Abstract
We have investigated the efficacy of modifying gene-specific antisense phosphorothioate oligodeoxyribonucleotides (PS-ODNs) by the addition of 5′ and 3′ hairpin extensions. As a model system, we have targeted the Mycobacterium tuberculosis 30/32-kDa mycolyl transferase protein complex genes encoding three highly related enzymes (antigens 85 A, B, and C). Whereas the addition of a hairpin extension at only one end of the PS-ODNs did not improve their inhibitory capacity, the addition of hairpin extensions at both ends enhanced their capacity to inhibit M. tuberculosis multiplication in comparison with unmodified PS-ODNs. A combination of three 5′-, 3′-hairpin-modified PS-ODNs (HPS-ODNs) targeting each of the three mycolyl transferase transcripts inhibited bacterial growth in broth culture by ≈1.75 log units (P < 0.0001) and in human THP-1 macrophages by ≈0.4 log units (P < 0.0001), which to our knowledge has not previously been demonstrated for any PS-ODN; reduced target gene transcription by ≥90%; caused ≈90% reduction in mycolyl transferase expression; and increased bacterial sensitivity to isoniazid by 8-fold. The growth-inhibitory effect of the HPS-ODNs was gene-specific. Mismatched HPS-ODNs had no growth-inhibitory capacity. This study demonstrates that 5′- and 3′-HPS-ODNs are highly efficacious against M. tuberculosis and supports the further development of antisense technology as a therapeutic modality against tuberculosis.
Keywords: antisense oligodeoxyribonucleotide therapy, phosphorothioate oligodeoxyribonucleotides, RNA–DNA hybrid stability, antituberculous antibiotics, species-specific targets
Tuberculosis, one of the leading causes of death worldwide, is caused primarily by the facultative intracellular bacterium Mycobacterium tuberculosis. Exacerbating the global problem of tuberculosis, the AIDS pandemic has resulted in an enormous population of HIV-infected persons highly susceptible to tuberculosis. Since the introduction of streptomycin in the 1950s, drugs have been available for the treatment of tuberculosis. However, with the exception of rifapentene, an analog of the preexisting drug rifampin, no new drugs have been licensed for use against tuberculosis in the past three decades, and strains of M. tuberculosis resistant to the major antituberculosis drugs have been emerging worldwide. This has given new urgency to the search for new therapeutic modalities against tuberculosis.
In previous studies, by using several M. tuberculosis gene transcripts as targets, we demonstrated that antisense technology is a feasible approach to combating this pathogen. In our first study, we established proof of concept by demonstrating that incubation of M. tuberculosis with antisense phosphorothioate oligodeoxyribonucleotides (PS-ODNs) directed against the M. tuberculosis glutamine synthetase I mRNA (glnA1 gene transcript) inhibited bacterial growth (1). In a second study, we identified the three 30/32-kDa mycolyl transferases of M. tuberculosis as excellent targets for antisense technology, and demonstrated that the effectiveness of PS-ODNs depends on (i) the genomic location of the target gene (the best targets were located at the beginning of a multigene operon or in a single transcription unit); (ii) the location of the target sequence within the gene (targeting the 5′ end of the transcript was most efficacious); and (iii) the propensity of the target region to form a stable secondary structure (a low propensity yielded a superior target) (2).
In this study, by using the three M. tuberculosis mycolyl transferase gene transcripts as a model target for the antisense approach, we have investigated the impact of modifying the ends of the PS-ODNs by the addition of hairpin and potential hairpin extensions. We hypothesized that the hairpin extensions might interfere with the functionality of the translation initiation start site. We demonstrate that the addition of such extensions at both the 5′ and 3′ ends of the PS-ODNs results in markedly increased efficacy.
Results
Transcript Target Sites and PS-ODN Selection.
M. tuberculosis expresses and secretes three closely related mycolyl transferases that catalyze the transfer of mycolic acid from one trehalose 6-monomycolate molecule to another, resulting in the formation of free trehalose and trehalose 6,6′-dimycolate; trehalose 6,6′-dimycolate is subsequently incorporated into the cell wall (3). The three 30/32-kDa enzymes, products of the M. tuberculosis genes fbpA, -B, and -C (3), are also known as antigens 85A, -B, and -C, but will be referred to herein as the 32A, 30, and 32B proteins, respectively.
The 30, 32A, and 32B protein gene transcripts are promising targets for antisense therapy. The three transcripts are encoded by three different, unlinked, single operon genes: the fbpA gene, which maps to position Rv3804c; the fbpB gene, which maps to position Rv1886c; and the fbpC gene, which maps to position Rv0129c of the M. tuberculosis genome (4). The three genes are constitutively expressed by M. tuberculosis throughout its growth in broth culture and in macrophages (5, 6). The three proteins encoded by fbpA, -B, and -C are expressed in a molar ratio of 2:3:1, which is mirrored at the mRNA level.
In our previous study (2) on the inhibitory capacity of mycolyl transferase-specific antisense PS-ODNs, we demonstrated that the mRNA nucleotides encoding the first 8 aa of each of the three enzymes, located at the initiation of translation, were the most effective target sites. In the present study, we evaluated a set of antisense PS-ODNs that had the same target sites but carried either one or two hairpin-like extensions of four or six base pairs in the form of fold-back structures at their 5′ and/or 3′ ends (Table 1). Critical to our rationale was recognition that precise tertiary interactions of the nascent initiation complex of proteins and aminoacylated tRNA occur at the ribosome, where the language of the gene is translated into that of the protein (7, 8). We hypothesized that the hairpin structures may breathe at times and thus initiate interactions with other macromolecular participants in the translational initiation complex, inhibiting either their assembly or functionality.
Table 1.
PS-ODNs targeting the M. tuberculosis 30, 32A, and 32B protein gene transcripts
| mRNA target 5′→3′ | PS-ODN 5′→3′ |
|---|---|
| 30-kDa protein | |
| 30:1–24 | AATCTTTCGGCTCACGTCTGTCAT |
| 30:HP2/HP1 | GCGCATATGCGCAATCTTTCGGCTCACGTCTGTCATGCGCGCGC |
| 30:1–24-MM | AtTCaTTtGGtTCtCGaCTcTCtT |
| 30:HP2/HP1-MM | GCGCATATGCGCAtTCaTTtGGtTCcCGaCTcTCtTGCGCGCGC |
| 32A-kDa protein | |
| 32A:1–24 | ACGAACCCTGTCAACAAGCTGCAT |
| 32A:HP2/HP1 | GCGCATATGCGCACGAACCCTGTCAACAAGCTGCATGCGCGCGC |
| 32A:1–24-MM | AgGAtCCtTGaCAtCAtGCaGCtT |
| 32A:HP2/HP1-MM | GCGCATATGCGCAtGAtCCaTGcCAgCAtGCaGCtTGCGCGCGC |
| 32B-kDa protein | |
| 32B:1–24 | TCGCACCTGTTCGAAGAACGTCAT |
| 32B:HP2/HP1 | GCGCATATGCGCTCGCACCTGTTCGAAGAACGTCATGCGCGCGC |
| 32B:1–24-MM | TgGCtCCaGTaCGtAGtACcTCtT |
| 32B:HP2/HP1-MM | GCGCATATGCGCTgGCtCCaGTaCGtAGtACcTCtTGCGCGCGC |
HP2/HP1, hairpin 2 at 5′ end and hairpin 1 at 3′ end; nucleotides comprising hairpins are in bold italics. MM, mismatched nucleotide(s), indicated by lowercase letters.
Effect on M. tuberculosis Proliferation of Mycolyl Transferase Complex-Specific Antisense PS-ODNs with Hairpin Extensions.
In a first set of experiments, we examined the inhibitory capacity of PS-ODNs containing one hairpin extension. These PS-ODNs, each targeting one of the transcripts of the mycolyl transferase complex, were administered individually or as a combination of all three. Each individual PS-ODN, added once at the start of the experiment, exerted a significant inhibitory effect on M. tuberculosis growth as evaluated by assaying cfu of M. tuberculosis during a 6-week growth period; however, the inhibitory capacity of these single hairpin-containing PS-ODNs, as well as the combination of all three of these PS-ODNs, was no greater than that of unmodified PS-ODNs. The position of the hairpin extension, i.e., 5′ or 3′, had no observable influence on a PS-ODN′s inhibitory capacity.
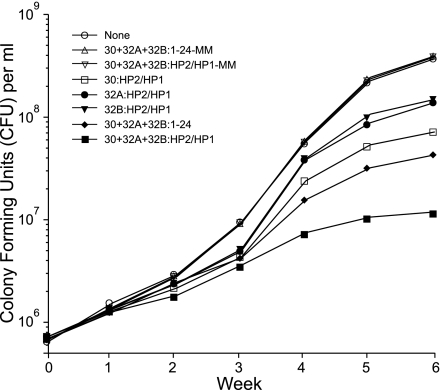
In a second set of experiments, we studied mycolyl transferase gene transcript-specific PS-ODNs containing hairpin extensions at both ends of the PS-ODN. Each of the 5′-, 3′-hairpin-modified PS-ODNs (HPS-ODNs) directed against an individual 30, 32A, or 32B protein gene transcript significantly inhibited M. tuberculosis growth compared with untreated cultures and cultures treated with mismatched PS-ODNs (Fig. 1). As previously observed with unmodified PS-ODNs, the HPS-ODN directed against the 30-kDa mycolyl transferase gene transcript, which encodes the most abundant of the three mycolyl transferases, was the most effective. Remarkably, the HPS-ODN targeting the 30-kDa protein was almost as effective as the combination of three unmodified PS-ODNs targeting all three mycolyl transferases. Most importantly, the combination of three HPS-ODNs targeting all three mycolyl transferases was significantly more potent than the combination of three unmodified PS-ODNs targeting the three mycolyl transferases.
Fig. 1.
Inhibition of M. tuberculosis growth by antisense HPS-ODNs specific for the 30/32-kDa protein gene complex. Duplicate bacterial cultures were grown for 6 weeks in 2 ml of 7H9 medium in the presence of antisense HPS-ODN(s) or mismatched HPS-ODNs (HPS-ODN-MM), which were added once at the start of the growth period at concentrations of 10 μM. For cultures with multiple HPS-ODNs, each of the matched or mismatched HPS-ODNs was added at a concentration of 10 μM. Cfus were enumerated weekly. The maximal standard deviation for each of the data points was 0.02 log cfu. The results shown are from a single experiment representative of three consecutive experiments with very similar results. In each of the three experiments, (i) differences in cfu between the group treated with three HPS-ODNs against the mycolyl transferases and each of the other groups, including the group treated with three unmodified PS-ODNs against the mycolyl transferases, were statistically significant (P < 0.0001 by ANOVA); (ii) differences in cfu between each of the groups treated with a single HPS-ODN against the 30-, 32A-, or 32B-kDa mycolyl transferase and groups treated with mismatched HPS-ODNs, mismatched unmodified PS-ODNs, or the untreated group were statistically significant (P < 0.0001 by ANOVA); and (iii) differences in cfu between the group treated with a single HPS-ODN against the 30-kDa protein and each of the groups treated with a single HPS-ODN against the 32A- or 32B-kDa protein were statistically significant (P < 0.0001 by ANOVA). HP1, hairpin 1; HP2, hairpin 2.
In three independent experiments with nearly identical results (a representative example is shown in Fig. 1), we observed growth inhibition by an individual HPS-ODN of up to 1 log unit and by the combination of all three HPS-ODNs of 1.5 to almost 2.0 log units, and in all cases, the HPS-ODNs were more potent than the unmodified PS-ODNs. Mismatched PS-ODNs, whether hairpin modified or not, had no inhibitory effect. In all of these experiments, the HPS-ODNs or PS-ODNs were added just once at the onset of the 6-week culture period. To date, the inhibitory effect of the HPS-ODNs on M. tuberculosis proliferation is the strongest we have observed for any antisense molecule after a single administration at the onset of the 6-week culture period.
Stability of HPS-ODNs in M. tuberculosis Cultures.
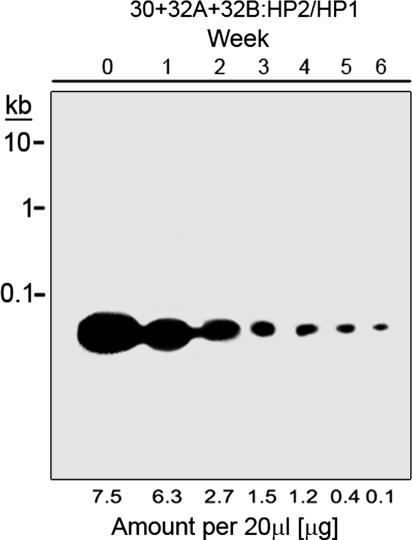
For PS-ODNs to be effective over a 6-week culture period, their concentration must remain sufficiently high to inhibit proliferation. The results shown in Fig. 1, where HPS-ODNs were added once at the beginning of the growth period, suggested that the HPS-ODNs were fairly stable in the culture medium. To evaluate their stability, we assayed their concentration in the culture medium weekly (Fig. 2). The fall-off in HPS-ODN concentration was somewhat biphasic, with an early growth phase half-life of ≈1.3 weeks and a late growth phase half-life of ≈0.5 weeks (Fig. 2).
Fig. 2.
Stability of HPS-ODNs in M. tuberculosis cultures. Triplicate bacterial cultures were grown for 6 weeks in 2 ml of 7H9 medium in the presence of antisense HPS-ODNs at a concentration of 10 μM each. The HPS-ODNs were added once at the start of the growth period. At weekly intervals, culture aliquots were electrophoresed on agarose gels, stained with ethidium bromide, and quantitated densitometrically. HP1, hairpin 1; HP2, hairpin 2.
Effect of Mycolyl Transferase-Specific HPS-ODNs on Protein Synthesis.
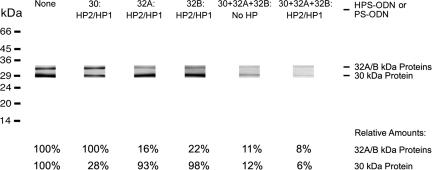
We hypothesized that the inhibitory effect of the mycolyl transferase-specific HPS-ODNs on bacterial proliferation resulted from their capacity to reduce expression of the three mycolyl transferase enzymes. The primary translation products of the mycolyl transferase protein gene complex contain leader peptides that, as the proteins are being processed, are cleaved off to release the mature enzymes into the extracellular milieu; consequently, ≥95% of these proteins are found in the culture filtrate of bacterial cultures. To explore the impact of the HPS-ODNs on mycolyl transferase expression, we assessed the detectable amount of each of the three mycolyl transferases in the culture filtrate of a 4-week-old bacterial culture, the earliest time point for a reliable analysis of mycolyl transferase expression in HPS-ODN-inhibited cultures. Analyzing the expression of the mycolyl transferase complex from a standardized number of M. tuberculosis by gel electrophoresis and immunoblotting, we found that the expression levels greatly decreased in the presence of HPS-ODNs (Fig. 3). Each HPS-ODN acted against its cognate gene transcript, thereby causing a decrease in expression of the encoded protein while leaving the expression of the other two components of the mycolyl transferase complex unchanged. Consistent with an earlier finding that the expression of the three constitutively expressed proteins correlates well with their transcription levels (5), we observed that the HPS-ODNs targeting the 32A or 32B proteins caused a greater decrease in protein expression (≈80%) than the HPS-ODN targeting the 30-kDa protein (≈70% decrease), presumably because the bacteria contain more 30-kDa protein gene-specific mRNA than 32A- or 32B-specific mRNA. Assuming similar uptake and hybridization kinetics for each of the three HPS-ODNs, we can conclude that the ratio of protein gene transcript-specific HPS-ODNs to target transcripts is a little less favorable for the 30-kDa protein gene than that for the other two protein genes. The combination of all three HPS-ODNs reduced the expression levels of the three proteins to ≤10% of their normal levels (Fig. 3).
Fig. 3.
Specific inhibition of protein synthesis by antisense HPS-ODNs. Quadruplicate bacterial cultures were grown for 6 weeks in 2 ml of 7H9 medium in the presence of single or multiple HPS-ODNs, as indicated, at concentrations of 10 μM for each HPS-ODN. The HPS-ODNs were added once at the start of the growth period. After 4 weeks, the 30/32-kDa complex proteins were analyzed in culture filtrates by immunoblotting and were quantitated densitometrically. Data below the immunoblot are the percentages of 30/32-kDa proteins present in each culture treated with HPS-ODN(s), compared with the amounts in an untreated culture. HP1, hairpin 1; HP2, hairpin 2.
As a control for specificity of inhibition of protein expression, we assayed the effect of the anti-mycolyl transferase-specific HPS-ODNs on the unrelated enzyme glutamine synthetase. Activity of the enzyme was assayed by the transfer reaction (9, 10) in bacterial cell extracts and culture filtrates. The glutamine synthetase activity was the same in untreated cultures and in cultures treated with HPS-ODNs for both the concentrated culture filtrates (0.25 nmol of γ-glutamylhydroxamate/15 min per 106 cell equivalents) and the cell extracts (0.80 nmol of γ-glutamylhydroxamate/15 min per 106 cell equivalents).
Because treatment with mycolyl transferase-specific HPS-ODNs depletes the M. tuberculosis culture of mycolyl transferases, we investigated whether the addition of exogenous mycolyl transferase to HPS-ODN-treated cultures could rescue the bacteria and allow normal growth. We estimated that the untreated cultures produced ≈1–2 μg of the 30-kDa mycolyl transferase over a 6-week period. We added 20 μg of purified recombinant 30-kDa protein (11), an amount in great excess of that produced by an untreated culture, to HPS-ODN-treated cultures; the protein was added once at the beginning of the 6-week culture period or weekly. When the 30-kDa protein was added only once at the start of the assay, it had no impact on the growth of the cultures treated with either a single HPS-ODN targeting only the 30-kDa mycolyl transferase or a combination of three HPS-ODNs targeting each of the mycolyl transferases. When the 30-kDa protein was added weekly to the cultures, there was a slight enhancement of growth both in cultures treated with the single HPS-ODN and cultures treated with the three HPS-ODNs in combination (data not shown). Hence, the cultures could only be slightly rescued by the addition of exogenous mycolyl transferase.
Effect of 30, 32A, and 32B Protein Gene-Specific HPS-ODNs on the Corresponding Gene Transcripts.
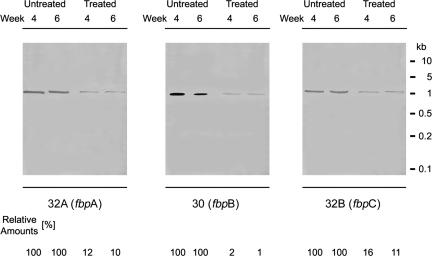
The reduction we measured for the expression of the three mycolyl transferases could be caused by a decrease in synthesis of the specific transcripts and/or by a decrease in mRNA translation. We performed Northern hybridizations for each individual transcript with total RNA isolated from 4- and 6-week-old cultures and biotinylated gene probes to distinguish between these two possibilities. By using RNA extracted from equal numbers of bacteria, we observed that the levels of mycolyl transferase-specific mRNA were greatly diminished in treated cultures compared with untreated cultures (Fig. 4), as was the case with the expression of the three mycolyl transferase proteins. However, in contrast to the results obtained for the protein expression levels, the 32A and 32B protein gene specific HPS-ODNs, when administered individually, reduced the levels of the corresponding transcripts to ≈10%, whereas the 30 protein gene specific HPS-ODN reduced the level of the corresponding transcript to ≈1%, although this transcript is normally present in concentrations equal to or higher than the 32A and 32 B protein gene transcripts. This was a remarkable result because it meant that the 30 protein gene transcript is present at a 10-fold lower concentration than the other two transcripts in the treated cultures and that this transcript had to be translated much more frequently than the other two transcripts to maintain a 30-kDa protein level that was ≈1.5 times the steady-state level of the 32A- plus 32B-kDa proteins in the HPS-ODN-treated cultures (Fig. 3). All hybridizations were very specific, because the 30, 32A, and 32B transcript probes did not detect other RNA species, treatment with ribonucleases abolished all signals, and unrelated probes (glnA1) did not hybridize to the 30, 32A, or 32B protein gene transcripts (data not shown). The level of the unrelated, constitutively expressed mpt64 gene transcript (Rv1980c, encoding a secreted 23.5-kDa protein), assayed as a control RNA, did not change under any of the culture conditions (data not shown).
Fig. 4.
Inhibition of 30, 32A, and 32B protein gene transcript synthesis. Quadruplicate bacterial cultures were grown for 6 weeks in 2 ml of 7H9 medium in the presence of antisense HPS-ODNs at concentrations of 10 μM for each HPS-ODN. The HPS-ODNs were added once at the start of the growth period. After 4 and 6 weeks of growth, total RNA was isolated from treated and untreated cultures, subjected to Northern blotting, and probed with biotin-labeled DNA fragments from the 5′ ends of the 30, 32A, and 32B protein gene coding regions. RNA–DNA hybrids were visualized after HRP reaction by exposure to x-ray film (Kodak) for 30 min at room temperature.
Isoniazid (INH) and HPS-ODNs Act Synergistically to Inhibit M. tuberculosis Growth.
Ordinarily, as a result of the biological activity of the mycolyl transferase complex, large amounts of trehalose dimycolate are incorporated into the mycobacterial cell wall. We hypothesized that the decreased expression of mycolyl transferase in the presence of HPS-ODNs would result in diminished incorporation of trehalose dimycolate into the bacterial cell wall and, consequently, increased sensitivity of the bacteria to antibiotics. To explore this hypothesis, we studied the susceptibility of HPS-ODN-treated and untreated cultures to INH, a leading antituberculous drug. Cultures either untreated or treated with HPS-ODNs for 6 weeks were washed and diluted to 1 × 106 cfu/ml and then treated for 2 weeks with various concentrations of INH. Whereas the minimal inhibitory concentration, defined here as the concentration of INH that prevented one doubling of the bacteria (0.3 log growth), for untreated cultures (or cultures pretreated with mismatched HPS-ODNs) was 0.03 μg/ml, the minimal inhibitory concentration for HPS-ODN pretreated cultures was 0.004 μg/ml, 8-fold lower. Hence, bacteria pretreated with HPS-ODNs exhibited much greater sensitivity to INH.
Moreover, the effect of the drugs in combination was synergistic rather than merely additive. This was demonstrated in an experiment in which the three mycolyl transferase-specific HPS-ODNs by themselves (at 10 μM each) inhibited M. tuberculosis growth by 1.2 log units, INH by itself (at 0.06 μg/ml) inhibited M. tuberculosis growth by 2.0 log units, and the combination of the three HPS-ODNs and INH at these same concentrations inhibited growth of M. tuberculosis by 4.7 log units, an amount of inhibition more than 1 log greater than that expected had the two drugs acted only additively.
HPS-ODNs Inhibit M. tuberculosis Growth in Human Monocytes.
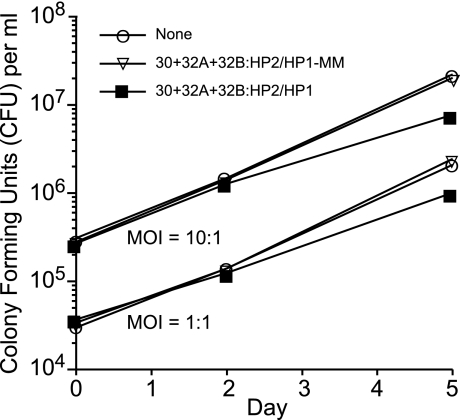
To determine whether HPS-ODNs could inhibit the growth of M. tuberculosis in human macrophages, we added the HPS-ODNs (10 μM of each of the three mycolyl transferase-specific HPS-ODNs) to THP-1 cells infected with M. tuberculosis at a multiplicity of infection (MOI) of 1 or 10 and assayed cfu over the subsequent 5 days. In a preliminary experiment, we determined that the HPS-ODNs at 10 or even 25 μM had no toxic or growth-inhibitory effect on THP-1 cultures. The three mycolyl transferase-specific HPS-ODNs in combination significantly inhibited M. tuberculosis multiplication within human monocytes. In two independent experiments, the growth reduction amounted to 0.3 ± 0.01 (mean ± SE) log cfu in macrophages infected at an MOI of 1 and 0.4 ± 0.02 log cfu in macrophages infected at an MOI of 10 (Fig. 5). In contrast, mismatched HPS-ODNs at 10 μM had no inhibitory effect on M. tuberculosis growing in THP-1 cells (MOI, 1 or 10). Although the inhibitory effect was modest, to our knowledge ODNs have not been demonstrated previously to be capable of inhibiting M. tuberculosis growth within human mononuclear phagocytes.
Fig. 5.
HPS-ODN inhibition of M. tuberculosis multiplication in human macrophages. THP-1 cells seeded in 24-well tissue culture plates at a concentration of ≈5 × 105 cells per well were infected with M. tuberculosis Erdman at an MOI of 1 or 10 and incubated for 5 days. All HPS-ODNs were added immediately after the 90-min infection period at a concentration of 10 μM. Triplicate cell cultures were lysed 3 h, 2 days, or 5 days after the infection for enumeration of cfu of M. tuberculosis. The maximal standard deviation for each of the data points at 5 days was 0.007 logs. Differences in cfu at 5 days between the group treated with three HPS-ODNs and either the untreated group or the group treated with mismatched HPS-ODNs were statistically significant at MOIs of both 1 and 10 (P < 0.0001 by ANOVA for all comparisons). In a second experiment that was similar to this one except that it lacked the group treated with mismatched HPS-ODNs and the cultures were assayed in duplicate, differences in cfu at 5 days between the group treated with HPS-ODNs and the untreated group were similar in magnitude to this experiment and statistically significant (P = 0.02 for MOI of 1 and P = 0.002 for MOI of 10). HP1, hairpin 1; HP2, hairpin 2.
Discussion
In this study, employing a previously developed model system consisting of antisense PS-ODNs directed against the three mycolyl transferases of M. tuberculosis, we have demonstrated that hairpin-modified antisense molecules are more potent inhibitors of mycobacterial proliferation and more potent inhibitors of mycolyl transferase expression than unmodified PS-ODNs. The enhancement in efficacy required hairpin extensions at both ends of the antisense molecule, because hairpin extensions at only one end of the molecule were no better than unmodified PS-ODNs.
Hairpins described in this study resemble “snap-back” structures that may open up at times and participate in bimolecular interactions with other HPS-ODNs, other RNA species, or other macromolecular moieties of the translation initiation complex, thereby enhancing the efficacy of the PS-ODNs described herein (12–15).
Since demonstrating that antisense PS-ODNs could inhibit M. tuberculosis, we have sought to improve the technology in two major ways. First, we have explored a variety of M. tuberculosis targets for the technology. These efforts culminated in our identifying the 5′ region of the transcripts of the mycolyl transferase complex genes as particularly good targets, as discussed further below. Second, we have sought to improve the efficacy of the PS-ODNs against their targets via a variety of approaches. Before this study, we assessed nearly 10 different modifications of PS-ODNs aimed at enhancing their uptake into bacteria, their intracellular stability, or their capacity to block the translation initiation complex. These modifications have included linking an antibiotic (amikacin, puromycin, ethambutol, or cycloheximide) to the PS-ODN, adding a series of one to five guanine nucleotides or two uracil nucleotides to the PS-ODN, adding a 2′ O-methyl group to selected nucleotides, or adding aliphatic side chains to selected nucleotides. However, none of these modifications significantly improved the efficacy of the PS-ODNs. In addition, in previous studies, we coadministered antibiotics with the PS-ODNs in an attempt to “soften up” the mycobacterial cell wall and thereby enhance the uptake of PS-ODNs; however, the effect of the antibiotics was only additive with that of the PS-ODNs and not synergistic (1). The addition of hairpin extensions at the 5′ and 3′ ends of the PS-ODNs enhances their potency substantially.
The mycolyl transferase gene transcripts are the best antisense target of nearly 30 different targets of M. tuberculosis that we have investigated. As a drug target, the mycolyl transferase complex has several advantages. First, from a genetic perspective, the DNA sequences encoding the mycolyl transferases do not show homology to any eukaryotic sequences; even the similarities to the few microbial species related to mycobacteria are sparse. Second, from a biochemical perspective, the enzymes perform a reaction that is specific for mycobacteria and there are no bypass reactions for the synthesis of the essential cell wall component trehalose dimycolate. Third, from a technical perspective, each of the three genes fulfills the criteria for an ideal antisense technology target: (i) each gene is located in a single cistron unit with its specific promoter and terminator regions, and (ii) each gene contains a region very near the 5′ end of its encoded transcript that has a very low propensity to assume a stable secondary structure.
Tuberculosis is an infectious disease that is treated with multiple antibiotics to prevent the emergence of resistant organisms. Antisense technology lends itself well to this strategy because it is conducive to the simultaneous administration of a combination of PS-ODNs directed against multiple targets. As with any drug, mycobacteria could potentially develop resistance to specific PS-ODNs by nucleotide changes in the target site. Moreover, mycobacteria could potentially develop a general resistance to PS-ODNs, for example, via mutations that prevent the uptake of PS-ODNs into the bacterial cell. Hence, PS-ODNs might best be administered in combination with conventional antibiotics. In this regard, it was encouraging to find in this study that HPS-ODNs not only increased the susceptibility of M. tuberculosis to the leading antituberculous drug INH but also acted synergistically with INH in inhibiting M. tuberculosis multiplication.
A milestone reached in the present study was the demonstration that HPS-ODNs could inhibit growth of M. tuberculosis inside macrophages. M. tuberculosis is an intracellular pathogen that resides and multiplies within a phagosome inside a host mononuclear phagocyte. Our study demonstrates that antisense molecules are capable of reaching the mycobacteria in their intracellular niche. In this regard, it is encouraging that in vivo studies have shown that PS-ODNs can be taken up by mononuclear phagocytes (16).
Important challenges remain for the application of antisense technology to the treatment of tuberculosis. For example, the efficacy of antisense molecules against M. tuberculosis in a relevant animal model has yet to be demonstrated. Because the capacity of HPS-ODNs to inhibit M. tuberculosis within macrophages in vitro is still modest, undoubtedly, greater improvement in their potency will be necessary to achieve in vivo efficacy.
Materials and Methods
PS-ODN Selection and Preparation.
All target sites encompassed the first eight codons of the transcripts of the fbpA (Rv3804c), fbpB (Rv1886c), and fbpC (Rv0129c) genes, referred to herein as the 32A, 30, and 32B protein genes, respectively. The PS-ODNs' DNA sequences and their target sites are described in Table 1. Antisense PS-ODNs were synthesized on a 394 DNA/RNA synthesizer (Applied Biosystems, Foster City, CA), by using standard phosphoro-amidite chemistry. Phosphorothioate bonds were introduced by oxidation with the Beaucage thiolating reagent (17), and assembled PS-ODNs were purified by HPLC and lyophilized. Mismatched PS-ODNs were synthesized in the same way except that the middle base of each base triplet was mismatched with regard to its complementary base in the mRNA strand. PS-ODN stock solutions were prepared just before use and added to mycobacterial cultures after sterilization through 0.45-μm HT Tuffryn membrane filters (Pall, Ann Arbor, MI).
Bacterial Inhibition Assays.
For the analyses of the inhibitory effects of the PS-ODNs on bacterial growth, M. tuberculosis cultures were set up in duplicates, triplicates, or quadruplicates as 2-ml broth cultures in polystyrene tubes (Fisher, Houston, TX) and maintained for 6 weeks. PS-ODNs were added to the medium at a final concentration of 10 μM just before the addition of the mycobacteria, M. tuberculosis strain Erdman (ATCC 35801; American Type Culture Collection, Manassas, VA), which was maintained in 7H9 medium (Difco, Detroit, MI) supplemented with 2% glucose at 37°C in a 5% CO2/95% air atmosphere as unshaken cultures. Growth of the bacteria was monitored by gently sonicating the cultures to break up bacterial clumps, removing small aliquots, washing the bacteria by centrifugation, plating serial dilutions of washed bacteria on 7H11 agar (Difco), and enumerating viable bacteria (cfu) after incubation for 2 weeks at 37°C in a 5% CO2/95% air atmosphere.
PS-ODN Stability Assay.
The stability of PS-ODNs added to M. tuberculosis cultures was analyzed by agarose gel electrophoresis. The cultures were sonicated immediately after inoculation and weekly thereafter, after which a 20-μl aliquot (1% of the total culture volume) was removed from each culture and loaded directly onto a 1.2% agarose gel. Samples were electrophoresed at 50 V in Tris/borate/EDTA (pH 8.3) until the bromophenol blue dye marker had migrated ≈70% of the distance between the loading well and the bottom of the gel. Gels were stained with ethidium bromide, photographed, digitized by using Photoshop 5.5 (Adobe Systems, San Jose, CA) software, and evaluated quantitatively by using NIH Image 1.62 software.
Expression of 30/32-kDa Complex Proteins.
Quadruplicate bacterial cultures of M. tuberculosis were set up as described above. After 4 and 6 weeks, 1 ml of each culture was centrifuged to pellet the bacteria, and each sample culture supernatant was concentrated in an Amicon (Beverly, MA) Diaflo unit to 100 μl. One-half of each concentrated culture filtrate was subjected to SDS/PAGE to analyze each protein profile; the remainder of each sample was used for several immunoblots for which the electrophoresed proteins (derived from the equivalent of 5 × 107 cfu of M. tuberculosis) were transferred to nylon membranes, probed with 30/32-kDa protein-specific immunoglobulins at dilutions of 1:100,000 and reacted with HRP-coupled secondary antibodies at a dilution of 1:50,000. The immunocomplexes were detected with an HRP substrate kit (Pierce, Rockford, IL) and exposure to x-ray film (Kodak, Rochester, NY) for 10–30 sec at room temperature. The images were digitized by using Photoshop 5.5 software (Adobe Systems), and protein-specific bands were quantitated densitometrically by using NIH Image 1.62 software.
Expression of the 30, 32A, and 32B Protein Gene Transcripts.
Total RNA was isolated from quadruplicate 4- and 6-week cultures by pelleting the bacteria and resuspending the cells in a solution of ice-cold 30 mM sodium acetate and 0.1% SDS and extracting RNA with hot, water-saturated phenol, followed by DNase I and proteinase K treatment, and isopropanol precipitations. This RNA isolation procedure, from a standardized number of 3 × 107 bacteria, yielded 5.25 μg of RNA, which was found to be free of DNA. For Northern blots, 2.5 μg of RNA (derived from the equivalent of 1.4 × 107 bacteria) was used per hybridization. The RNA was mixed with glyoxal/dimethyl sulfoxide/sodium phosphate buffer, heated to 50°C, electrophoresed on 1% agarose gels, and transferred in 10× SSC (1× SSC is 150 mM NaCl/15 mM Na citrate) to nitrocellulose membranes. For RNA probing, membranes were incubated separately at 65°C with denatured biotin-labeled (random priming kit; Amersham Biosciences, Piscataway, NJ) probes which encompassed the first 160–170 nucleotides of the coding regions of the three fbp genes, where the three DNA sequences lack long stretches of identity and share an overall homology of only 55–60% (fbpA: atg to SbfI site; fbpB: atg to SbfI site; fbpC: atg to PstI site). RNA–DNA hybrids were detected by reaction with streptavidin-coupled HRP (Bio-Rad, Hercules, CA), incubation with enzyme substrate (Pierce), and exposure to x-ray film (Kodak) for 30 min at room temperature.
Effect of Pretreatment of M. tuberculosis with HPS-ODNs on Bacterial Sensitivity to INH.
M. tuberculosis cultures were set up and maintained for 6 weeks in the presence or absence of HPS-ODNs as described above, after which the bacteria were washed, subdivided, and incubated for an additional 2 weeks in the presence of 0.002, 0.004, 0.008, 0.016, 0.03, 0.06, 0.12, or 0.25 μg/ml INH. cfu were assayed in duplicate as described above, and the minimal inhibitory concentration of INH was determined in cultures pretreated and not pretreated with HPS-ODNs.
Effect of INH and HPS-ODNs in Combination on M. tuberculosis Growth.
INH, HPS-ODNs, or both inhibitors were added once to M. tuberculosis cultures, in duplicate, at the start of a 4-week incubation period. HPS-ODNs were added at 10 μM, and INH was added at concentrations ranging in 2-fold increments from 0.00025 to 0.06 μg/ml. At 0, 2, and 4 weeks, aliquots were removed from the cultures, washed, serially diluted, plated on 7H11 agar plates, and incubated for 2 weeks at 37°C in a 5% CO2/95% air atmosphere to enumerate cfu.
Inhibition of Mycobacterial Growth in Human Monocytes.
Human acute monocytic leukemia line THP-1 (TIB 202; American Type Culture Collection) cells were seeded into wells of Costar (Cambridge, MA) 24-well tissue culture plates at a concentration of ≈5 × 105 cells per well, differentiated over a period of 36 h at 37°C in a 5% CO2/95% air atmosphere, infected with M. tuberculosis Erdman at a MOI of 1 or 10 in the presence of human serum for 90 min, washed, and incubated for 5 days with or without HPS-ODNs at a concentration of 10 μM. The M. tuberculosis bacteria used to infect the THP-1 cultures were first cultured in broth, and then on 7H11 plates for 10 days at 37°C in a 5% CO2/95% air atmosphere before being collected, washed, and diluted to the concentration required. At 3 h, 2 days, and 5 days after infection, the THP-1 cell cultures were lysed by the addition of 0.1% SDS, and culture aliquots were serially diluted, plated on 7H11 agar, and incubated for 2 weeks at 37°C in a 5% CO2/95% air atmosphere for the enumeration of cfu. cfu from culture aliquots taken 3 h after the infection were used to determine the infection rate (8–12%) and the data point at time “0 days.”
Statistics.
Means were compared across treatment groups within an experiment by using factorial ANOVA methods.
Acknowledgments
We thank Jeffrey Gornbein and Rebecca Radbod for conducting statistical analyses. This work was supported by National Institutes of Health Grants AI055352 and AI060872 and by a grant from The G. Harold and Leila V. Mathers Foundation.
Abbreviations
- HPS-ODN
hairpin-modified PS-ODN
- INH
isoniazid
- MOI
multiplicity of infection
- PS-ODN
phosphorothioate oligodeoxyribonucleotide.
Footnotes
The authors declare no conflict of interest.
References
- 1.Harth G, Zamecnik PC, Tang JY, Tabatadze D, Horwitz MA. Proc Natl Acad Sci USA. 2000;97:418–423. doi: 10.1073/pnas.97.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harth G, Horwitz MA, Tabatadze D, Zamecnik PC. Proc Natl Acad Sci USA. 2002;99:15614–15619. doi: 10.1073/pnas.242612299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 4.Cole ST, Brosch R, Parkhill L, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, et al. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Harth G, Lee BY, Wang J, Clemens DL, Horwitz MA. Infect Immun. 1996;64:3038–3047. doi: 10.1128/iai.64.8.3038-3047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee BY, Horwitz MA. J Clin Invest. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen DW, Zamecnik PC. Biochem Biophys Acta. 1962;55:865–874. doi: 10.1016/0006-3002(62)90899-5. [DOI] [PubMed] [Google Scholar]
- 8.Yassin A, Fredrick K, Mankin AS. Proc Natl Acad Sci USA. 2005;102:16620–16625. doi: 10.1073/pnas.0508444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woolfolk CA, Shapiro B, Stadtman ER. Arch Biochem Biophys. 1966;116:177–192. doi: 10.1016/0003-9861(66)90026-9. [DOI] [PubMed] [Google Scholar]
- 10.Harth G, Clemens DL, Horwitz MA. Proc Natl Acad Sci USA. 1994;91:9342–9346. doi: 10.1073/pnas.91.20.9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harth G, Lee BY, Horwitz MA. Infect Immun. 1997;65:2321–2328. doi: 10.1128/iai.65.6.2321-2328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.SantaLucia J, Jr, Hicks D. Annu Rev Biophys Biomol Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 13.Hyeon C, Thirumalai D. Proc Natl Acad Sci USA. 2005;102:6789–6794. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li PTX, Bustamante C, Tinoco I., Jr Proc Natl Acad Sci USA. 2006;103:15847–15852. doi: 10.1073/pnas.0607202103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyeon C, Thirumalai D. Proc Natl Acad Sci USA. 2005;102:6789–6794. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Q, Zhou R, Temsamani J, Zhang Z, Rosky A, Agrawal S. Antisense Nucleic Acid Drug Dev. 1998;8:451–458. doi: 10.1089/oli.1.1998.8.451. [DOI] [PubMed] [Google Scholar]
- 17.Padmapriya AA, Tang J, Agrawal S. Antisense Res Dev. 1994;4:185–199. doi: 10.1089/ard.1994.4.185. [DOI] [PubMed] [Google Scholar]