Abstract
MicroRNAs (miRNAs) are a family of ≈22-nt noncoding RNAs that can posttranscriptionally regulate gene expression. Several miRNAs are specifically expressed in hematopoietic cells. Here we show that one such miRNA, miR-150, is mainly expressed in the lymph nodes and spleen and is highly up-regulated during the development of mature T and B cells; expression of miR-150 is sharply up-regulated at the immature B cell stage. Overexpression of miR-150 in hematopoietic stem cells, followed by bone marrow transplantation, had little effect on the formation of either mature CD8- and CD4-positive T cells or granulocytes or macrophages, but the formation of mature B cells was greatly impaired. Furthermore, premature expression of miR-150 blocked the transition from the pro-B to the pre-B stage. Our results indicate that miR-150 most likely down-regulates mRNAs that are important for pre- and pro-B cell formation or function, and its ectopic expression in these cells blocks further development of B cells.
Keywords: bone marrow transplantation, hematopoiesis, hematopoietic stem/progenitor cell, lymphopoiesis
B and T cells are derived from hematopoietic stem cells, which reside in the bone marrow of adult mice and the liver of embryos. In particular, the bone marrow is the major site of B cell development in adult mice. B cells are derived from a common lymphoid progenitor (CLP) cell (lin−c-kitlowSca-1low IL-7R+) (1), which is limited in its ability to develop into T lymphocytes but is fully capable of developing into B cells. B cell development is divided into several successive steps as judged by surface markers and rearrangement of immunoglobin genes. The first B lineage-restricted cells are termed pro-B cells, which initiate rearrangement at the Ig heavy-chain (IgH) locus: DH to JH joining at the early pro-B cell stage, followed by VH to DJH joining at the late pro-B cell stage (2, 3). The immunoglobin heavy chain (μ chain) then assembles with a surrogate light chain (SLC) and forms the pre-B cell receptor (pre-BCR) to mediate expansion of pre-B cells. When the SLC is replaced by a successfully rearranged immunoglobin light chain (IgL) to form IgM, the cells become immature B cells (3–5).
Many cytokines and transcription factors play critical regulatory roles in B lymphopoiesis (6). Fms-like tyrosin kinase-3 (flt3) and IL-7 receptor (IL-7R)-mediated signaling are required for the production of pro-B cells (7–11), whereas the transcription factor E2A, early B cell factor (EBF), Myb, Foxp1, and Pax5 all have critical effects on the early stages of B lymphopoiesis (12–15). However, many critical questions pertaining to the regulation of B cell fate determination and early B cell development remain unanswered.
MicroRNAs (miRNAs) are 20- to 24-nt noncoding RNAs found in diverse plants and animals (16). In animals, they are processed from characteristic hairpin structures of longer primary transcripts by the sequential action of Drosha and Dicer, two RNase III-type nucleases that act in the nucleus and cytoplasm, respectively (17). The mature miRNAs are then incorporated into a silencing complex containing an Argonaute protein, in which they can pair to the messages of protein-coding genes to direct their posttranscriptional repression (18–20). Extensive base pairing of the miRNA and the target mRNA leads to mRNA cleavage, whereas imperfect matches often result in translational inhibition and mRNA destabilization (21–23).
miRNAs play important roles in hematopoiesis. For example, miR-181a is detected in lineage− (lin−) undifferentiated cells in the bone marrow and is up-regulated in B cells (B220+). Ectopic expression of miR-181a in hematopoietic stem cells leads to an increase in the production of CD19+ B cells and to a decrease of CD8+ T cells (24). miR-15 and miR-16 genes are often deleted or expressed at reduced levels in B cell chronic lymphocytic leukemias (CLL), suggesting that these miRNAs might be tumor-suppressor genes (25–27). However, miR-17-92 and miR-155 are highly expressed in B cell lymphomas (28, 29), and miR-17-92 can function as an oncogene when ectopically expressed with c-Myc (28). Recent studies also found a unique miRNA expression signature in chronic lymphocytic leukemia (30). miRNA expression profiles vary significantly across different cancer types, and almost all miRNAs were down-regulated in mixed lineage leukaemia (MLL) (31). In addition, the interaction between miR-223 and two transcription factors, NFI-A and C/EBPα, appears to play an important regulatory role in granulocyte formation (32).
In this study, we found that one lymphopoietic-specific miRNA, miR-150, is highly expressed in mature B and T cells. To study the role of miR-150 in hematopoiesis, we ectopically expressed this miRNA; hematopoietic stem and progenitor cells were retrovirally infected with miR-150 and then introduced into lethally irradiated mice by bone marrow transplantation. Overexpression of miR-150 in hematopoietic progenitor/stem cells led to significantly reduced mature B cell (MB) levels in the circulation and the spleen and lymph nodes, with little or no change in the population levels of T cells and myeloid cells. Further studies demonstrated that overexpression of miR-150 in hematopoietic stem cells blocked B lymphopoiesis by inhibiting the transition from the pro-B to the pre-B cell stage.
Results
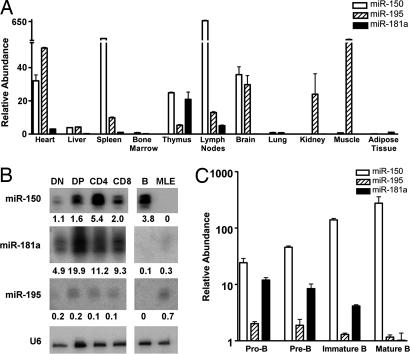
We first used miRNA arrays to examine the miRNAs' expression profile and found a group of miRNAs that were highly expressed in several differentiated hematopoietic cell types (data not shown). Expression of these miRNAs in various hematopoietic tissues was then evaluated by quantitative RT-PCR assays. Of the miRNAs we detected, we chose to focus on two hematopoietic cell-specific miRNAs, miR-150 and miR-181a. miR-195 was used as a control miRNA because it was expressed in many types of mouse cells (Fig. 1A). Mature miR-150 was most abundantly expressed in the lymph nodes. It was also highly expressed in the spleen and detectable in the thymus, heart, and brain; expression was not detectable in other tissues. Expression levels of miR-181a were very high in the thymus; were lower in the heart, lymph nodes, and bone marrow, and at very low or undetectable levels in most of the other tissues examined, consistent with results in ref. 24.
Fig. 1.
Expression profile of selected miRNAs. (A) miRNA expression in various tissues. Total RNAs were isolated from tissues collected from 4- to 6-week-old C57BL/6 mice and treated with DNase I to remove trace amounts of DNA contamination. Equal amounts of total RNAs from each sample were subjected to reverse transcription using specifically designed primers for each miRNA, followed by quantitative PCR. Relative expression levels of each mature miRNA were normalized to U6 expression levels. (B) miRNA expression in thymic T cell precursors detected by Northern blot analysis. To examine the expression of miR-150, miR-181a, and miR-195 in various thymic T cells, the total RNA samples were loaded onto a 10% denaturing polyacrylamide gel and hybridized with different probes complementary to the indicated mature RNAs. The relative intensities of each band was normalized to the U6 loading control by using Multigage software and are indicated below each lane. (C) miRNA expression in B cell precursors detected by quantitative RT-PCR. Various stages of B cell precursors were isolated from bone marrow by FACS: pro-B cells (B220+CD43+IgM−), pre-B cells (B220+CD43−IgM−), and IMBs (B220+CD43−IgM+). MBs (B220+IgM+IgD+) were isolated from spleen (SI Fig. 5). Total RNAs were extracted from each population by using TRIzol reagent, and miRNA expression was measured by quantitative RT-PCR using the ABI microRNA assay kit (Applied Biosystems, Foster City, CA). The relative abundances displayed are an average of triplicates of quantitative PCR (log scale). All results were normalized to U6 levels that were detected by using the ABI miRNA U6 assay kit (Applied Biosystems).
RNA blots showed that expression of miR-150 increased during T cell development. When normalized to U6 small nuclear RNA (snRNA) levels, miR-150 expression was low in the precursor double negative (DN) cells, moderate in double positive (DP) cells and CD8 single positive cells (SP), and high in CD4 cells. Expression of miR-181a was also up-regulated during T cell development, although to a lower extent. In contrast, the expression of miR-195 was essentially unchanged throughout the stages of T cell development.
In parallel, we used FACS to isolate mature splenic B cells (B220+IgM+IgD+) and three bone marrow B cell progenitor populations: pro-B cells (B220+CD43+IgM−), pre-B cells (B220+CD43−IgM−), and immature B cells (IMB) (B220+CD43−IgM+) [supporting information (SI) Fig. 4]. Quantitative RT-PCR showed that expression of miR150 was up-regulated throughout B cell differentiation (Fig. 1C). Expression was lowest in pro- and pre-B cells and up-regulated during the immature and MB stages. In contrast, the levels of miR-195 did not change during B cell development, and miR-181a expression was down-regulated.
Ectopic expression of miR-181a in hematopoietic stem cells leads to an increase in B cell production and a decrease in CD8 T cell production (24). To test the effects of miR-150 and miR-195 overexpression, we first developed a retroviral vector able to induce expression of the mature miR-150 and miR-195 miRNAs (SI Fig. 5). To this end, various lengths of sequences flanking the stem-loops of miR-150 and miR-195 were cloned into the retroviral vector MDH, and production of mature miRNA was examined 48 h after transfection of 293T cells with the retroviral construct. Two retroviral constructs, miR-150-130nt and miR-195–90nt, which expressed primary transcripts that were efficiently processed (SI Fig. 5), were carried forward for in vivo studies.
Next, fetal liver hematopoietic stem/progenitor cells (C57BL/6 CD45.2+Lin−) were infected with these retroviruses and injected into lethally irradiated recipient mice (C57BL/6 CD45.1+). Mice injected with bone marrow cells infected with a vector containing no miRNA (MDH) were used as a control. Because the retroviral constructs also expressed a GFP gene, infected cells and their progeny could be distinguished from the other donor-derived cells by selecting for donor-derived (45.2-positive) cells in peripheral blood, spleen, or lymph nodes that did or did not express GFP. By comparing the donor-derived GFP+ and GFP− populations, we were thus able to visualize the effect of miRNA overexpression on the production of different cell lineages from hematopoietic stem and progenitor cells.
We collected peripheral blood samples at 4 weeks for short-term (Fig. 2A and SI Fig. 6) and 16 weeks for long-term transplantation repopulation analysis (Fig. 2B). Repopulation at 4 weeks primarily represents the progeny of transplanted late stem cells and lymphoid and myeloid progenitor cells, whereas repopulation after 4 months or longer represents the progeny of long-term repopulating hematopoietic stem cells.
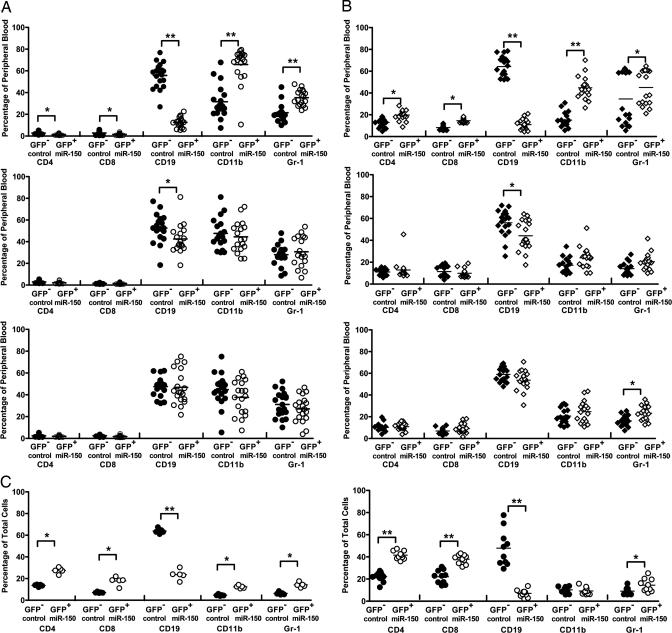
Fig. 2.
Effects on short-term and long-term repopulation caused by overexpression of miR-150 and miR-195. (A and B) Lin− fetal liver cells from day 13.5–14.5 C57BL/6 mice (CD45.2) were prepared as described in Methods, spin-infected with miR-150 (Top), miR-195 (Middle), or empty MDH retroviral supernatant (Bottom), and transplanted into irradiated CD45.1 recipients. To evaluate the effects of short-term and long-term repopulation, peripheral blood samples were collected at 4 (A) or 16 (B) weeks after transplantation. Donor-derived (CD45.2) cell lineages were examined by using antibodies against specific cell lineage surface markers followed by FACS analysis: B cells (CD19+), T cells (CD4+ or CD8+), and myeloid cells (CD11b+ or Gr-1+). The percentage of each cell lineage in either the GFP− or GFP+ population from each mouse was defined as that specific cell lineage proportion in the peripheral blood. The mean value for each group was the average of all cell proportions from 20 mice in two independent transplantations. (C) The long-term repopulation effects of the transplantation in the spleen (Right) and lymph (Left) nodes of miR-150 mice were examined as described. Statistics from each cell lineage were calculated by the Mann–Whitney test. Significance was defined as follows: ∗, P < 0.05; ∗∗, P < 0.001.
Overexpression of miR-150 affected both lymphoid and myeloid differentiation. At both 4 and 16 weeks after transplantation, the number of GFP+ donor-derived B lymphocytes in the peripheral blood was reduced ≈4- to 6-fold, whereas the number of GFP+ donor-derived granulocytes (Gr-1+) and monocytes (CD11b+) increased slightly. There was also a slight increase in mature CD4 and CD8 T populations after 16 weeks. Similar changes were observed in the spleen and lymph nodes after 16 weeks: Expression of miR-150 severely reduced the numbers of B cells while slightly increasing numbers of CD4 and CD8 T cells, with little or no increase in numbers of granulocytes (Gr-1+) and monocytes (CD11b+) (Fig. 2C). The slight increases in the proportions of non-B cells we observed could be caused by the large decrease in numbers of B cells in peripheral blood, spleen, and bone marrow and not by a specific effect of ectopic expression of miR-150 in these non-B cells. Neither overexpression of miR-195 nor the control MDH vector had any effect on the number of donor-derived lymphoid or myeloid cells after both 4 weeks and 16 weeks of transplantation.
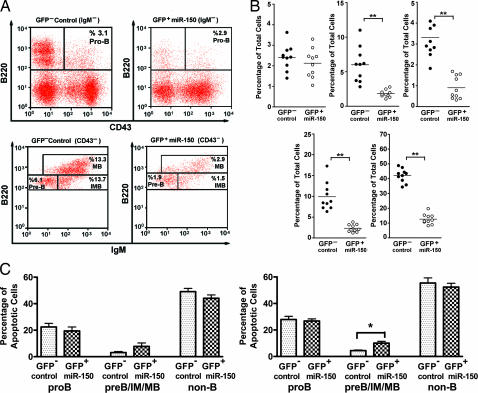
Because a significant decrease in the MB production was observed in peripheral blood, spleen, and lymph nodes of mice that ectopically expressed miR-150, we hypothesized that this phenotype resulted from defects in B cell development. To test this hypothesis, we examined B cell precursors in the bone marrow of mice 16 weeks after transplantation. Ectopic expression of miR-150 did not affect the number of pro-B cells (B220+CD43+IgM−) but led to significantly decreased numbers of B cells beyond this stage, including pre-B cells (B220+CD43−IgM−), IMBs (B220+CD43−IgM+), and MBs (B220hiCD43−) (Fig. 3A).
Fig. 3.
Impaired B cell development in mice ectopically expressing miR-150. Bone marrow cells were collected at 16 weeks from mice that were transplanted with hematopoietic stem/progenitor cells overexpressing miR-150. The cell population was then analyzed by staining with antibodies against B220, IgM, and CD43, followed by FACS analysis. (A) Donor-derived (CD45.2) B cell population from one mouse: pro-B cells (B220+IgM−CD43+), pre-B cells (B220+ IgM− CD43−), IMBs (B220+IgM+CD43−), and MBs (B220highCD43−). The proportion of each population was defined as the percentage of the entire GFP− (Left) or GFP+ (Right) cell population from each mouse. Pro-B cells from the GFP− control population and the GFP+ miR-150-expressing population from one mouse are displayed. (B) A total of 10 mice from two independent transplantations were examined for all four bone marrow B cell precursors and MBs (B220+IgM+IgD+). Shown are bone marrow pro-B cells (Upper Left), bone marrow pre-B cells (Upper Center), bone marrow IMBs (Upper Right), bone marrow MBs (Lower Right), and spleen MBs (Lower Left). (C) For the apoptosis assay, red blood cell-depleted bone marrow cells were cultured in RPMI 1640 complete medium for 0 h (Left) to 3 h (Right) and stained for B220, CD43, and Annexin V. The percentage of Annexin V cells in pro-B cells (B220+CD43+) or pre-B cells/IMBs/MBs (B220+CD43−) from GFP− or GFP+ populations were analyzed by the Mann–Whitney test. Significance was defined as follows: ∗, P < 0.05; ∗∗, P < 0.001.
To confirm that impaired B cell production was not simply an effect of higher sensitivity to apoptosis, we examined Annexin V-positive cells in both GFP+ and GFP− bone marrow populations from mice that were transplanted with hematopoietic stem cells overexpressing miR-150. Overexpressed miR-150 caused no change in apoptosis in the pro-B (B220+CD43+) and non-B cell (B220−) populations, and a slightly but significantly higher rate of apoptosis was observed in the pre-B cells/IMBs/MBs (B220+CD43−) (Fig. 3C; P = 0.0952 for 0-h culture, P = 0.0159 for 3-h culture). These results support the idea that miR-150 overexpression blocks B cell development at the pro-B cell stage in part by causing a slightly increased rate of apoptosis. Most likely, the reduced number of cells observed beyond this stage is also due to miR-150 regulation of certain target genes essential for early B cell development.
Discussion
We found that miR-150 was expressed at very high levels in the lymph nodes and was also present at high levels in the spleen and the thymus. Furthermore, miR-150 expression increased strikingly during successive stages of both B and T cell maturation in the bone marrow and thymus, respectively, suggesting that it may participate in B and/or T lymphopoiesis. Indeed, prematurely expressing miR-150 led to severe defects in B cell development in the bone marrow, along with moderately enhanced T lymphopoiesis and myelopoiesis from hematopoietic stem and progenitor cells. Consistent with this inhibition of B cell production, dramatically reduced MB levels were also observed in the spleen and the lymph nodes 16 weeks after transplantation.
The donor-derived B cell populations 4 weeks after transplant represent the progeny mainly of short term hematopoietic stem cells and later lineage-restricted progenitors, whereas the donor-derived B cell populations seen 16 weeks after transplant represent the progeny mainly of long-term repopulating hematopoietic stem cells. A detailed analysis of B cell maturation showed no abnormality in the numbers of donor-derived pro-B cells overexpressing miR-150 both 4 and 16 weeks after bone marrow transplantation. In contrast, at both time points, there were significantly reduced numbers of donor-derived pre-B cells, IMBs, and MBs overexpressing miR-150. Thus, very few cells overexpressing miR-150 were able to progress from the pro-B cell stage to the pre-B stage and beyond. Overexpressing miR-150 caused a slight increase of apoptosis in IMBs, as judged by the percentage of apoptotic B cells in the bone marrow either immediately after sample preparation, representing the original rate of cell death in vivo, or after a 3-h culture, representing continued apoptosis.
Thus over expression of miR-150 in hematopoietic stem and progenitor cells did not affect B cell lineage commitment, as evidenced by unaltered pro-B cell numbers both 4 and 16 weeks after transplantation. Nor did overexpression of miR-150 affect apoptosis of pro- and pre-B cells. Overexpression of miR-150 did block the differentiation of pro-B cells to pre-B cells. Presumably, miR-150 normally down-regulates late in B cell development some mRNA(s) important for development or function of the earlier pro- or pre-B cells. In this scenario, premature repression of these messages from early ectopic expression of miR-150 in pro- or pre-B cells could inhibit the development or function of these cells, thereby interfering with their ability to progress to the next stage of maturation. The increased apoptosis in IMBs caused by miR-150 overexpression could be triggered by failure of these cells to pass a cell division checkpoint that is part of their normal differentiation.
Surveying the predicted targets of miR-150 (33), we found several genes that play important roles in B cell development, including Myb and Foxp1. Two highly conserved 8-nt sites in the 3′ UTR of Myb mRNA and one conserved 7-nt site in the Foxp1 3′ UTR are complementary to the miR-150 “seed” region (33).
Myb is highly expressed in pro-B and pre-B cells, then down-regulated >80% in IMBs (15), concomitant with the increase in miR-150 expression we observed. Foxp1, together with PAX5, is required for IgH gene rearrangement and is highly expressed in pro-B and pre-B cells (34, 14). Mice with deleted Myb or Foxp1 genes display severe B cell development defects at the pro-B to pre-B transition (14, 15), a phenotype similar to what we observed in mice ectopically expressing miR-150 in hematopoietic stem and progenitor cells.
Even though the expression of miR-150 was low in thymic double-negative cells and high in the more differentiated single positive cells, overexpressing miR-150 in hematopoietic stem and progenitor cells did not significantly alter T cell development, as judged by the levels of donor-derived CD4 and CD8 T cells. Thus the high expression of miR-150 we observed in the naïve single positive cells thus suggests that this miRNA may play a role in T cell activation and function rather than in thymic T cell development.
We conclude that miR-150, a hematopoiesis-specific miRNA, exerts critical regulation on B lymphopoiesis. Ectopic expression of miR-150 in hematopoietic stem cells leads to severe defects in B cell development and decreased expression of this miRNA was detected in mixed lineage leukemia (MLL) (31), suggesting that regulated miR-150 expression is necessary for B cell development.
Materials and Methods
Mouse and Tissue Preparation.
CD45.1 and CD45.2 C57BL/6 mice were purchased from either The Jackson Laboratory (Bar Harbor, ME) or the National Cancer Institute and were maintained at the animal facility of the Whitehead Institute for Biomedical Research. All animal experiments were performed with the approval of the Massachusetts Institute of Technology Committee on Animal Care.
Bone Marrow Transplantation and Repopulation Assay.
Retrovirally infected hematopoietic stem/progenitor cells were prepared as described in refs. 35 and 36. In general, fetal livers from day 13.5 or day 14.5 C57BL/6 mice were collected and stained with biotin-conjugated anti-CD3e, anti-CD5, anti-B220, anti-CD11b, anti-Gr-1, and anti-Ter119 (BD Pharmingen, San Diego, CA). After 20 min of incubation on ice, the cells were washed with cold PBS, reacted with streptavidin-conjugated magnetic beads (Miltenyi Biotec, Auburn, CA), and then removed by using AutoMacs magnetic separation. Lin− cells were then plated at 4 × 105 cells per ml to 5 × 105 cells per ml in a six-well plate and spin-infected for 1 hour with the desired retroviral supernatant. Cells were cultured overnight in Iscoves' modified Dulbecco medium (IMDM) containing 15% FBS, 30 ng/ml stem cell factor (SCF), 20 ng/ml Flt3L, and 10 ng/ml IL-6. Cells were then resuspended with retroviral supernatant for a second round of spin infection at 1,600 × g and at 37°C. The cells were subsequently washed and resuspended in PBS with 2% FBS at a concentration of 1 × 107 cells per ml. For each miRNA, 10 recipient CD45.1 mice were lethally irradiated (10 Gy) and then retro-orbitally injected with 1 × 106 cells. Our results are from 20 mice from two individual injections.
Immunostaining and Flow Cytometry Analysis.
All antibodies used in immunostaining were purchased from BD Pharmingen unless otherwise indicated.
For Northern blot analysis, thymocytes were prepared from 4- to 6-week-old mice and stained with fluorescence-tagged antibodies: phycoerythrin (PE)-anti-CD4 (GK1.5), FITC-anti-CD8 (53-6.7), biotin-anti-CD11b (M1/70), CD11c (HL3), and B220 (RA3-6B2). After secondary staining with streptavidin-allophycocyanin (APC), the cells were sorted by FACS to collect the double positive (DP), CD4+CD8−, and CD4−CD8+ T cell populations. Double negative (DN) cells were purified by negative selection (B220−CD11c−CD11b−CD4−CD8−).
Peripheral blood samples or single-cell suspensions from the spleen, bone marrow, or lymph nodes were collected. From transplanted mice, CD45.1+ (recipient) cells were depleted with biotin-anti-CD45.1 (A20; eBioscience, San Diego, CA) in combination with streptavidin magnetic beads (Miltenyi Biotec). Red blood cells were lysed with ammonium chloride solution, and the remaining cells were stained with APC-anti-CD45.2 (104; eBioscience) and the following lineage surface markers: PE-anti-CD4 (GK1.5), PE or APC-anti-CD8 (53-6.7) for T cells; PE-anti-CD19 (1D3) or PE-Cy5.5 anti-B220 (RA3-6B2; eBioscience) for B cells; PE-anti CD11b (M1/70) for monocytes; and PE-Cy5.5 anti-Gr-1 (RB6-8C5; eBioscience) or PE-anti-Gr-1 (RB6-8C5) for granulocytes. The proportion of each cell lineage was defined as the percentage of cells displaying a specific lineage marker either in the GFP+ population (cells that ectopically express the miRNAs of interest) or in the uninfected GFP− control population. Cells from the spleen and lymph nodes were also stained with PE-Cy5.5-anti-B220, PE-IgD (11-26c.2a) and APC-IgM (II/41; eBioscience) for MBs (B220+IgM+IgD+).
To study the various B cell developmental stages, bone marrow cells were collected from transplanted mice and stained with PE-Cy5.5-anti-B220, PE-CD43 (eBioscience), and APC-IgM antibodies. B cell progenitors were defined by the presence of these antibody combinations: pro-B cells (B220+CD43+IgM−), pre-B cells (B220+CD43−IgM−), IMBs (B220+CD43−IgM+), and bone marrow MBs (B220hiCD43−).
In Vitro Apoptosis Assay.
Single-cell suspensions from the bone marrow were prepared after CD45.1 and erythrocyte depletion. Cells were stained with PE-Cy5.5-anti-B220, APC-anti-CD43, and Annexin V (Annexin V PE Apoptosis Detection kit; BD Pharmingen) after 0- and 3-h incubation in RPMI 1640 medium supplemented with 10% FBS. All samples were then analyzed by FACS.
Supplementary Material
Acknowledgments
We thank Drs. Jianzhu Chen and Herman Eisen for their intellectual support; Michael J. Axtell and I-Hung Shih for help with the miRNA array; Chengcheng Zhang for help with hematopoietic stem cell preparation and bone marrow transplantation; Tony Chavarria for help with blood sample collection; Qibin Leng for help with T cell preparation; and G. Paradis at the Massachusetts Institute of Technology Flow Cytometry Core Facility for cell sorting and analysis. This work was supported by U.S. National Institutes of Health Grant R01 DK068348 (to H.F.L. and D.P.B.).
Abbreviations
- APC
allophycocyanin
- IMB
immature B cell
- MB
mature B cell
- miRNA
microRNA
- PE
phycoerythrin.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702409104/DC1.
References
- 1.Kondo M, Weissman IL, Akashi K. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 2.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karasuyama H, Rolink A, Shinkai Y, Young F, Alt FW, Melchers F. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 4.Torres RM, Flaswinkel H, Reth M, Rajewsky K. Science. 1996;272:1804–1808. doi: 10.1126/science.272.5269.1804. [DOI] [PubMed] [Google Scholar]
- 5.Li YS, Wasserman R, Hayakawa K, Hardy RR. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 6.Busslinger M. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 7.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, et al. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeurer MJ, Lotze MT. Int Rev Immunol. 1998;16:309–322. doi: 10.3109/08830189809042999. [DOI] [PubMed] [Google Scholar]
- 9.Henney CS. Immunol Today. 1989;10:170–173. doi: 10.1016/0167-5699(89)90175-8. [DOI] [PubMed] [Google Scholar]
- 10.Mertsching E, Meyer V, Linares J, Lombard-Platet S, Ceredig R. Int Rev Immunol. 1998;16:285–308. doi: 10.3109/08830189809042998. [DOI] [PubMed] [Google Scholar]
- 11.Veiby OP, Lyman SD, Jacobsen SE. Blood. 1996;88:1256–1265. [PubMed] [Google Scholar]
- 12.Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 13.Lin H, Grosschedl R. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 14.Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, Tucker PW, Rao A. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- 15.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. Immunity. 2005;23:275–286. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Kim VN. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 18.Tang G. Trends Biochem Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Hutvagner G, Zamore PD. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 21.Yekta S, Shih IH, Bartel DP. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 22.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 23.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Chen CZ, Li L, Lodish HF, Bartel DP. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 27.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 28.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Croce CM. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 32.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 33.Lewis BP, Burge CB, Bartel DP. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 34.van Zelm MC, van der Burg M, de Ridder D, Barendregt BH, de Haas EF, Reinders MJ, Lankester AC, Revesz T, Staal FJ, van Dongen JJ. J Immunol. 2005;175:5912–5922. doi: 10.4049/jimmunol.175.9.5912. [DOI] [PubMed] [Google Scholar]
- 35.Zhang CC, Steele AD, Lindquist S, Lodish HF. Proc Natl Acad Sci USA. 2006;103:2184–2189. doi: 10.1073/pnas.0510577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong W, Zhang J, Lodish HF. Blood. 2005;105:4604–4612. doi: 10.1182/blood-2004-10-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.