Abstract
Dendritic spines are small, actin-rich protrusions on the surface of dendrites that receive the majority of excitatory synaptic inputs in the brain. The formation and remodeling of spines, processes that underlie synaptic development and plasticity, are regulated in part by Eph receptor tyrosine kinases. However, the mechanism by which Ephs regulate actin cytoskeletal remodeling necessary for spine development is not fully understood. Here, we report that the Rac1 guanine nucleotide exchange factor Tiam1 interacts with the EphB2 receptor in a kinase-dependent manner. Activation of EphBs by their ephrinB ligands induces the tyrosine phosphorylation and recruitment of Tiam1 to EphB complexes containing NMDA-type glutamate receptors. Either knockdown of Tiam1 protein by RNAi or inhibition of Tiam1 function with a dominant-negative Tiam1 mutant blocks dendritic spine formation induced by ephrinB1 stimulation. Taken together, these findings suggest that EphBs regulate spine development in part by recruiting, phosphorylating, and activating Tiam1. Tiam1 can then promote Rac1-dependent actin cytoskeletal remodeling required for dendritic spine morphogenesis.
Keywords: synapses, Rho GTPase, actin cytoskeleton, NMDA receptor
Most excitatory synaptic transmission in the brain occurs at dendritic spines, small actin-rich protrusions that extend from dendritic shafts (1). Spines are dynamic structures that undergo actin-dependent changes in size, shape, and number during development and in response to physiological stimuli, including hormonal fluctuations, neuronal activity, and learning (2). Alterations in spine morphology and density are also associated with a number of neurological disorders such as Down's syndrome and X-linked mental retardation (3). Because changes in spine morphology affect synaptic function (4), understanding the molecular mechanisms that regulate spine morphogenesis should provide insight into the processes of synaptic development and plasticity as well as brain function and disease.
In recent years, it has become clear that Eph receptor tyrosine kinases play critical roles in regulating spine morphogenesis (5). Ephs constitute the largest family of receptor tyrosine kinases, which can be subdivided into two classes (EphAs and EphBs) based on sequence similarity and ligand preference. EphAs bind predominantly to glycosylphosphatidylinositol-linked ephrinA ligands, whereas EphBs interact with transmembrane ephrinB ligands. During nervous system development, Ephs regulate cell migration, axon guidance, and topographic mapping (6). In addition, Ephs localize to synapses, where they control spine morphogenesis as well as synapse development, function, and plasticity (5). In particular, EphBs appear to promote spine formation and maturation (7–9), whereas EphAs induce spine retraction (10). Although it is clear that Ephs play a critical role in spine morphogenesis, the downstream pathways through which they exert their effects are not clear. It is possible that EphBs promote spine development in part by cooperating with NMDA-type glutamate receptors, which play a central role in regulating synapse development and plasticity (11). This idea is supported by the observation that ephrinB activation of EphBs induces the formation of a complex between EphB and NMDA receptors, resulting in the tyrosine phosphorylation of the NMDA receptor subunit NR2B and the potentiation of calcium influx through the NMDA receptor (12–14). In addition, accumulating evidence indicates that Eph receptors regulate spine morphogenesis by modulating the activity of Rho family GTPases (8, 10, 15, 16).
Rho GTPases, including Rac1, RhoA, and Cdc42, are key regulators of the actin cytoskeleton that play critical roles in spine morphogenesis. Rac1 promotes the development and maintenance of spines, whereas RhoA inhibits spine formation, stabilization, and elongation (17). Rho family GTPases regulate spine morphogenesis by functioning as molecular switches, cycling between an active GTP-bound state and an inactive GDP-bound state. In their active GTP-bound state, Rho GTPases interact with effector molecules, leading to downstream signal propagation. Rho GTPase activity is tightly controlled by guanine nucleotide exchange factors (GEFs), which activate Rho GTPases by catalyzing the exchange of GDP for GTP (18), and GTPase-activating proteins, which inhibit Rho GTPases by stimulating GTP hydrolysis (19). In addition to activating Rho GTPases, GEFs have also recently been shown to contribute to the signaling specificity of their target GTPases by interacting with scaffolding proteins that link Rho GTPases to specific downstream effector molecules (20–22).
We previously identified the Rac1-specific GEF Tiam1 as a critical mediator of NMDA receptor-dependent spine development (23). Tiam1 is a large, multidomain protein that is highly expressed in the nervous system (24) and whose Drosophila homolog, Still life, has been implicated in synaptic development (25, 26). We showed that Tiam1 is present in spines and is necessary for proper spine and synapse development (23). Tiam1 interacts with the NMDA receptor and is required for NMDA receptor-dependent spine formation. Tiam1 appears to link the NMDA receptor to spine development by activating particular Rac1-dependent signaling pathways that control actin cytoskeletal remodeling and protein synthesis (23). Tiam1 has also recently been shown to cooperate with the polarity protein PAR-3 in regulating spine morphogenesis (27).
Because EphBs form a complex with NMDA receptors and positively modulate their function (12–14), we hypothesized that Tiam1 might also play a role in regulating EphB-dependent spine morphogenesis. We show here that Tiam1 specifically interacts with EphB2. This interaction requires EphB2 kinase activity and is mediated by the PH-CC-Ex domain [consisting of a pleckstrin homology domain followed by a coiled-coiled (CC) domain and an adjacent region (Ex)] of Tiam1, which is critical for Tiam1 membrane localization and function (28). EphrinB activation of EphB receptors induces the phosphorylation and recruitment of Tiam1 to EphB complexes containing NMDA receptors. Furthermore, disruption of Tiam1 function with RNAi or a dominant-negative mutant of Tiam1 blocks ephrinB-induced spine formation. Taken together, our results suggest that EphB receptors regulate spine development in part by recruiting, phosphorylating, and activating Tiam1, which leads to Rac1-dependent actin remodeling required for spine formation. By functioning downstream of both EphB and NMDA receptors, Tiam1 may act as a convergence point to help integrate these activity-dependent and -independent signaling pathways during the development and remodeling of synaptic connections.
Results
Association of Tiam1 with EphB2.
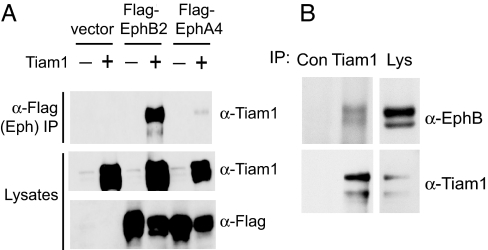
To investigate whether Tiam1 plays a role in EphB-mediated spine development, we first examined the possibility that Tiam1 might interact with EphB receptors. Tiam1 was tested for its ability to associate with EphB2 by transiently transfecting human embryonic kidney (HEK) 293T cells with expression vectors encoding Tiam1 and FLAG-tagged EphB2 or EphA4 and then immunoprecipitating the Eph receptors with an anti-FLAG antibody. When overexpressed in 293T cells, Ephs are constitutively active (12), presumably as a result of high expression levels, which leads to receptor oligomerization and activation. We found that Tiam1 efficiently coimmunoprecipitates with EphB2 but not with EphA4 (Fig. 1A), indicating that Tiam1 can specifically interact with EphB2 in cells.
Fig. 1.
Tiam1 interacts with EphB2. (A) Extracts of 293T cells transfected with Tiam1 in combination with empty vector, FLAG-EphB2, or FLAG-EphA4 were immunoprecipitated (IP) with anti-FLAG antibodies and then immunoblotted with anti-Tiam1 antibodies. The expression levels of Tiam1, EphB2, and EphA4 were confirmed by immunoblotting (Bottom). (B) Association of endogenous Tiam1 and EphB2. Lysates from P15 rat brain synaptosomes were immunoprecipitated with anti-Tiam1 antibodies or with control (anti-ZAP70) antibodies and then immunoblotted with anti-EphB2 or anti-Tiam1 antibodies. The presence of EphB2 and Tiam1 in the lysate (Lys) was confirmed with anti-EphB2 and anti-Tiam1 antibodies.
Because Tiam1 and EphB2 both localize to synapses and play a role in synaptic development, we next asked whether endogenous Tiam1 and EphB2 associate with one another in the brain at a time when synaptic connections are forming. Tiam1 was immunoprecipitated from synaptosomes purified from postnatal day 15 (P15) rat brains, and the immunoprecipitates were probed with anti-EphB2 antibodies. We found that EphB2 coimmunoprecipitates with Tiam1 but not with an unrelated protein (ZAP70) (Fig. 1B), demonstrating that endogenous Tiam1 and EphB2 specifically interact in the brain.
Colocalization of Tiam1 with EphB Complexes in Response to EphrinB Stimulation.
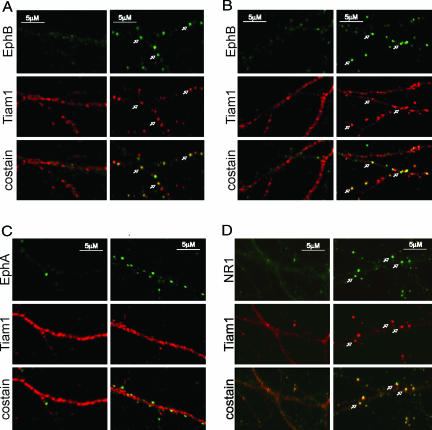
To investigate whether Tiam1 is recruited to EphB complexes in neurons after EphB receptor activation, we examined whether Tiam1 and EphB2 colocalize in embryonic day 18 (E18) rat hippocampal neurons (6 days in vitro; DIV6) upon exposure of neurons to aggregated ephrinB2-Fc (referred to as ephrinB2 stimulation). As reported previously (12), EphB receptors become tyrosine-phosphorylated and form clusters along dendrites in response to ephrinB stimulation (Fig. 2A). We found that ephrinB2 stimulation also induces the clustering of Tiam1 and that these Tiam1 clusters colocalize with EphB receptor clusters (Fig. 2A). Similar results were obtained when neurons were treated with ephrinB1-Fc (Fig. 2B). In contrast, the redistribution of Tiam1 was not detected when neurons were subjected to control (Fc) treatment (Fig. 2A) or stimulated with ephrinA1-Fc (Fig. 2C). This result indicates that Tiam1 is recruited to EphB complexes after EphB receptor activation in neurons.
Fig. 2.
EphrinB stimulation induces the colocalization of Tiam1 with EphB complexes. (A) Tiam1 clusters and colocalizes with EphBs upon ephrinB2 stimulation. E18 rat hippocampal neurons (DIV6) were treated with aggregated ephrinB2-Fc (Right) or control Fc (Left) for 60 min and were then fixed and stained for Tiam1 (red) and EphB (green). The white arrows indicate places where Tiam1 and EphB receptors colocalize. (B) EphrinB1 stimulation induces Tiam1-EphB colocalization. Neurons were stimulated with clustered ephrinB1-Fc (Right) or control Fc (Left) and then fixed and stained for Tiam1 (red) and EphB (green). (C) EphrinA1 stimulation fails to induce Tiam1–EphA colocalization. Neurons were stimulated with aggregated ephrinA1-Fc (Right) or control Fc (Left), fixed, and stained for Tiam1 (red) and EphA (green). (D) Tiam1 colocalizes with NMDA receptors upon ephrinB2 stimulation. E18 rat hippocampal neurons (DIV6) were stimulated with aggregated ephrinB2-Fc (Right) or control Fc (Left), fixed, and stained for Tiam1 (red) and the NR1 subunit of the NMDA receptor (green). The white arrows indicate places where Tiam1 and NR1 colocalize.
EphB activation has previously been shown to induce EphB/NMDA receptor complex formation in neurons, which is thought to contribute to excitatory synapse development and function (12, 13). Because Tiam1 interacts with both EphBs (Fig. 1) and NMDA receptors (23), we examined whether EphB activation results in the recruitment of Tiam1 to EphB complexes that contain NMDA receptors. We costained ephrinB-treated hippocampal neurons with anti-NR1 and anti-Tiam1 antibodies and found that Tiam1 colocalizes with the NR1 subunit of the NMDA receptor when neurons are exposed to ephrinB2-Fc (Fig. 2D) and ephrinB1-Fc (23) but not when cells are exposed to Fc alone. Taken together, these results indicate that EphB receptor activation induces the recruitment of Tiam1 to EphB complexes containing the NMDA receptor.
Characterization of the Tiam1–EphB2 Interaction.
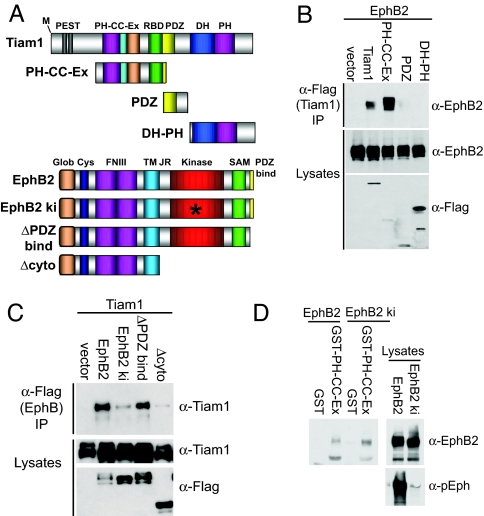
To characterize further the nature of the association between Tiam1 and EphB2, we defined the domains of each protein that are required for the interaction. Tiam1 contains several protein–protein interaction domains, including two PEST sequences, a PH-CC-Ex domain, a Ras-binding domain, a PDZ domain, and a catalytic Dbl homology (DH)-PH domain (Fig. 3A). To map the region of Tiam1 that binds to EphB2, we investigated the ability of EphB2 to coimmunoprecipitate with different FLAG-tagged Tiam1 constructs expressed in 293T cells. We found that the PH-CC-Ex domain of Tiam1 was both necessary and sufficient for binding to EphB2 (Fig. 3B). In contrast, EphB2 was unable to interact with Tiam1 constructs containing only the PDZ or DH-PH domain of Tiam1. The PH-CC-Ex domain has previously been shown to be essential for recruiting Tiam1 to the plasma membrane where it is active (28). It is possible that one way in which the PH-CC-EX domain recruits Tiam1 to the membrane is by binding to activated EphB receptors.
Fig. 3.
Characterization of the Tiam1–EphB2 interaction. (A) Schematic representation of Tiam1 and EphB2 constructs used in this work. RBD, Ras-binding domain. (B) The Tiam1 PH-CC-Ex domain mediates the interaction with EphB2. EphB2 was cotransfected into 293T cells alone or in combination with one of the following FLAG-tagged Tiam1 constructs: full-length Tiam1, a PH-CC-Ex construct and a Ras-binding domain, a PDZ domain, or the catalytic DH-PH domain. The Tiam1 constructs were immunoprecipitated with anti-FLAG antibodies, and EphB2 coprecipitation was assessed by using anti-EphB2 antibodies. The expression levels of each construct were evaluated by immunoblotting. (C) Tiam1 interacts with the cytoplasmic domain of EphB2 in a kinase-dependent manner. Tiam1 was transiently transfected into 293T cells with one of the following FLAG-tagged EphB2 receptor constructs: full-length EphB2, a kinase-inactive mutant (EphB2 ki), an EphB2 receptor lacking the PDZ-binding domain (ΔPDZ bind), or an EphB2 receptor lacking the entire cytoplasmic domain (Δcyto). The EphB2 receptor constructs were immunoprecipitated (IP) with anti-FLAG antibodies and then immunoblotted with anti-Tiam1 antibodies. The expression levels of each construct were confirmed by immunoblotting. (D) The isolated Tiam1 PH-CC-Ex domain interacts with EphB2 in a kinase-independent manner. Control GST or a GST fusion protein of the Tiam1 PH-CC-Ex domain immobilized on GSH beads was incubated with 293T cell lysate expressing EphB2 or EphB2 ki. Bound proteins were analyzed by immunoblotting with an anti-EphB2 antibody. The levels and activity of the EphB2 receptor constructs in the lysate were assessed with anti-EphB2 and anti-pEphB2 antibodies.
To determine the region of EphB2 that mediates its interaction with Tiam1, FLAG-tagged mutant EphB2 receptor constructs (Fig. 3A) were tested for their ability to associate with Tiam1 in 293T cells. As mentioned previously, EphB2 is constitutively active when overexpressed in 293T cells. We found that Tiam1 coimmunoprecipitated with full-length active EphB2 and an EphB2 receptor mutant lacking the PDZ-binding domain (ΔPDZ bind) but failed to coimmunoprecipitate with the EphB2 receptor mutants lacking kinase activity (EphB2 ki) or the cytoplasmic domain (Δcyto) (Fig. 3C). This result indicates that Tiam1 interacts with the cytoplasmic domain of EphB2 and that this interaction requires EphB2 kinase activity.
The ability of activated EphB2 to interact with Tiam1 could be due to EphB2 autophosphorylation, resulting in a conformation change and/or the generation of phosphotyrosine-binding sites within the EphB2 cytoplasmic domain that allow Tiam1 or a Tiam1-associated protein to bind. Alternatively, by phosphorylating Tiam1, EphB2 may trigger a conformational change in Tiam1, thereby facilitating the Tiam1–EphB2 interaction. To begin to distinguish between these possibilities, we asked whether the interaction between the isolated PH-CC-Ex domain of Tiam1 and EphB2 was kinase-dependent. GST and a GST fusion protein of the PH-CC-Ex domain of Tiam1 produced in Escherichia coli were bound to GSH beads and then incubated with the lysate of 293T cells overexpressing active wild-type or kinase-inactive EphB2. We found that the isolated PH-CC-Ex domain of Tiam1 effectively binds to both EphB2 and EphB2 ki, whereas GST alone fails to interact with either EphB2 construct (Fig. 3D). Given that the interaction between EphB2 and full-length Tiam1 requires EphB2 kinase activity, this result suggests that active EphB2 may induce tyrosine phosphorylation of full-length Tiam1, leading to a conformational change that reveals the Tiam1 PH-CC-Ex domain, allowing it to bind to EphB2.
EphB Activation Induces Tiam1 Tyrosine Phosphorylation.
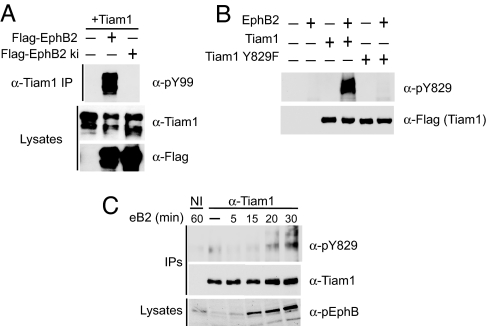
We next examined whether EphBs phosphorylate and modulate Tiam1 function. Tyrosine phosphorylation of Tiam1 has recently been shown to regulate Tiam1 by enhancing its GEF activity toward Rac1 (29, 30). To investigate whether Tiam1 is a substrate for EphB2 or an EphB2-associated kinase, we coexpressed Tiam1 in 293T cells with a FLAG-tagged wild-type or kinase-inactive EphB2 receptor and then immunoprecipitated Tiam1 and assessed Tiam1 tyrosine phosphorylation by using a phosphotyrosine-specific antibody (pY99). We found that Tiam1 was tyrosine-phosphorylated only when coexpressed with wild-type EphB2 (Fig. 4A), suggesting that EphB2 or a kinase that associates with activated EphB2 phosphorylates Tiam1.
Fig. 4.
EphrinB stimulation induces Tiam1 tyrosine phosphorylation. (A) HEK 293T cells were cotransfected with Tiam1 and FLAG-tagged wild-type or kinase-inactive EphB2 receptor, and then Tiam1 was immunoprecipitated (IP) and assessed for tyrosine phosphorylation by using a phosphotyrosine-specific antibody (pY99). Tiam1 and EphB2 receptor expression levels were confirmed with anti-Tiam1 and anti-FLAG antibodies, respectively. (B) Generation of the site-specific phospho-Tiam1 antibody, pY829. FLAG-tagged wild-type and Y829F Tiam1 were expressed in 293T cells alone or in combination with EphB2. The Tiam1 constructs were immunoprecipitated with anti-FLAG antibodies and then examined for phosphorylation on tyrosine-829 by using the phospho-specific Tiam1 antibody, pY829. The failure of the anti-pY829 Tiam1 antibody to recognize Tiam1 Y829F coexpressed with EphB2 suggests that the antibody is site-specific. (C) Tyrosine phosphorylation of Tiam1 in neurons upon ephrinB2-Fc stimulation. E18 hippocampal neurons (DIV4) were treated with clustered ephrinB2-Fc or control Fc for the indicated times, and then Tiam1 was immunoprecipitated with an anti-Tiam1 antibody. Tyrosine phosphorylation of Tiam1 was analyzed by using the anti-pY829 Tiam1 antibody. Immunoprecipitated Tiam1 was visualized with the anti-Tiam1 antibody, and EphB2 receptor autophosphorylation was detected in the lysate by using anti-phospho-EphB2 antibodies.
A detailed analysis of the amino acid sequence of Tiam1 (http://scansite.mit.edu) revealed the existence of several putative tyrosine phosphorylation sites on Tiam1. We generated a phospho-specific antibody to the most highly predicted site (tyrosine-829) and found that this antibody (pY829) recognizes Tiam1 when it is phosphorylated on tyrosine-829 after coexpression with EphB2 in 293T cells (Fig. 4B). This phospho-specific antibody failed to recognize Tiam1 coexpressed with EphB2 when tyrosine-829 was converted to phenylalanine, suggesting that the antibody is site-specific.
To examine whether activated EphBs induce Tiam1 phosphorylation in neurons, we stimulated hippocampal neurons with clustered ephrinB2-Fc or Fc alone for the indicated times and then immunoprecipitated Tiam1 from neuronal homogenate with an anti-Tiam1 antibody. Tyrosine phosphorylation of Tiam1 was then analyzed by using the anti-pY829 Tiam1 antibody. We found that ephrinB2-Fc treatment of hippocampal neurons increases the phosphorylation of Tiam1 on tyrosine-829, which peaks at ≈30 min, the time when EphB activation is maximal (Fig. 4C). Interestingly, phosphorylation of Tiam1 on tyrosine-829 by the TrkB receptor has recently been reported to be essential for Tiam1 activation and brain-derived neurotrophic factor-induced neurite formation (30). Our results indicate that Tiam1 is tyrosine-phosphorylated on tyrosine-829 in neurons in response to EphB receptor activation, which likely results in a change in Tiam1 activity and/or localization.
Role for Tiam1 in EphB-Dependent Spine Development.
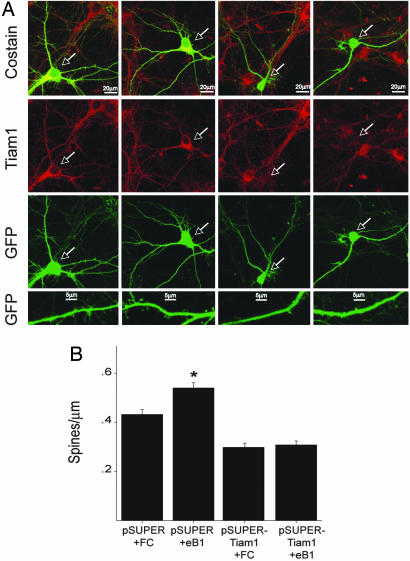
Taken together, our results suggest the following possibility. When EphB receptors are activated by ephrinBs after cell–cell contact, Tiam1 is phosphorylated and recruited to EphB complexes containing NMDA receptors. Tiam1 may then activate Rac1, leading to actin cytoskeletal remodeling required for spine development. To investigate whether Tiam1 plays a role in EphB-dependent spine morphogenesis, we assessed the effects of knocking down Tiam1 expression on ephrinB-induced spine development. Knockdown of Tiam1 protein was accomplished by using the plasmid-based pSUPER RNAi system (Fig. 5A) that specifically inhibits Tiam1 expression in neurons without affecting the levels of other neuronal proteins (23). We demonstrated previously that selectively blocking Tiam1 function with either RNAi knockdown or dominant-negative Tiam1 mutants reduces spine density and inhibits NMDA receptor-dependent spine formation (23). To assess the role of Tiam1 in EphB-dependent spine development, we transfected E18 hippocampal neurons (DIV5) with pSUPER or pSUPER-Tiam1 RNAi in combination with an enhanced green fluorescent protein (EGFP) expression vector. Nine days after transfection (DIV14), neurons were stimulated with clustered ephrinB1-Fc or Fc for 4 h and then fixed and imaged for dendritic spine density. We found that stimulating control pSUPER-expressing hippocampal neurons with clustered ephrinB1 promotes new spine development (Fig. 5), as has been reported previously (8). Quantification revealed that the average spine density of ephrinB1-Fc-treated pSUPER-expressing neurons (0.54 ± 0.02 spines per μm) was significantly larger than that of control Fc-treated pSUPER-expressing neurons (0.43 ± 0.02 spines per μm; P < 0.0001). Tiam1 appears to be required for this ephrinB1-induced increase in spine density because knockdown of Tiam1 expression significantly reduced spine density in Fc-treated neurons expressing pSUPER-Tiam1 RNAi (0.30 ± 0.02 spines per μm; P < 0.0001) and blocked ephrinB1-induced spine growth in pSUPER-Tiam1 RNAi-expressing neurons stimulated with ephrinB1 (0.31 ± 0.02 spines per μm). These results suggest that Tiam1 plays a role in EphB receptor-mediated spine development.
Fig. 5.
RNAi knockdown of Tiam1 expression blocks ephrinB1-induced spine development. (A) Dissociated hippocampal neurons (DIV5) were transfected with an EGFP expression vector together with the pSUPER control vector (columns 1 and 2) or the pSUPER-Tiam1 RNAi vector (columns 3 and 4). Nine days later (DIV14), neurons were stimulated for 4 h with clustered ephrinB1-Fc (eB1) (columns 2 and 4) or control Fc (columns 1 and 3) and then fixed and subjected to immunofluorescence. Tiam1 expression was specifically reduced in neurons transfected with the pSUPER-Tiam1 RNAi: vector (as indicated by the white arrows) but not in neighboring untransfected cells or in neurons transfected with the control pSUPER vector. Suppression of Tiam1 expression inhibited ephrinB1-induced spine development, as can be seen in the bottom row of panels. (B) Quantification of the effect of decreased Tiam1 expression on ephrinB1-induced spine development. ∗, P < 0.001, Student's t test.
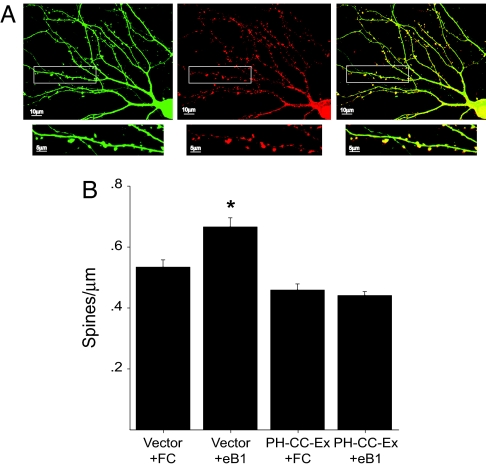
To confirm by an independent approach a role for Tiam1 in EphB receptor-dependent spine development, we used a dominant-negative mutant of Tiam1 to inhibit Tiam1 function. The isolated Tiam1 PH-CC-Ex domain has previously been shown to act in a dominant-negative manner (28, 31), presumably by binding to Tiam1-interacting proteins and blocking the recruitment of endogenous Tiam1 to the plasma membrane where it is active. Because the Tiam1 PH-CC-Ex domain mediates the binding of Tiam1 to EphB2, overexpression of this domain in neurons might also be expected to disrupt the endogenous Tiam1–EphB2 interaction. These effects will likely occur at spines because we find that the isolated PH-CC-Ex domain of Tiam1 localizes to spines when expressed in hippocampal neurons (Fig. 6A). To determine the effects on EphB-dependent spine development of blocking Tiam1 function with the PH-CC-Ex domain, we transfected E18 hippocampal neurons (DIV5) with an EGFP expression vector in combination with a plasmid encoding the myc-tagged Tiam1 PH-CC-Ex domain or an empty vector as control. Nine days after transfection (DIV14), the neurons were stimulated with clustered ephrinB1-Fc or Fc alone, fixed, and imaged for spine density. Consistent with our RNAi studies, we found that the increase in spine density induced by ephrinB1 stimulation of control neurons is blocked in neurons expressing the Tiam1 PH-CC-Ex domain (Fig. 6B). Control neurons stimulated with Fc alone possessed ≈0.53 ± 0.02 spines per μm, whereas control neurons treated with clustered ephrinB1-Fc had a significantly larger spine density (0.67 ± 0.03 spines per μm; P < 0.001). In contrast, ephrinB1 stimulation failed to increase the spine density of neurons overexpressing the Tiam1 PH-CC-Ex domain (0.46 ± 0.02 versus 0.44 ± 0.1 spines/μm). These findings indicate that disrupting Tiam1 function with the isolated Tiam1 PH-CC-Ex domain blocks ephrinB-induced spine growth, and confirms a role for Tiam1 in EphB-mediated spine morphogenesis.
Fig. 6.
Overexpression of the Tiam1 PH-CC-Ex domain inhibits EphB-dependent spine development. (A) Hippocampal neurons expressing EGFP and myc-tagged PH-CC-Ex domain were fixed and stained for myc. The Tiam1 PH-CC-EX domain (Center) appears to preferentially localize to spines (Right). EGFP (Left). (B) The Tiam1 PH-CC-Ex domain blocks ephrinB1-induced spine development. E18 hippocampal neurons (DIV5) were transfected with an EGFP expression vector together with a myc-tagged PHn-CC-Ex expression vector or an empty control vector. After an additional 9 days in culture (DIV14), neurons were stimulated with aggregated ephrinB1-Fc (eB1) or Fc alone, fixed, and imaged for spine density. ∗, P < 0.001, Student's t test.
Discussion
Eph receptor tyrosine kinases and their ephrin ligands play critical roles in spine morphogenesis and synapse development and plasticity. However, the mechanisms by which Ephs regulate these processes are not fully understood. In this work, we identify the Rac1-GEF Tiam1 as an important mediator of EphB-dependent spine development. We show that Tiam1 specifically associates with EphB2 in a kinase-dependent manner and that this interaction is mediated by the Tiam1 PH-CC-Ex domain. EphrinB activation of EphB receptors induces the tyrosine phosphorylation and recruitment of Tiam1 to EphB complexes containing NMDA receptors. Disrupting Tiam1 function with RNAi or overexpression of the Tiam1 PH-CC-Ex domain blocks ephrinB-induced spine development. Taken together, our results suggest that EphBs regulate spine development in part by recruiting, phosphorylating, and activating Tiam1. Tiam1 can then promote the Rac1-dependent actin cytoskeletal remodeling required for spine morphogenesis
Two other GEFs implicated in spine morphogenesis, the Rac1-GEF Karlirin-7 and the Cdc42-GEF Intersectin, have also been reported to act downstream from EphBs (8, 15). Why would multiple Rho family GEFs be required at synapses to regulate EphB-dependent spine development? It is possible that these different GEFs have discrete tissue or synaptic distributions or are expressed at different times during development and plasticity (32). Within the same spine, different GEFs might regulate distinct downstream signaling pathways and/or cellular functions. This differential regulation could be accomplished if, in addition to stimulating Rho GTPases, GEFs help specify which downstream pathways are activated. Indeed, by associating with particular protein complexes, Tiam1 is able to select the signaling pathways that are induced by Rac1 (20, 21). Thus, by activating a variety of different downstream pathways, Tiam1, Kalirin-7, and Intersectin may coordinately regulate various Rac1- and Cdc42-dependent processes that are critical to EphB-dependent spine development and morphogenesis.
The finding that Tiam1 plays a role in both EphB and NMDA receptor-dependent spine development suggests that Tiam1 may act as a convergence point for EphB and NMDA receptor signaling. During CNS development, the initial establishment of neuronal circuits relies on processes such as neuronal migration and axon guidance, which are largely independent of neuronal activity (33). Ephs play critical roles in regulating these activity-independent processes (34). Neuronal activity then guides the specification and maturation of synaptic connections by means of NMDA receptors, which mediate calcium entry into cells and activate signaling pathways important for synapse development and plasticity (35, 36). Modulation of NMDA receptor signaling by EphBs may allow activity-dependent and -independent signals to converge (13). By functioning downstream from both EphB and NMDA receptors, Tiam1 might help integrate these activity-dependent and -independent signals during the development and remodeling of synaptic connections.
Our findings indicate that by modulating Tiam1 function, EphB receptors can induce Rac1-dependent signaling pathways, resulting in actin cytoskeletal remodeling required for spine morphogenesis. The precise regulation of Rho GTPase signaling is critical for the normal development and remodeling of spines (17). Furthermore, mutations in genes involved in Rho GTPase signaling are the cause of various forms of mental retardation that are associated with spine abnormalities (37). Given that the structure of spines is intimately related to their function (4), these spine abnormalities are thought to impair neuronal connectivity and synaptic plasticity, resulting in cognitive deficits (3). Further elucidation of the mechanisms by which Tiam1 and other Rho family regulatory proteins are regulated in spines may therefore help to illuminate the underlying causes of mental retardation and other neurological disorders.
Materials and Methods
For additional details, see supporting information (SI) Materials and Methods.
Antibodies and Reagents.
Anti-Tiam1 pY829 antibodies were generated against the phosphopeptide CPQPEEDIpYELL and then affinity-purified on a column of covalently coupled peptide. Antibodies against EphB2 and pEphB were generated previously in the laboratory (12). The following antibodies were purchased: anti-Tiam1, anti-pY99, and anti-myc9E10 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-FLAG M2 (Sigma, St. Louis, MO), and anti-NR1 (BD PharMingen, San Diego, CA). EphB2, EphA4, Tiam1, and pSUPER-Tiam1 RNAi constructs were described previously (12, 23). Tiam1 Y829F was created by site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA).
Cell Cultures and Transfections.
Hippocampal neurons were isolated from E18 Long–Evans rat embryos (Charles River Laboratories, Wilmington, MA) as described previously (38). Neurons were plated at 50,000–100,000 cells per well on a 24-well plate on coverslips coated with 20 μg/ml poly-d-lysine/3 μg/ml laminin and cultured in neurobasal medium supplemented with B27 (Invitrogen, Carlsbad, CA)/2 mM glutamine/100 units/ml penicillin/100 μg/ml streptomycin. HEK 293T cells were cultured in DMEM supplemented with 10% FBS (Invitrogen)/glutamine/penicillin/streptomycin. 293T cells and hippocampal neurons (DIV5) were transfected by using the calcium phosphate method (38).
Preparation of Aggregated Ephrin-Fc Fusion Proteins.
EphrinB1-Fc, ephrinB2-Fc, ephrinA1-Fc, and Fc control (R&D Systems, Minneapolis, MN) were preclustered with anti-human Fcγ antibody (Jackson Laboratory, Bar Harbor, ME) at 10 μg/ml (50 μg/ml for spine experiments) for 1 h and then used at 0.5 μg/ml (2.5 μg/ml for spine experiments) to stimulate neurons.
Immunoprecipitations and Western Blot Analysis.
Immunoprecipitations were performed by using lysate prepared from 293T cells, dissociated neurons, or synaptosomes purified from P15 rat pups as described previously (23). For details, see SI Materials and Methods.
Immunocytochemistry.
Neurons were stimulated at 37°C with clustered ephrin and then fixed and stained as described previously (23). For details, see SI Materials and Methods.
Image Analysis and Quantification.
Images were acquired by using an LSM 510 confocal microscope with a ×63 oil immersion lens (Zeiss, Thornwood, NY). For a detailed description of image analysis and quantification, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the M.E.G. laboratory for reagents and advice. We acknowledge the Children's Hospital Mental Retardation Developmental Disabilities Research Center (MRDDRC) Imaging core, supported by the National Institutes of Health (NIH) Grant P030-HD18655, for assistance with confocal microscopy and the support of the F. M. Kirby Foundation to the Children's Hospital Neurobiology Program. This work was supported by NIH Grant NS-045500 (to M.E.G.), a Damon Runyon Cancer Research Postdoctoral Fellowship (to K.F.T.), NIH Training Grant NS-07484 (to K.F.T.), a Children's Hospital Postdoctoral Career Development Award (to K.F.T.), and a Howard Hughes Medical Institute Predoctoral Fellowship (to J.B.B.).
Abbreviations
- CC-Ex
coiled-coil domain with an adjacent sequence
- DH
Dbl homology domain
- DIV
days in vitro
- En
embryonic day n
- GEF
guanine nucleotide exchange factor
- PH
pleckstrin homology domain
- Pn
postnatal day n.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702044104/DC1.
References
- 1.Harris KM, Kater SB. Annu Rev Neurosci. 1994;17:341–371. doi: 10.1146/annurev.ne.17.030194.002013. [DOI] [PubMed] [Google Scholar]
- 2.Yuste R, Bonhoeffer T. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- 3.Kaufmann WE, Moser HW. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 4.Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H. Trends Neurosci. 2003;26:360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi Y, Pasquale EB. Curr Opin Neurobiol. 2004;14:288–296. doi: 10.1016/j.conb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Pasquale EB. Nat Rev Mol Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 7.Ethell IM, Irie F, Kalo MS, Couchman JR, Pasquale EB, Yamaguchi Y. Neuron. 2001;31:1001–1013. doi: 10.1016/s0896-6273(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 8.Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- 9.Henkemeyer M, Itkis OS, Ngo M, Hickmott PW, Ethell IM. J Cell Biol. 2003;163:1313–1326. doi: 10.1083/jcb.200306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Nat Neurosci. 2003;6:153–160. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- 11.Cull-Candy S, Brickley S, Farrant M. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 12.Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 13.Takasu MA, Dalva MB, Zigmond RE, Greenberg ME. Science. 2002;295:491–495. doi: 10.1126/science.1065983. [DOI] [PubMed] [Google Scholar]
- 14.Grunwald IC, Korte M, Wolfer D, Wilkinson GA, Unsicker K, Lipp HP, Bonhoeffer T, Klein R. Neuron. 2001;32:1027–1040. doi: 10.1016/s0896-6273(01)00550-5. [DOI] [PubMed] [Google Scholar]
- 15.Irie F, Yamaguchi Y. Nat Neurosci. 2002;5:1117–1118. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- 16.Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, Ip NY. Nat Neurosci. 2007;10:67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- 17.Govek EE, Newey SE, Van Aelst L. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt A, Hall A. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 19.Moon SY, Zheng Y. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 20.Buchsbaum RJ, Connolly BA, Feig LA. Mol Cell Biol. 2002;22:4073–4085. doi: 10.1128/MCB.22.12.4073-4085.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchsbaum RJ, Connolly BA, Feig LA. J Biol Chem. 2003;278:18833–18841. doi: 10.1074/jbc.M207876200. [DOI] [PubMed] [Google Scholar]
- 22.Jaffe AB, Hall A, Schmidt A. Curr Biol. 2005;15:405–412. doi: 10.1016/j.cub.2004.12.082. [DOI] [PubMed] [Google Scholar]
- 23.Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Ehler E, van Leeuwen F, Collard JG, Salinas PC. Mol Cell Neurosci. 1997;9:1–12. doi: 10.1006/mcne.1997.0602. [DOI] [PubMed] [Google Scholar]
- 25.Sone M, Hoshino M, Suzuki E, Kuroda S, Kaibuchi K, Nakagoshi H, Saigo K, Nabeshima Y, Hama C. Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- 26.Sone M, Suzuki E, Hoshino M, Hou D, Kuromi H, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C. Development (Cambridge, UK) 2000;127:4157–4168. doi: 10.1242/dev.127.19.4157. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Macara IG. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- 28.Stam JC, Sander EE, Michiels F, van Leeuwen F, Kain HE, van der Kammen RA, Collard JG. J Biol Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- 29.Servitja JM, Marinissen MJ, Sodhi A, Bustelo XR, Gutkind JS. J Biol Chem. 2003;278:34339–34346. doi: 10.1074/jbc.M302960200. [DOI] [PubMed] [Google Scholar]
- 30.Miyamoto Y, Yamauchi J, Tanoue A, Wu C, Mobley WC. Proc Natl Acad Sci USA. 2006;103:10444–10449. doi: 10.1073/pnas.0603914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. EMBO J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshizawa M, Sone M, Matsuo N, Nagase T, Ohara O, Nabeshima Y, Hoshino M. Gene Expr Patterns. 2003;3:375–381. doi: 10.1016/s1567-133x(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 33.Tessier-Lavigne M, Goodman CS. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 34.Flanagan JG, Vanderhaeghen P. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- 35.Katz LC, Shatz CJ. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 36.Constantine-Paton M, Cline HT. Curr Opin Neurobiol. 1998;8:139–148. doi: 10.1016/s0959-4388(98)80017-2. [DOI] [PubMed] [Google Scholar]
- 37.Ramakers GJ. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- 38.Xia Z, Dudek H, Miranti CK, Greenberg ME. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.