Abstract
G protein-coupled receptors (GPCRs) belonging to class A contain several highly conserved (>90%) amino acids in their transmembrane helices. Results of mutational studies of these highly conserved residues suggest a common mechanism for locking GPCRs in an inactive conformation and for their subsequent activation upon ligand binding. Recently, a second set of sites in the transmembrane helices has been identified in which amino acids with small side chains, such as Gly, Ala, Ser, Thr, and Cys, are highly conserved (>90%) when considered as a group. These group-conserved residues have not been recognized as having essential structural or functional roles. To determine the role of group-conserved residues in the β2-adrenergic receptor (β2-AR), amino acid replacements guided by molecular modeling were carried out at key positions in transmembrane helices H2–H4. The most significant changes in receptor expression and activity were observed upon replacement of the amino acids Ser-161 and Ser-165 in H4. Substitution at these sites by larger residues lowered the expression and activity of the receptor but did not affect specific binding to the antagonist ligand dihydroalprenolol. A second site mutation, V114A, rescued the low expression of the S165V mutant. Substitution of other group-conserved residues in H2–H4 by larger amino acids lowered receptor activity in the order Ala-128, Ala-76, Ser-120, and Ala-78. Together these data provide comprehensive analysis of group-conserved residues in a class A GPCR and allow insights into the roles of these residues in GPCR structure and function.
Keywords: rhodopsin, G protein-coupled receptors, helix packing, site-directed mutagenesis, molecular modeling
G protein-coupled receptors (GPCRs) contain seven transmembrane helices and mediate signal transduction in response to a wide variety of extracellular stimuli. The large GPCR family is subdivided into five classes on the basis of sequence conservation. Although the β2-adrenergic receptor (β2-AR) and the visual pigment rhodopsin are well-characterized members of class A GPCRs, rhodopsin is the only GPCR for which high-resolution structural information has been obtained by protein crystallography (1, 2) and by NMR spectroscopy (3, 4). The most extensively studied ligand-activated GPCR is β2-AR, which mediates physiological responses to adrenaline and noradrenaline and, therefore, plays a significant role in the regulation of the cardiovascular system.
The class A GPCRs are made up of >1,000 unique sequences that are subdivided into several subclasses, which include the opsin, amine, peptide, and olfactory receptors. The class A receptors have been analyzed extensively in regard to structure and function with a focus on those amino acids with sequence identities of >90%, such as the conserved Asn at position 1.50‖ (100%) in H1, Asp at position 2.50 (93%) in H2, and the signature D/ERY and NPxxY motifs in helices H3 and H7, respectively. In addition to these polar residues, there are several conserved hydrophobic amino acids at positions 2.46 (98%), 4.50 (98%), 6.50 (98%), and 5.50 (91%). In all there are ≈20 sites in class A GPCR sequences that have amino acid identities of >75%.
Recently, a second set of sites in the transmembrane helices has been identified in which small (Gly, Ala) and weakly polar (Ser, Thr, Cys) amino acids are highly conserved (>90%) when considered as a group (6). Structural analysis of helical membrane proteins has shown that small and weakly polar amino acids are highly likely to mediate helix–helix interactions. Thus, the group-conserved amino acids with small side chains in the class A GPCRs are likely to play a role in stabilizing receptor structure (e.g., in determining the proper fold of the protein) and in receptor function (e.g., in facilitating rigid body motion of the transmembrane helices that are thought to be involved in the activation mechanism).
Mutational studies on rhodopsin suggest that the group-conserved residues have both structural and functional roles. For example, the amino acids at positions 4.53 and 4.57 are small residues that are highly conserved in the class A GPCR family. These sites contain alanine in rhodopsin, and their mutation to leucine prevents folding to form a functional receptor (7). In contrast, the amino acid at position 3.36, glycine, is strictly conserved in the opsin subclasses of GPCRs. Mutations at this site to amino acids with larger molecular volumes lead to activity in the dark (8). In rhodopsin, the retinal chromophore intercalates between Gly-121 (3.36) in H3 and Trp-265 (6.48) in H6. The close packing of H3 and H6 locks the receptor off in the dark, and motion of Trp-265 (6.48) induced by retinal isomerization leads to motion of H6 and receptor activation. Thus, the small residue at position 3.36 plays both a structural and functional role in the opsin subclass of GPCRs.
In this paper, we focus on the roles of the group-conserved amino acids in H2, H3, and H4 in β2-AR structure and function. The identification of the group-conserved residues in class A GPCRs (6), along with recent mutational studies of these residues in rhodopsin (P.J.R., M.E., P.C., V.H., U.L.R., H.G.K., and S.O.S., unpublished data), has led to the conclusion that transmembrane helices H2–H4 form a stable core, whereas receptor activation involves rigid body motion of helices H5–H7. Our strategy is to replace the group-conserved residues with amino acids containing both small and large molecular volumes and to study the effect of these replacements on receptor expression, activity, and binding of the antagonist dihydroalprenolol (DHA).
In general, the conservative mutations, serine-to-alanine or alanine-to-serine, did not change activity of β2-AR in response to agonist binding, and in many cases, these mutations actually increased the level of receptor expression. The most dramatic effects were observed for nonconservative mutations of Ser-161 and Ser-165, comprising an SxxxS motif in H4. Although both valine and leucine substitutions at positions 161 and 165 lower the expression and activity of the receptor, they do not affect binding of DHA. This result suggests a structural role for Ser-161 and Ser-165 in β2-AR. These findings contrast with those observed for mutations of Ser-203 and Ser-207 in H5, which comprise another conserved SxxxS motif in the β-adrenergic receptor subfamily and are known to have a functional role in ligand binding (9, 10). Using mutation at a second site, V114A, we were able to rescue expression of the poorly expressed S165V mutant to wild-type level. In contrast to a previous report (9), we observe that Ser-120 on H3 can be mutated to alanine, and the resulting receptor is expressed at wild-type levels. Finally, replacement of other group conserved sites in H2–H4 with valine and leucine lowered receptor activity in the order Ala-128, Ala-76, Ser-120, and Ala-78.
Results
Expression and Ligand Binding Properties of Group-Conserved Mutants in H2–H4.
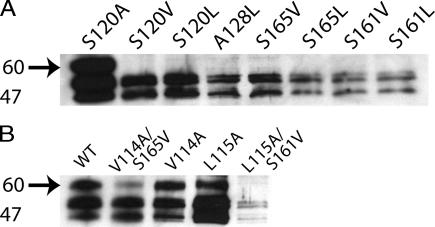
One main structural difference between aryloxypropanolamine β2-AR antagonists, such as alprenolol, and arylethanolamine β2-AR agonists, such as isoproterenol, is that the distance between the aryl moiety and the amino group is shorter in the agonists, by a –OCH2– linkage. In the current study, the ligand binding properties of the β2-AR mutants were measured by using the antagonist DHA (Tables 1 and 2. The level of receptor expression is qualitatively gauged by immunoblots and quantified by saturation binding assays using DHA. In general, the binding of DHA to β2-AR with mutations at the group-conserved sites was similar to the wild-type receptor. In fact, in contrast to the finding in a previous report (9), we observed that mutations of Ser-120 in H3 are tolerated with regard to expression (Fig. 1A) and the ability of the mutant receptors to bind ligand (Table 1). Only the S161L and S165L mutants lacked the ability to bind ligand in a specific manner. The S161V and the S165V mutant receptors were very poorly expressed (as reflected by the low Bmax values in Table 1 and the weak intensity in the immunoblot analysis in Fig. 1A) and were used for the isolation of compensatory mutants (see below).
Table 1.
Summary of ligand binding properties of wild-type β2-AR and mutant receptors
| Receptor | Transmembrane helix | Kd, nM | 95% confidence intervals | Bmax, pmol/mg | |
|---|---|---|---|---|---|
| Wild type | 2.1 | 1.70–2.40 | 18 ± 2.2 | ||
| A76S | II | 1.2 | 0.92–1.39 | 12 ± 0.5 | |
| A76L | II | 1.4 | 1.10–1.67 | 14 ± 0.4 | |
| A76V | II | 1.4 | 1.00–1.28 | 9.1 ± 0.2 | |
| A78S | II | 1.5 | 1.14–1.73 | 17 ± 1.1 | |
| A78L | II | 0.9 | 0.47–1.39 | 9.0 ± 0.6 | |
| A78V | II | 1.4 | 1.14–1.73 | 19 ± 0.4 | |
| S120A | III | 2.3 | 1.77–2.96 | 28 ± 1.2 | |
| S120L | III | 1.5 | 0.81–2.14 | 4.3 ± 0.3 | |
| S120V | III | 1.6 | 0.58–2.62 | 4.9 ± 0.5 | |
| A128S | III | 2.2 | 1.58–2.92 | 28 ± 1.4 | |
| A128L | III | 3.2 | 1.00–5.39 | 4.8 ± 0.5 | |
| A128V | III | 1.6 | 1.31–1.83 | 16 ± 0.5 | |
| S161A | IV | 2.7 | 1.98–3.55 | 33 ± 2.0 | |
| S161L | IV | ND | ND | ||
| S161V* | IV | 2.0 | 0.80–3.14 | 2.7 ± 0.4 | |
| S165A | IV | 4.7 | 3.54–5.98 | 15 ± 0.8 | |
| S165L | IV | ND | ND | ||
| S165V | IV | 1.2 | 0.68–1.71 | 6.7 ± 0.4 |
The values are expressed as the mean ± SE (n = 3–5 experiments). ND, not detected (no significant specific binding detected under the assay conditions).
*High nonspecific binding (15–20% of total binding).
Table 2.
Summary of ligand binding properties of single and compensatory mutant receptors
| Receptor | Transmembrane helix | Kd, nM | 95% confidence intervals | Bmax, pmol/mg |
|---|---|---|---|---|
| Wild type | 2.1 | 1.70–2.40 | 18 ± 2.2 | |
| V114A | III | 19 | 17.0–21.1 | 39 ± 1.6 |
| S165V/V114A | IV/III | 9.0 | 8.64–9.38 | 17 ± 0.6 |
| S165V | IV | 1.2 | 0.68–1.71 | 6.7 ± 0.4 |
| L115A | III | 2.4 | 2.06–2.89 | 24 ± 1.2 |
| S161V/L115A* | IV/III | 4.8 | 0.81–8.90 | 4.5 ± 0.7 |
| S161V* | IV | 2.0 | 0.80–3.14 | 2.7 ± 0.4 |
The values are expressed as the mean ± SE (n = 3–5 experiments).
*High nonspecific binding (15–20% of total binding).
Fig. 1.
Immunoblot analysis of β2-AR expressed in COS-1 cells. (A) S120A and low-expressing β2-AR mutants (≈5 μg of solubilized membrane protein was loaded). (B) Wild-type β2-AR, V114A, L115A, and the compensatory mutants (≈2.5 μg of solubilized membrane protein was loaded). The arrows indicate the position of the fully glycosylated receptor. Mobility of molecular mass standards (kDa) is indicated next to the gel.
An interesting observation from the saturation binding data is that Bmax is higher, in general, in the mutants containing small amino acid replacements. For example, the mutants S120A, A128S, S161A, V114A, and L115A show Bmax values in the range of 24–39 pmol/mg, whereas the mutants A76S, A78S, and S165A show Bmax values similar to that of wild-type β2-AR. Immunoblotting showed heterogeneous expression of the wild-type and the mutant β2-ARs, as indicated by the presence of three predominant bands in the molecular mass range of 45–65 kDa, with none of the low-expressing receptors producing the ≈65 kDa band (Fig. 1A). Photocrosslinking of β2-AR expressed in COS-1 cells was previously used to show that the band at ≈65 kDa corresponds to the completely glycosylated receptor (11).
Gαs Mediated Signaling of the Group-Conserved Mutants.
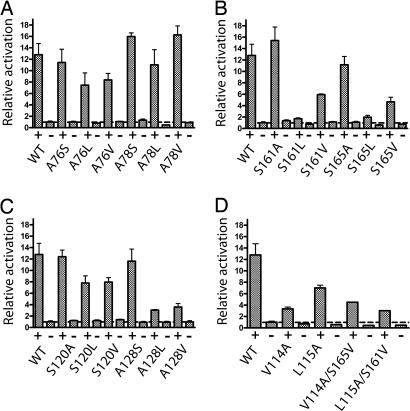
The effect of β2-AR mutations on G protein activation was measured by isoproterenol-stimulated cAMP accumulation in HEK293S cells. Transiently transfected HEK293S cells were induced with 10 μM (−)-isoproterenol for 30 min, and the Gαs-mediated cAMP production was measured as described (12). Fig. 2 shows that the agonist-induced activity of the group-conserved mutants containing small amino acid replacements (e.g., A76S, A128S) was either similar to or higher than that of the wild-type β2-AR. In contrast, there was generally a loss of activity in mutants containing larger amino acid replacements. The H3 mutants A128L and A128V showed 3- to 4-fold lower agonist-stimulated cAMP production. The H4 mutants S161L and S165L showed the most dramatic effects with almost no isoproterenol-stimulated cAMP accumulation, whereas the S161V and S165V mutants showed a 3-fold lower relative activity than wild-type receptor (Fig. 2B). These results indicate a unique role of the two group-conserved residues, Ser-161 and Ser-165, in the packing of helix H4. Structural perturbations in this region of the receptor directly effect receptor expression (Fig. 1A) and agonist-dependent activity (Fig. 2B). In general, the mutations in the group-conserved amino acids to other small amino acids did not change the level of agonist-independent activity. The A78S and S161A mutants, as well as all of the Ser-120 mutants, exhibited slightly higher levels of agonist-independent activity relative to the wild-type receptor (Fig. 2 A–C).
Fig. 2.
Gαs-mediated signaling activity of wild-type (WT) and group-conserved mutant β2-ARs as measured by cAMP accumulation assay. Shown are the basal activity (−) and activity after stimulation (+) with 10 μM isoproterenol. Results are normalized to the basal activity of wild-type receptors. The relative activation rates of β2-AR mutants present in H2 (A), H3 (C), H4 (B), and the compensatory mutants (D) are shown. Results are from at least three independent experiments performed in duplicate.
Design and Characterization of Compensatory Mutants.
Ser-161 and Ser-165 lie on the same face of helix H4 and are part of an SxxxS motif that has been characterized as a common mechanism for close packing of transmembrane helices (13). Substitution of these residues with larger amino acids may affect the proper packing of the helices due to steric interactions. If this is the case, introduction of a second mutation at an appropriate site on an opposing helix may compensate for the steric clash and restore correct packing. Using the ligand-free β2-AR model as a template (Fig. 3), possible compensatory mutants were designed by selecting residues within 4 Å of Ser-161 and Ser-165 for mutagenesis. In the ligand-free β2-AR model, Ser-161 is close to Leu-115 and Val-114 is close to Ser-165.
Fig. 3.
Molecular model of ligand-free β2-AR. (A) Helices H2–H4 are proposed to form the packing core of the receptor. The positions of Ala-78 (2.49), Asp-113 (3.32), Val-114 (3.33), Ser-161 (4.53), and Ser-165 (4.57) are highlighted. Asp-113 provides the counter ion for the positively charged amine in the ligand. The electrostatic interaction between the ligand and Asp-113 parallels the interaction between the protonated retinal Schiff base in rhodopsin and its Glu-113 counter ion. (B) Helices H5–H7 are shown with approximately the same receptor orientation as in A. Ser-120 interacts with Asn-318 on H7 and is in close proximity to Phe-282 on H6. Ala-128 is packed against Val-214 and Val-218 on H5. Both models are viewed from the extracellular surface of the receptor.
S161V (4.53)/L115A (3.34).
It was shown previously in rhodopsin that the L119A mutation rescues the chromophore-binding defect caused by the A164V mutation (14). Because Leu-119 (3.34) in rhodopsin corresponds to Leu-115 (3.34) in β2-AR [supporting information (SI) Fig. 4], the double mutant S161V/L115A was constructed to test whether the helix–helix interactions observed in rhodopsin also occur in β2-AR. Data from saturation binding experiments using DHA with this mutant indicated that the L115A mutation did not enhance expression of the S161V mutant to any significant extent, although the L115A mutant itself showed wild-type levels of expression (Fig. 1B and Table 2).
V114A (3.33)/S165V (4.57).
In rhodopsin, the F115A mutation rescues the defect in chromophore formation caused by the A168L mutation (P.J.R., M.E., P.C., V.H., U.L.R., H.G.K., and S.O.S., unpublished data). Phe-115 (3.30) in rhodopsin corresponds to Ser-111 (3.30) in β2-AR, whereas Ala-168 (4.57) in rhodopsin corresponds to Ser-165 (4.57) in β2-AR (SI Fig. 4). Given the small size of serine, it seemed unlikely that the reduced expression level of the β2-AR S165V mutant would be compensated for by an S111G mutation. In agreement with this expectation, the double mutant S111G/S165V showed the phenotype characteristic of the single mutant S165V and not that of the wild-type receptor (data not shown). However, the double mutant V114A/S165V was expressed at wild-type levels (Table 2), and immunoblotting showed that a part of it migrated as a fully glycosylated receptor (Fig. 1B). Thus the V114A mutation compensates for the reduced expression level of the S165V mutant. The V114A mutation reduced the binding affinity of the receptor for DHA from a Kd of 2.1 nM for the wild-type receptor to 19 nM for the mutant. The double mutant V114A/S165V exhibited a Kd of 9 nM, a value that is intermediate between those found for the wild-type receptor and the V114A mutant. Thus, the S165V mutant in turn compensates, at least partially, for the decreased affinity of the V114A mutant for the antagonist DHA.
Agonist Competition Assays.
Val-114 (3.33) is a highly conserved amino acid (96%) in the adrenergic receptor family (α and β) and is in close proximity to Ser-165 in the model of the ligand-free receptor. The V114A mutation decreased the affinity of β2-AR for [3H]DHA by 9-fold but sustained high-level expression (Table 1 and Fig. 1B). These observations are consistent with Val-114 having a functional role by forming part of the ligand-binding site for DHA and a structural role by being involved in close H3-H4 packing (Fig. 3). To investigate the possibility that Val-114 is important for ligand binding, we carried out competition radioligand binding assays using the catecholamine agonists, isoproterenol, epinephrine and norepinephrine, which differ only in the number of methyl substituents attached to the amine. The V114A mutation reduced by 200- to 300-fold the affinity for isoproterenol and epinephrine while causing only a 3-fold decrease in affinity for norepinephrine compared with the wild type β2-AR (SI Table 3). These results suggest that Val-114 is present in the ligand-binding pocket of the receptor and influences the position of bound ligand (see Discussion). The weak affinity of the V114A mutant for isoproterenol (SI Table 3) likely explains the finding that the agonist-stimulated activity of the V114A mutant as measured by cAMP accumulation is 4-fold lower than that of the wild type β2-AR under our assay conditions (Fig. 2D).
Molecular Modeling of β2-AR.
Molecular models of β2-AR were constructed to facilitate structural comparison of rhodopsin and β2-AR and to illuminate possible roles for the group-conserved sites in the two receptors. The sequence alignment is presented in SI Fig. 4. The structure of the ligand-free β2-AR was based on homology with the crystal structure of rhodopsin, except for the second extracellular loop (EL2) that was constructed de novo. EL2 forms a part of the retinal binding site in rhodopsin and likely forms part of the ligand-binding site in the β2-AR. However, there is virtually no homology between the EL2 sequences in these receptors. Noda et al. (11) concluded on the basis of alanine mutations and DTT sensitivity that two unique disulfide bonds constrain the position of EL2 in β2-AR, one between Cys-106 and Cys-191 and the other between Cys-184 and Cys-190. Furse and Lybrand (15) found that a de novo model containing EL2 having these two disulfide links was favored over an EL2 model constructed on the basis of homology with rhodopsin. Fig. 3 shows the ligand-free β2-AR structure. In A, we highlight the position of group-conserved residues Ala-78 (2.49), Ser-161 (4.53), and Ser-165 (4.57). This figure also shows the proximity of the H2-H4 core to the ligand-binding site and the positions of highly conserved binding site residues Val-114 (3.33) and Asp-113 (3.32) on H3. One of the functions of conserved Val-114 may be to orient the Asp-113 side chain for interaction with ligand. In B, we highlight the interactions of H3 with helices H5, H6, and H7, which are thought to move upon receptor activation. Group-conserved residues Ser-120 and Ala-128 are oriented toward H7 and H5, respectively. A structural model of β2-AR containing the bound antagonist alprenolol (SI Fig. 5) was based on the model of the ligand-free β2-AR. The alprenolol was manually docked into the binding site; the orientation of the antagonist was guided primarily by position of the amine (near Asp-113) and the aromatic ring (near Ser-203 and Ser-207).
Discussion
Structural Role of the Group-Conserved Residues in the β2-AR.
Identification of the group-conserved residues in class A GPCRs led to the conclusion that the transmembrane helices H1–H4 form a stable core containing the specific residues 1.46, 2.47, 2.49, 4.53, and 4.57 that mediate helix interactions within this core (6). In β2-AR, residue 1.46 is an isoleucine, which is not a group-conserved amino acid, suggesting that the packing core involves only helices H2–H4. Fig. 3A illustrates that close packing of H2, H3, and H4 in β2-AR allows an interhelical hydrogen bond to form between Trp-158 (4.50) and Ser-74 (2.45). These residues are highly conserved (97% and 88%, respectively) in the amine subfamily of class A GPCRs. In rhodopsin, substitution with leucine at each of the three group-conserved sites within the H2–H4 core (i.e., positions 2.49, 4.53, and 4.57) is detrimental to chromophore regeneration with 11-cis retinal and formation of a stable pigment. Interestingly, substitution of position 2.47 in the H1–H2 interface with leucine is tolerated in rhodopsin without effecting the ability to form the rhodopsin pigment or the conversion to the active metarhodopsin II intermediate (P.J.R., M.E., P.C., V.H., U.L.R., H.G.K., and S.O.S., unpublished results). In β2-AR, replacement of Ala-78 (2.49) with either serine or valine is tolerated. However, A78L mutation decreases the level of expression as illustrated by a low Bmax value, consistent with the idea that Ala-78 contributes to the H2–H4 core packing. The observation that all of the expressed and folded mutants at positions 2.47 and 2.49 had similar affinities for [3H]DHA eliminates a role for these residues in ligand binding.
Recent studies in membrane proteins have shown that sequence motifs containing small (Gly, Ala) and weakly polar (Ser, Thr, Cys) residues can mediate the interaction of α-helical transmembrane helices. In β2-AR, group-conserved residues Ser-161 and Ser-165 in H4 form an SxxxS packing motif; however, the structural or functional significance of this motif is unknown. Our mutational studies show that the β2-AR mutants containing the smaller amino acids (i.e., alanine) at positions Ser-161 (4.53) and Ser-165 (4.57) are expressed at high levels and bind ligand with near wild-type affinity, whereas substitution with valines lowers the expression of the mutant receptors. The bulkier amino acid leucine is not tolerated well at either position, with S161L and S165L mutants showing no significant specific binding to the antagonist DHA. Furthermore, the successful rescue of the poorly expressed S165V mutant by the V114A mutation supports the argument that helices H3 and H4 are in close proximity in this region of β2-AR. These results suggest that Ser-161 and Ser-165 in β2-AR have a structural role in the tight packing of helices H3 and H4.
Comparison of Roles of SxxxS Motifs in the β2-AR.
It has been previously shown that Ser-203 (5.42), Ser-204 (5.43), and Ser 207 (5.46) conserved in H5 of β2-AR are directly involved in agonist binding and receptor activation (9, 10). Our model of the inactive receptor (ligand-free or alprenolol-bound) places Ser-203 and Ser-207 in the interface with transmembrane helix H3, hydrogen bonding with Thr-118 on H3 and Thr-164 on H4 (SI Fig. 5). This placement is consistent with the observation that SxxxS motifs often mediate helix interactions by forming interhelical hydrogen bonds (13). It is also consistent with extensive mutational studies of Ser-207 by Ambrosio et al. (16) suggesting that this serine is not exposed to the polar binding crevice in the ligand-free receptor. We propose that the initial binding of the agonist to β2-AR leads to rotation of H5 so that the two serines in H5 are exposed to the ligand-binding site and interact with the hydroxyl-groups in the catechol ring. The same mechanism appears to be present in the visual pigment rhodopsin. Ser-207 in β2-AR corresponds to His-211 in rhodopsin, whereas Thr-118 (3.37) corresponds to Glu-122 in rhodopsin. Glu-122 in rhodopsin forms an interhelical hydrogen bond with the backbone carbonyl of His-211 in the inactive receptor. In the active receptor, H5 changes orientation and a new hydrogen bonding interaction is formed between the side chains of Glu-122 and His-211 (3).
The two conserved serines on H4 (Ser-161 and Ser-165), which also correspond to an SxxxS motif, do not appear to play functional roles in the β2-AR. Rather, they are involved primarily in helix packing and thereby maintaining the stability of the protein. Thus, the two conserved SxxxS motifs (S203xxxS207 and S161xxxS165) have different roles in the β2-AR.
Role of Group Conserved Residues in Receptor Activation.
Transmembrane helix H3 in rhodopsin and β2-AR plays a central role in receptor activation (Fig. 3). In addition to the group-conserved residues that facilitate the formation of the H2–H4 core, there are group-conserved residues on H3 (Ser-120 and Ala-128) that are oriented away from the H2–H4 core toward H5–H7. Helices H5–H7 are generally thought to change orientation upon GPCR activation and consequently, the group-conserved sites on H3 facing these helices may be involved in receptor function.
Ser-120 (3.39) is one of the highly conserved serines in the amine family (100% conserved among 387 sequences of amine family receptors). However, its role in receptor function was not known. Strader et al. (9) showed that the S120A mutation in hamster β2-AR resulted in no protein detectable by immunoblot analysis. To elucidate the role of Ser-120 (3.39) in β2-AR structure-function, we mutated it to alanine, leucine, and valine. Surprisingly, S120A behaves exactly like wild-type β2-AR in terms of ligand binding and the amount of protein observed on immunoblots. S120L and S120V are also tolerated based on the binding of the antagonist [3H]DHA, although the levels of receptor expression are 4- to 5-fold lower than that of the wild-type β2-AR (Table 1). Thus, Ser-120 (3.39) in β2-AR might be serving a role similar to that of Ala-124 rather than that of Gly-121 in rhodopsin, in which the A124L mutant is tolerated but not the G121L mutant. In our ligand-free β2-AR model (Fig. 3B), Ser-120 is situated in the interface with H6 and H7. The serine side chain is in close proximity to Phe-282 on H6 and also interacts with Asn-318 on H7. Mutation of Phe-282 in β2-AR has been shown to promote constitutive activity of the receptor (17), whereas Asn-318 is predicted to interact with Trp-286 (6.48). Motion of tryptophan at position 6.48 is generally thought to be associated with receptor activation (4, 18). Furthermore, Ser-120 in β2-AR is present in the part of H3, which is also referred to as the “switch region,” which was suggested to be involved in active state isomerization (19). Not surprisingly, our results from the cAMP assays suggest that the Ser-120 mutants display a slightly higher level of agonist-independent basal activity compared with the wild type β2-AR, however, the mechanism underlying this slight increase in activity needs further study.
Ala-128 (3.47) in β2-AR corresponds to Ala-132 in rhodopsin and is 1 aa away from the highly conserved E/DRY motif at the cytoplasmic end of H3 in GPCRs. In both receptors, mutation of alanine to serine, valine, and leucine are well tolerated, although the A128L β2-AR mutant shows reduced expression levels. Interestingly, both A128V and A128L mutants show very low levels of agonist-induced activity (Fig. 2C). In our ligand-free β2-AR model, Ala-128 (3.47) is present in the H3–H5 interface positioned between Val-214 and Val-218 on H5. As indicated above, it is thought that agonist binding results in motion of H5. Our mutational data suggest that the larger side chains at this position (3.47) might prevent H5 from adopting an active orientation.
Role of Val-114 (3.33) in Ligand Binding.
Val-114 (3.33) in β2-AR is found to be highly conserved (96%) in the adrenergic receptor family (including the α and β receptors) and is at a critical position relative to H4, H5, and the ligand-binding site(s) (Fig. 3). Val-114 (3.33) is located between Asp-113 (3.32), an amino acid essential for electrostatic interaction with the amine-group common to the ligands in the amine receptor subfamily (20), and the serine residues of the two conserved SxxxS motifs. As a result, the side chain of Val-114, is positioned to be in van der Waals contact with the bound ligand. On the basis of the results presented above, the methyl groups on the receptor agonists (SI Table 3) contribute to the binding energy of the ligand in the wild-type receptor. Mutation of Val-114 to alanine decreases agonist binding with the largest effect being on the ligands with amine methyl groups. Given the central position of Val-114, the V114A mutation must change the structure of the ligand-binding site. The methyl groups on the amine ligand may directly interact with Val-114 in the wild-type receptor or the β-branched side chain of Val-114 may orient the Asp-113 side chain. It is also possible that Val-114 may serve to orient the ligand to maximize hydrophobic interactions with other nonpolar amino acids and hydrogen bonding contacts with polar amino acids in the binding site. For instance, the low activity of the V114A mutant with bound isoproterenol may reflect the loss of interaction with the conserved serines on H5. Interestingly, we find that the double mutant S165V/V114A is able to rescue, at least partially, the affinity of β2-AR for the antagonist DHA.
Together, the results presented above indicate that an understanding of the mechanism of activation of the β2-AR and other class A GPCRs will require knowledge of the interplay between highly conserved signature residues, the group-conserved residues, and those residues that are specific to a receptor subfamily or type of ligand.
Methods
Synthetic oligonucleotides were purchased from Invitrogen (Carlsbad, CA). The β2-AR antagonists, [3H]DHA and [3H]CGP 12177, were purchased from Amersham (Little Chalfont, U.K.). Protease inhibitors and common chemicals were purchased either from Sigma (St. Louis, MO) or Invitrogen. Restriction enzymes were purchased from NEB (Boston, MA). Buffers used were as follows: PBS buffer, 137 mM NaCl, 2.7 mM KCl, 1.8 mM KH2PO4, 10 mM Na2HPO4 (pH 7.4); Buffer A (lysis buffer), 10 mM Tris·HCl, pH 7.4, containing protease inhibitors (1 mM EDTA, 10 μg/ml benzamidine, 10 μg/ml leupeptin, 20 μg/ml soybean trypsin inhibitor, 5 μg/ml aprotinin, and 0.2 mM phenylmethylsulfonyl fluoride); Buffer B (storage buffer), 50 mM Tris·HCl, pH 7.4, 12.5 mM MgCl2, containing protease inhibitors as in Buffer A; Buffer C (binding buffer), 75 mM Tris·HCl, pH 7.4, 12.5 mM MgCl2, containing protease inhibitors as in Buffer A.
Construction of Mutant β2-AR Genes.
Amino acid substitutions were introduced into the synthetic β2-AR gene carried by the pMT4 expression vector (21) by using the Quick-Change mutagenesis kit (Stratagene, La Jolla, CA). Compensatory mutations for the S161V and S165V mutants were introduced by using the S161V or S165V mutant, respectively, as the template DNA. DNA sequences of the mutated genes were verified by dideoxynucleotide chain-termination sequencing (MIT Biopolymers Laboratory, Cambridge, MA).
Cell Culture and Immunoblot Analysis.
The wild-type β2-AR gene was expressed in COS-1 cells by using a DEAE-dextran-based transient transfection method (22). For transient transfection of HEK293S cells, the plasmid pMT4 was cotransfected with pRSVTag plasmid, a gift from Jeremy Nathans (The Johns Hopkins University, Baltimore, MD), and Lipofectamine 2000-(Invitrogen) mediated transfection was used as described by the manufacturer. Membranes were prepared as described in ref. 21. The protein concentration in the resuspended membrane pellet was determined by using a modified DC protein assay kit from Bio-Rad Laboratories (Hercules, CA). Solubilized protein (2.5 or 5 μg) was resolved by 10% SDS/PAGE and electroblotted onto a nitrocellulose membrane. β2-AR was visualized by immunodetection with the monoclonal antibody rho-1D4.
Radioligand Binding Assays.
These were carried out as previously described (21). Competition binding assays were performed by using 2 nM [3H]DHA and different concentrations of unlabeled agonists (10−2 to 10−9 M) and the reactions were carried out for 2 h at room temperature. Binding was terminated by filtering under vacuum on GF/A filters (Millipore, Bedford, MA). Filter-bound radioactivity was measured by using a liquid scintillation counter. Equilibrium dissociation constants (Kd) were determined from saturation isotherms. Radioligand binding data obtained from competition curves were analyzed by nonlinear regression analysis to determine the EC50 values and Ki values by using PRISM software version 4.03 (GraphPad Software, San Diego, CA).
cAMP Assays.
Functional characterization of β2-AR was carried out by using a commercially available cAMP assay system (Amersham Biosciences) following the directions supplied by the manufacturer, other details are as described in ref. 12. The cAMP values were normalized to the total membrane protein in each assay.
Molecular Modeling of the β2-Adrenoreceptor.
A homology model of β2 adrenergic receptor was constructed by using Modeller (version 8v2) (23). The high-resolution (2.2 Å) crystal structure of rhodopsin (1) was used as a template with the sequences (Opsd_bovin and Adrb2_mesau) aligned according to GPCR database (http://www.gpcr.org/7tm) (24). The EL2 was built de novo by using loop modeling facility in Modeller (24) incorporating restraints due to two disulfide bonds (26). The resulting structure was further refined by the use of the all-atom force-field ff94 (27) and Amber simulation package by running short (200 ps) molecular dynamics with weak positional restraints (0.5 kcal/mol per Å) on α-carbons in the transmembrane region, followed by complete minimization. Alprenolol was manually docked into the binding site and refined by short molecular dynamics (100 ps) and minimization.
Supplementary Material
Acknowledgments
The assistance of Ms. Judy Carlin in the preparation of this manuscript is gratefully acknowledged. This work was supported by National Institutes of Health Grants EY11716 (to H.G.K.) and GM41412 (to S.O.S.) and National Science Foundation Grant EIA-0225609 (to H.G.K.). P.C. is supported by an American Heart Association Postdoctoral fellowship.
Abbreviations
- β2-AR
β2-adrenergic receptor
- EL2
extracellular loop-2
- DHA
dihydroalprenolol
- GPCR
G protein-coupled receptor.
Footnotes
The authors declare no conflict of interest.
The nomenclature used to describe amino acid positions in GPCRs follows the convention established by Ballesteros and Weinstein (5).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702024104/DC1
References
- 1.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 3.Patel AB, Crocker E, Reeves PJ, Getmanova EV, Eilers M, Khorana HG, Smith SO. J Mol Biol. 2005;347:803–812. doi: 10.1016/j.jmb.2005.01.069. [DOI] [PubMed] [Google Scholar]
- 4.Patel AB, Crocker E, Eilers M, Hirshfeld A, Sheves M, Smith SO. Proc Natl Acad Sci USA. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballesteros JA, Weinstein H. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 6.Liu W, Eilers M, Patel AB, Smith SO. J Mol Biol. 2004;337:713–729. doi: 10.1016/j.jmb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Hwa J, Garriga P, Liu X, Khorana HG. Proc Natl Acad Sci USA. 1997;94:10571–10576. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han M, Lin SW, Smith SO, Sakmar TP. J Biol Chem. 1996;271:32330–32336. doi: 10.1074/jbc.271.50.32330. [DOI] [PubMed] [Google Scholar]
- 9.Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RA. J Biol Chem. 1989;264:13572–13578. [PubMed] [Google Scholar]
- 10.Liapakis G, Ballesteros JA, Papachristou S, Chan WC, Chen X, Javitch JA. J Biol Chem. 2000;275:37779–37788. doi: 10.1074/jbc.M002092200. [DOI] [PubMed] [Google Scholar]
- 11.Noda K, Saad Y, Graham RM, Karnik SS. J Biol Chem. 1994;269:6743–6752. [PubMed] [Google Scholar]
- 12.Kim JM, Hwa J, Garriga P, Reeves PJ, RajBhandary UL, Khorana HG. Biochemistry. 2005;44:2284–2292. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- 13.Adamian L, Liang J. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- 14.Stojanovic A, Hwang I, Khorana HG, Hwa J. J Biol Chem. 2003;278:39020–39028. doi: 10.1074/jbc.M303625200. [DOI] [PubMed] [Google Scholar]
- 15.Furse KE, Lybrand TP. J Med Chem. 2003;46:4450–4462. doi: 10.1021/jm0301437. [DOI] [PubMed] [Google Scholar]
- 16.Ambrosio C, Molinari P, Fanelli F, Chuman Y, Sbraccia M, Ugur O, Costa T. J Biol Chem. 2005;280:23464–23474. doi: 10.1074/jbc.M502901200. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, Lin F, Xu M, Riek RP, Novotny J, Graham RM. Biochemistry. 2002;41:6045–6053. doi: 10.1021/bi012189c. [DOI] [PubMed] [Google Scholar]
- 18.Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA. J Biol Chem. 2002;277:40989–40996. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 19.Perez DM, Karnik SS. Pharmacol Rev. 2005;57:147–161. doi: 10.1124/pr.57.2.2. [DOI] [PubMed] [Google Scholar]
- 20.Strader CD, Sigal IS, Register RB, Candelore MR, Rands E, Dixon RA. Proc Natl Acad Sci USA. 1987;84:4384–4388. doi: 10.1073/pnas.84.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chelikani P, Reeves PJ, Rajbhandary UL, Khorana HG. Protein Sci. 2006;15:1433–1440. doi: 10.1110/ps.062080006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oprian DD, Molday RS, Kaufman RJ, Khorana HG. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 24.Horn F, Bettler E, Oliveira L, Campagne F, Cohen FE, Vriend G. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiser A, Do RK, Sali A. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dohlman HG, Caron MG, Deblasi A, Frielle T, Lefkowitz RJ. Biochemistry. 1990;29:2335–2342. doi: 10.1021/bi00461a018. [DOI] [PubMed] [Google Scholar]
- 27.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.