that regulates the length of sensory cilia in C. elegans, similar to what has been reported for DYF-5 homologues in Chlamydomonas (LF4) and Leishmania (LmxMPK9) (17, 18): dyf-5 loss-of-function results in cilia elongation, whereas overexpression results in shorter cilia. In addition, fluorescence and EM showed that, in the absence of DYF-5, the cilia are more dispersed and that many cilia are not properly aligned into the amphid channel and sometimes turn back to the base, although some cilia do enter the channel.
Within the cilia, we find three defects. First, six components of the IFT machinery are not properly localized in dyf-5(lf) animals. Interestingly, heterotrimeric kinesin II localizes to the distal segments of dyf-5(lf) animals. This is an intriguing finding, because this motor cannot enter this segment of the cilia in wild-type animals. In addition, all six IFT proteins tested accumulate approximately halfway and in the distal regions of the cilia, which correlates with the accumulation of protein seen in these regions by using EM. Second, in dyf-5(lf) animals, OSM-3-containing particles travel at a different speed than particles containing either kinesin II, complex-A and -B proteins, or dynein, suggesting that OSM-3 travels separately. In addition, this suggests that DYF-5 functions in a different process than the BBS-7 and BBS-8 proteins, mutations of which result in separation of complex-A and -B particles (10). Interestingly, OSM-3 can still mediate IFT in dyf-5(lf) animals but only in the absence of functional kinesin II. This suggests that inactivation of dyf-5 does not fully block docking of OSM-3 onto IFT particles, but rather changes the affinity between either of the two kinesins and the IFT particles. Third, the speed of OSM-3 in dyf-5(lf) animals was markedly reduced, from 1.2 μm/s to 0.6 μm/s. Our results suggest that this reduced speed is not a result of a combined speed of kinesin II and OSM-3, but rather an effect on OSM-3 itself.
But how does mutation of dyf-5 mediate these three effects on IFT, and how do these effects correlate with cilia morphology and length? A possible explanation for the effects on the localization of IFT proteins is that DYF-5 functions at sites where the IFT particles switch between different motor complexes, such as the transition from the middle to the distal segment and at the distal tip. Failure in removing kinesin II from IFT particles would explain its entry into the distal segments in dyf-5(lf) animals. In addition, a failure in switching from anterograde to retrograde transport would explain the aggregation of IFT proteins in the distal segments. However, such a model does not provide an explanation for the effects that dyf-5(lf) has on the speed of OSM-3. The motility of kinesins can be modulated at several levels. For example, the processivity of these motors, and hence their speed, may be reduced by interference with the properties of the kinesin itself, such as ATP binding and hydrolysis or allosteric inhibition of directional movement (39, 40). Also, posttranslational modifications of the microtubules on which the kinesins walk have been reported to affect the motility of kinesins (41). Perhaps DYF-5 functions in a similar manner, changing the affinity of both kinesins for its normal targets, either the IFT particles or the microtubules, resulting in the effects we observe. Interestingly, the entry of kinesin II in the distal segments or the presence of unattached OSM-3 by themselves do not cause the elongation of the cilia of dyf-5(lf) animals, because removal of kinesin II or OSM-3 in dyf-5(lf); kap-1 or dyf-5(lf); osm-3 animals did not suppress cilia elongation. It will be very interesting to further unravel the mechanisms by which DYF-5 regulates IFT, cilia morphology, and length.
The DYF-5 kinase is conserved in evolution. Using BLAST searches, we have identified putative DYF-5 homologues in various organisms that have cilia: in mammals, Drosophila, Chlamydomonas, Leishmania, and Trypanosoma (results not shown). Interestingly, we could not find an obvious homologue in Plasmodium, which has cilia, but the cilia are assembled in the cytoplasm in a process that does not require IFT (42). Mammals have three putative DYF-5 homologues that form a small subfamily of MAP kinases, the MAK kinases (22–25). The functions of these MAK, MOK, and ICK/MRK proteins remain to be determined. However, given the evolutionary conservation of the IFT machinery, the DYF-5/MAK kinases and their functions in C. elegans, Chlamydomonas, and Leishmania, we expect that one or more of the mammalian MAK kinases play a role similar to that of DYF-5 in the control of cilia length and the docking and undocking of kinesin-2 motors from IFT particles.
Materials and Methods
Strains and Constructs.
All strains were grown at 20°C. Strains and alleles used: Bristol N2 (wild type), dyf-5(mn400)I, dyf-5(ok1170)I, dyf-5(ok1177)I, kap-1(ok676)III, osm-3(p802)IV, dyf-5XS(gjIs828), and dyf-5XS(gjIs831). dyf-5::gfp reporter constructs were generated by using a fusion PCR protocol (43).
Microscopy dye-filling was performed by using DiI (Molecular Probes, Eugene, OR) (19). Immunofluorescence using monoclonal rat antibody against tubulin (ab6160; Abcam, Cambridge, U.K.) and monoclonal mouse antibody against OSM-5 (a kind gift from B. Yoder and C. Haycraft) was performed as described (44).
EM.
Animals were fixed in 3% glutaraldehyde for 16 h, followed by postfixation in 1% osmiumtetroxide for 2 h. Epon-embedded animals were cut and stained with uranyl acetate and lead nitrate.
Live Imaging of IFT Particles.
Live imaging of the GFP-tagged IFT particles was carried out as described (8). Kymographs were generated in ImageJ with the Kymograph plugin, written by J. Rietdorf. For additional information, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank E. Severijnen, T. de Vries Lentsch, and R. Koppenol for technical assistance; M. Barr (University of Wisconsin, Madison, WI), A. Fire (Stanford University, Stanford, CA), C. Haycraft, and B. Yoder (University of Alabama at Birmingham, Birmingham, AL) G. Ou and J. Scholey (University of California, Davis, CA), the Caenorhabditis Genetics Center, and the C. elegans Gene Knockout Consortium for C. elegans strains and constructs; M. Leroux for discussions; and the members of the G.J. laboratory for critical reading of the manuscript. This work was supported by the Center for Biomedical Genetics and the PKD Foundation. G.J. is a Royal Netherlands Academy of Sciences fellow.
Abbreviations
- IFT
intraflagellar transport
- lf
loss-of-function.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606974104/DC1.
References
- 1.Rosenbaum JL, Witman GB. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 2.Scholey JM. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 3.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. J Cell Biol. 2004;164:255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin H, Burnette DT, Bae YK, Forscher P, Barr MM, Rosenbaum JL. Curr Biol. 2005;15:1695–1699. doi: 10.1016/j.cub.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 5.Ou G, Qin H, Rosenbaum JL, Scholey JM. Curr Biol. 2005;15:R410–411. doi: 10.1016/j.cub.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 6.White JG, Southgate E, Thomson JN, Brenner S. Philos Trans R Soc London Ser B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 7.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 8.Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, Scholey JM. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 9.Pan X, Ou G, Civelekoglu-Scholey G, Blacque OE, Endres NF, Tao L, Mogilner A, Leroux MR, Vale RD, Scholey JM. J Cell Biol. 2006;174:1035–1045. doi: 10.1083/jcb.200606003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Nature. 2005;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- 11.Evans JE, Snow JJ, Gunnarson AL, Ou G, Stahlberg H, McDonald KL, Scholey JM. J Cell Biol. 2006;172:663–669. doi: 10.1083/jcb.200509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall WF, Rosenbaum JL. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dentler W. J Cell Biol. 2006;170:649–659. doi: 10.1083/jcb.200412021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan J, Snell WJ. Dev Cell. 2005;9:431–438. doi: 10.1016/j.devcel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Tan LW, Dentler WL, Lefebvre PA. J Cell Biol. 2003;163:597–607. doi: 10.1083/jcb.200307143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen RL, Tam LW, Lefebvre PA. Genetics. 2005;169:1415–1424. doi: 10.1534/genetics.104.027615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengs F, Scholz A, Kuhn D, Wiese M. Mol Microbiol. 2005;55:1606–1615. doi: 10.1111/j.1365-2958.2005.04498.x. [DOI] [PubMed] [Google Scholar]
- 18.Berman SA, Wilson NF, Haas NA, Lefebvre PA. Curr Biol. 2003;13:1145–1149. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- 19.Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- 20.Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL. Genetics. 1995;139:171–188. doi: 10.1093/genetics/139.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen N, Mah A, Blacque OE, Chu J, Phgora K, Bakhoum MW, Newbury CRH, Khattra J, Chan S, Go A, et al. Genome Biol. 2006;7:R126. doi: 10.1186/gb-2006-7-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsushime H, Jinno A, Takagi N, Shibuya M. Mol Cell Biol. 1990;10:2261–2268. doi: 10.1128/mcb.10.5.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Togawa K, Yan YX, Inomoto T, Slaugenhaupt S, Rustgi AK. J Cell Physiol. 2000;183:129–139. doi: 10.1002/(SICI)1097-4652(200004)183:1<129::AID-JCP15>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 24.Yang T, Jiang Y, Chen J. Biomol Eng. 2002;19:1–4. doi: 10.1016/s1389-0344(02)00002-3. [DOI] [PubMed] [Google Scholar]
- 25.Miyata Y, Akashi M, Nishida E. Genes Cells. 1999;4:299–309. doi: 10.1046/j.1365-2443.1999.00261.x. [DOI] [PubMed] [Google Scholar]
- 26.Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, et al. Curr Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 27.Colosimo M, Brown A, Mukhopadhyay S, Gabel C, Lanjuin A, Samuel A, Sengupta P. Curr Biol. 2004;14:2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 28.Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Development (Cambridge, UK) 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- 29.Pazour GJ, Agrin N, Leszyk J, Witman GB. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stolc V, Samanta MP, Tongprasit W, Marshall WF. Proc Natl Acad Sci USA. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swoboda P, Adler HT, Thomas JH. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- 32.Ward S, Thomson N, White JG, Brenner S. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- 33.Ware RW, Clark D, Crossland K, Russell RL. J Comp Neurol. 1975;162:71–110. [Google Scholar]
- 34.Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH. Nat Genet. 1999;21:414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- 35.Qin H, Rosenbaum JL, Barr MM. Curr Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- 36.Haycraft CJ, Swoboda P, Taulman PD, Thomas JH, Yoder BK. Development (Cambridge, UK) 2001;128:1493–1505. doi: 10.1242/dev.128.9.1493. [DOI] [PubMed] [Google Scholar]
- 37.Haycraft CJ, Schafer JC, Zhang Q, Taulman PD, Yoder BK. Exp Cell Res. 2003;284:251–263. doi: 10.1016/s0014-4827(02)00089-7. [DOI] [PubMed] [Google Scholar]
- 38.Schafer JC, Haycraft CJ, Thomas JH, Yoder BK, Swoboda P. Mol Biol Cell. 2003;14:2057–2070. doi: 10.1091/mbc.E02-10-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carter NJ, Cross RA. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 40.Kwok BH, Kaptein LC, Kim JH, Peterman EJG, Schmidt CF, Kapoor TM. Nat Chem Biol. 2006;9:480–485. doi: 10.1038/nchembio812. [DOI] [PubMed] [Google Scholar]
- 41.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Witman GB. Curr Biol. 2003;13:R796–R798. doi: 10.1016/j.cub.2003.09.047. [DOI] [PubMed] [Google Scholar]
- 43.Hobert O. BioTechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 44.Finney M, Ruvkun G. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.