Abstract
An actomyosin motor located underneath the plasma membrane drives motility and host-cell invasion of apicomplexan parasites such as Plasmodium falciparum and Plasmodium vivax, the causative agents of malaria. Aldolase connects the motor actin filaments to transmembrane adhesive proteins of the thrombospondin-related anonymous protein (TRAP) family and transduces the motor force across the parasite surface. The TRAP–aldolase interaction is a distinctive and critical trait of host hepatocyte invasion by Plasmodium sporozoites, with a likely similar interaction crucial for erythrocyte invasion by merozoites. Here, we describe 2.4-Å and 2.7-Å structures of P. falciparum aldolase (PfAldo) obtained from crystals grown in the presence of the C-terminal hexapeptide of TRAP from Plasmodium berghei. The indole ring of the critical penultimate Trp-residue of TRAP fits snugly into a newly formed hydrophobic pocket, which is exclusively delimited by hydrophilic residues: two arginines, one glutamate, and one glutamine. Comparison with the unliganded PfAldo structure shows that the two arginines adopt new side-chain rotamers, whereas a 25-residue subdomain, forming a helix–loop–helix unit, shifts upon binding the TRAP-tail. The structural data are in agreement with decreased TRAP binding after mutagenesis of PfAldo residues in and near the induced TRAP-binding pocket. Remarkably, the TRAP- and actin-binding sites of PfAldo seem to overlap, suggesting that both the plasticity of the aldolase active-site region and the multimeric nature of the enzyme are crucial for its intriguing nonenzymatic function in the invasion machinery of the malaria parasite.
Keywords: actin, Apicomplexa, cell invasion machinery, gliding motility, induced fit
Malaria is one of the most devastating parasitic diseases worldwide, amounting to 300–500 million cases and 1–2 million deaths per year (1). Different species of Plasmodium infect the human host, the most important ones being Plasmodium falciparum and Plasmodium vivax. Increased occurrence of multidrug-resistant Plasmodium strains reflects the need and development for new effective antimalarials (2, 3). Human infection starts when anopheline mosquitoes inject sporozoites into the skin during a blood meal. The sporozoites gain access to the blood stream and ultimately invade hepatocytes where they develop and multiply. Upon rupture of the infected hepatocytes, merozoites are released and rapidly enter erythrocytes, where they undergo schizogony and propagate the blood cycle of the infection that causes the symptoms of malaria.
A multiprotein complex located in the narrow space between the plasma membrane and the microtubule-supported inner membrane complex (IMC) empowers both substrate-dependent gliding motility and host-cell invasion in Plasmodium (reviewed in refs. 4 and 5). This machinery consists of short-length actin filaments (6–8) moved by an unconventional class XIV myosin A (MyoA) (9) that is tightly anchored to the IMC by means of its association with the MyoA-tail-interacting protein (10–12) and the GAP45–GAP50 complex (13, 14). Our recent structure of MTIP bound to the MyoA-tail (15) showed interaction between two invasion machinery proteins. The current study provides insight at the atomic level of the second inter-protein interaction occurring in this fascinating and essential multiprotein assembly of the malaria parasite.
The transmembrane surface adhesins responsible for contacting the host-cell receptor(s) belong to the TRAP family, present across the phylum Apicomplexa (16, 17). Surprisingly, the connection between the actin filaments and the cytoplasmic tail of TRAP-like proteins is not direct but mediated by fructose 1,6-bisphosphate (F1,6P) aldolase (18, 19). This interaction requires the presence of acidic residues and an essential, penultimate, Trp residue (20, 21) that characterizes the cytoplasmic tail of TRAP family proteins. It is through the aldolase connection that the motor force is translated into capping and redistribution of TRAP-like molecules along the parasite surface (4, 22). When TRAP-like molecules engage cellular receptors, a moving junction is formed between the host-cell membrane and the parasite, and invasion follows, as originally described in Plasmodium merozoites (23). All of the components of this machinery, as well as their overall arrangement, are present in different Plasmodium invasive stages (sporozoites, merozoite, and the mosquito vector-restricted ookinetes) and are evolutionary conserved across the phylum Apicomplexa (5, 20, 24–26). Because it is obligatory for Plasmodium and most Apicomplexa to invade various host cells to complete their life cycle, the machinery is a very attractive target for drug development (15, 27).
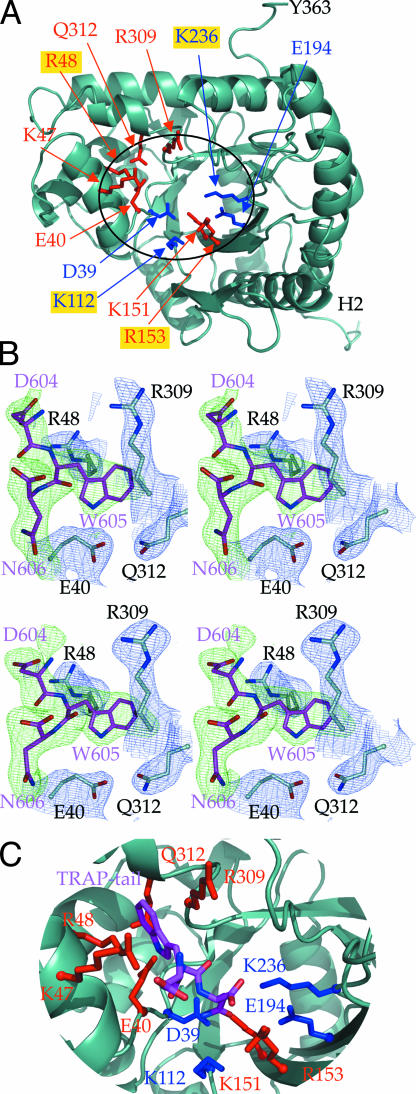
P. falciparum aldolase (PfAldo) belongs to the class I aldolases, which are present in most organisms and are highly conserved at the amino acid sequence and structural level (28). The crystal structure of PfAldo was initially solved in our laboratory to 3 Å resolution in an unliganded state (29). Crystallographic studies of class I aldolases from several other species have been reported. Most relevant for our work are those of the human aldolases where unliganded structures of the enzyme isoforms found in muscle (HsA) (30), liver (HsB) (31), and brain tissue (HsC) (32) have been described, and also that of HsA in complex with the substrate F1,6P (30). Additional crystallographic studies of covalent reaction intermediates and aldolases trapped in complex with the substrate F1,6P, the products dihydroxyacetone phosphate and glyceraldehyde-3-phosphate, or competitive inhibitors have helped in understanding the reaction mechanism (30, 33–36). In class I aldolases, the reversible cleavage reaction of F1,6P is mediated by Schiff-base formation of a lysine with the substrate. This lysine, which corresponds to Lys-236 in PfAldo (Lys-229 in HsA), together with additional key active-site residues of PfAldo, is depicted in Fig. 1A.
Fig. 1.
Structure of P. falciparum aldolase and bound TRAP-tail. (A) Structural overview of one PfAldo subunit (light blue ribbon). Residues binding to the TRAP-tail are highlighted in red; residues involved in binding of F1,6P in the active site are shown in blue. Key substrate binding residues are as follows: Pf-Asp-39 (HsA-Asp-33), Pf-Lys-112 (HsA-Lys-107), Pf-Lys-151 (HsA-Lys-146), Pf-Arg-153 (HsA-Arg-148), and Pf-Glu-194 (HsA-Glu-187). The main chain nitrogens of Pf-Ser-278 (HsA-Ser-271), Pf-Gly-279 (HsA-Gly-272) bind to the 1-phosphate group of F1,6P (30). Labels of PfAldo residues equivalent to rabbit aldolase amino acids implicated by site directed mutagenesis studies (43) to be involved in actin binding (i.e., R48, K112, R153, and K236) are highlighted with a gold background. (B) Difference electron densities for the DWN C-terminal residues of the TRAP-tail in two independent PfAldo:TRAP-tail structures. The σA weighted (Fo − Fc) difference densities, shown in green and contoured at 2σ, were calculated by omitting all TRAP-tail residues during the refinement procedure. The C-terminal TRAP-tail residues 604–606 are shown as sticks in magenta. The corresponding (2Fo − Fc) electron density maps of subunit D in both structures is contoured at 1σ in blue. (Upper) The 2.4-Å data set. (Lower) The 2.7-Å data set (see SI Materials and Methods and SI Table 1). (C) Close-up of the active site and TRAP-binding site with important residues highlighted. The TRAP-tail residues are shown in magenta, TRAP-binding residues in red, and active-site residues in blue.
Here we report the structure of PfAldo in complex with the C-terminal TRAP-tail of the rodent malaria parasite P. berghei. Our findings reveal a most surprising binding mode for the critical penultimate tryptophan residue near the PfAldo active site, involving an induced fit mechanism, which includes a rigid body motion of a 25 residue subdomain. The position of the TRAP-tail residues is in good general agreement with recently reported molecular modeling studies (37), which were carried out entirely independently from the current study [supporting information (SI) Fig. 6]. Based on our results we propose that molecular flexibility and the multimeric nature of aldolase are both critical for its interactions with both TRAP and actin in the invasion machinery.
Results
Overall Structure of the TRAP-Tail-Bound PfAldo.
Even though the 25 C-terminal residues of TRAP play a role in the binding to aldolase, biochemical data indicated that the extreme C terminus of TRAP is a crucial component of this interaction (21). Based on these results, we used a TRAP-tail hexapeptide (Ac-EDNDWN) corresponding to residues 601–606 of P. berghei TRAP for cocrystallization with PfAldo.
Two structures of PfAldo complexed with the TRAP-tail peptide were solved to 2.4 and 2.7 Å resolution (see SI Materials and Methods) by using the unliganded PfAldo structure [PDB ID code (29)] as a molecular replacement model. The refined tetrameric structures have an Rwork/Rfree combination of 20.1/25.0% and 20.8/26.9%, with excellent geometry (see SI Table 1).
Clearly defined difference electron density was observed in the two PfAldo structures in subunit D. The other three subunits in both structures displayed less well defined TRAP-tail density. The difference density observed in subunit D corresponds to the C-terminal residues DWN of the TRAP-tail with the indole ring of the penultimate W605, and the carboxylate and carboxamide of the C-terminal N606, fitting well into the difference density in the higher resolution structures (Fig. 1B). The DWN tripeptides in the two structures superimpose within ≈1.0 Å (see SI Fig. 7); therefore, we describe in the rest of the article the 2.4 Å resolution structure. The most striking feature is the binding mode of the penultimate TRAP-W605, previously shown to be essential for the PfAldo-TRAP interaction (19, 21). Its indole ring is buried in a hydrophobic groove formed solely by four hydrophilic residues of which three are charged: PfAldo R48, E40, R309, and Q312. The side chain of R48 and main chain plus Cβ atoms of R309 are the major constituents of the indole-binding pocket, with the indole nitrogen forming a hydrogen bond with the carboxyl group of E40 (Fig. 2A). The C-terminal TRAP-N606 interacts with K151 via its carboxamide group, and with R153 using its C-terminal carboxyl group. The side chain of TRAP-D604 does not seem to make contacts with aldolase but its negative charge is likely to contribute electrostatically to the free energy of binding because of the large overall positive charge of PfAldo in this region. The three residues upstream of D604 of the TRAP hexapeptide bound do not display interpretable density, which suggests significant conformational flexibility. However two of these side chains are negatively charged and occur in the neighborhood of positively charged PfAldo side chains of K46, K47, R48, K53, and R309. Therefore alteration in the charge of these side chains are likely to affect affinity in agreement with biochemical studies (19–21).
Fig. 2.
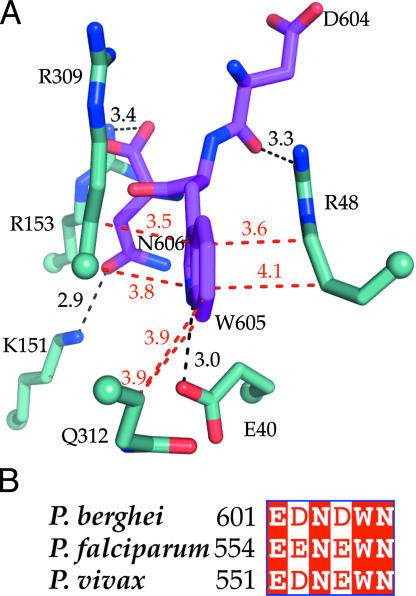
Key interactions of the TRAP-tail with P. falciparum aldolase. (A) TRAP-tail residues D604, W605, and N606 (magenta) interacting with PfAldo residues (light blue). Cα positions are marked as spheres. Selected hydrophobic contacts are shown with red lines and distances in angstroms. The hydrogen bond between the indole nitrogen of TRAP-W605 and the carboxylate of E40, and the interactions between TRAP-N606 with R153 and K151, and of the backbone oxygen of TRAP-D604 with R48, are depicted in black. (B) Sequence alignment of the C-terminal TRAP-tail residues of three plasmodial species. Identical residues are shaded in red. All TRAP residues involved in contacts with aldolase are well conserved in several Plasmodium species.
The Indole Anchor-Binding Pocket.
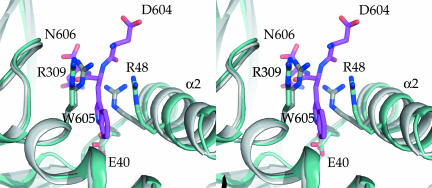
Comparing the PfAldo:TRAP complex with the unliganded PfAldo structure (29) reveals that TRAP-tail binding is accompanied by critical conformational changes. Of the four indole-binding residues, E40 and Q312 undergo relatively small changes, by 0.6 and 0.2 Å for their side chains, respectively. In contrast, the side chain of the indole-binding R309 moves considerably, up to ≈3.1 Å for the Cζ atom, whereas the side chain of R48 shifts by 3.0 Å and its main chain by 1.5 Å. The latter motion is correlated with a rigid body shift of ≈1.1 Å of ≈25 residues of the helix α2–loop–helix α3 region comprising residues T42 to D66 (Fig. 3). Evidently, the TRAP-tryptophan indole creates its own binding pocket, which is absent in the unliganded PfAldo structure (see SI Fig. 8). The overall movements of side and main chains induced by the intercalating TRAP-W605 result in a distinctly larger active-site groove when compared with the unliganded state of PfAldo. Therefore, TRAP-tail binding by PfAldo constitutes a classical example of “induced fit.”
Fig. 3.
Conformational changes of P. falciparum aldolase upon TRAP-tail binding. Shown is a stereoview of the TRAP-binding region. The TRAP-bound structure is shown in light blue, the unliganded structure in gray (29), and the TRAP-tail C-terminal tripeptide DWN in magenta. Key residues binding the penultimate Trp-indole ring are highlighted with sticks. Note the rigid body motion of helix α2, which is part of the helix–loop–helix T42-D66 subdomain shift upon TRAP-binding (see The Indole Anchor-Binding Pocket).
TRAP and Substrate-Binding Regions Overlap.
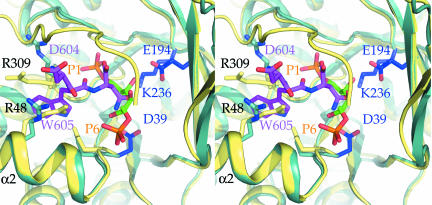
Several structures have revealed how the substrate F1,6P is bound to class I aldolases from a variety of species (30, 33–35), and it is of interest to compare the F1,6P-binding site with that of the TRAP-tail. The superposition reveals that F1,6P binding is incompatible with simultaneous binding of the TRAP-tail because of several clashes between the two entities bound. For instance, the C4 and C5 atoms of the substrate F1,6P are essentially at the same position as the TRAP-N606 side chain (Fig. 4). This positional overlap is in excellent agreement with previous biochemical data, which demonstrated decreased enzymatic activity of PfAldo upon addition of a 34- and 25-mer peptide spanning the C terminus of TRAP, and inhibition of TRAP-binding by aldolase substrates such as F1,6P, dihydroxyacetone phosphate, or glyceraldehyde-3-phosphate (19, 21, 37). Peptide inhibition studies of PfAldo by the TRAP-tail hexapeptide from P. berghei, EDNWN and P. falciparum, EENEWN yielded IC50 values of ≈40 μM and ≈300 μM, respectively. This inhibition of activity by TRAP hexapeptides is in complete agreement with the observed binding site of the P. berghei TRAP-tail in our structure.
Fig. 4.
TRAP-tail binding interferes with substrate binding. Shown is a stereoview of the PfAldo:TRAP-tail complex superimposed onto the structure of human aldolase A complexed with F1,6P (30) (PDB ID code 4ALD). Key residues enabling the accommodation of the penultimate Trp-indole ring are highlighted as sticks. The TRAP-tail is shown in magenta, residues involved in substrate binding in blue, and human aldolase A in yellow. The substrate F1,6P are shown as sticks in green, with the two phosphate groups in orange. The C-terminal TRAP-tail residue partially occludes the substrate-binding site.
Mutation Studies.
We mutated several residues of PfAldo in the neighborhood of the TRAP-tail-binding site and evaluated the TRAP-binding capacity of each mutant (Fig. 5) using previously described methods (19, 21). Mutation of the indole-contacting R48 to either D or G, completely abolished TRAP-binding, which correlates well with its extensive contact with TRAP-W605 (Fig. 2A). The E40G mutant displays essentially no TRAP-binding, which is in agreement with the fact that the interaction partner for the indole nitrogen of TRAP-W605 has been removed in this mutant. Significant effects are also observed when two other indole-contacting residues, R309 and Q312, are individually mutated to serine, leading in each case to a 20-fold reduction in TRAP-binding. These two residues mainly mediate hydrophobic contacts with TRAP-W605 via their Cβ atom and R309 also with its Cα atom.
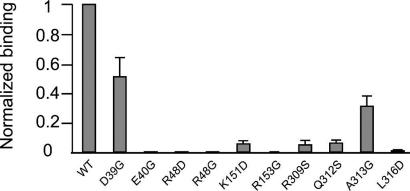
Fig. 5.
Effects of PfAldo residue substitutions on TRAP-binding. The percentage of TRAP-binding compared with WT PfAldo, as evaluated by methods described elsewhere (19), is plotted on the vertical axis. The aldolase mutants studied are plotted along the horizontal axis. The excellent correlation between the effect of the substitutions and the structure of the PfAldo:TRAP-tail complex is discussed in Mutation Studies.
Mutation of R153 to glycine removes its interaction with the carboxyl group of TRAP-N606 which agrees with decreased affinity of the R153G substitution. Changing K151 to an aspartate removes an interaction with the side chain of TRAP-N606 which explains the decreased TRAP-binding of this mutant. The substitution of L316 by an aspartate results in decreased affinity for TRAP because the Cδ2 of L316 is in van der Waals contact with the Cβ of R48. This change in side chain at position 316 alters therefore the position of the critical R48. Other substitutions, e.g., of D39 and A313 to glycine, have a less dramatic effect on TRAP-binding, which correlates well with the fact that these residues are further away from the TRAP-tail. The mutagenesis results seem to be in excellent agreement with our structure (Figs. 1, 2A, and 5).
Discussion
Comparison with Other Studies.
The binding site of the three C-terminal residues of the TRAP-tail to PfAldo observed in our structures (Figs. 1B and 2A) corresponds well with results from other investigations. In particular the demonstration that mutations of the penultimate Trp residue of TRAP-like molecules to structurally closely related Tyr and Phe residues disrupts binding to aldolase (21) is in excellent agreement with the binding mode observed. The smaller phenol side chain of a Tyr cannot form the hydrogen bond with E40, whereas a Phe would have a very unfavorable interaction with this glutamate side chain. This substitution is also in agreement with our mutagenesis studies, which show that the E40G PfAldo mutation essentially leads to a complete loss of TRAP-tail-binding affinity (Fig. 5). This result is all the more remarkable because the E40G mutation removes a negative charge from the TRAP-tail-binding site, which would be expected to promote binding of the negatively charged TRAP-tail, but the opposite is the case thereby confirming the importance of E40 in interacting with the indole.
The MWMD tail of MIC2, the homolog of TRAP in Toxoplasma gondii (18), can be modeled without problems in the T. gondii aldolase structure: (i) the first Met of the MIC2 tail points toward the solution; and, (ii) a favorable interaction between the second Met of the MIC2 tail and the aliphatic part of the side chain of R148 in T. gondii aldolase, corresponding to PfAldo R153, occurs after some minor side-chain shifts (see SI Fig. 9). Our observed binding mode of the DWN TRAP-tail tripeptide is compatible with the EWDD tail of the Wiskott–Aldrich Syndrome protein peptide binding to aldolase (21). When maintaining the position of the W-indole anchor, the N-terminal D to E substitution, and the addition of the C-terminal D, can be readily accommodated in our structure (data not shown). It therefore seems that the aldolase active-site region is capable of binding related amino acid sequences from different proteins with the indole ring of the binding partner serving as an anchor (Figs. 1 and 2A) whereas multiple negatively charged carboxylates upstream and downstream are engaged in favorable electrostatic interactions with the numerous positive charges of the active-site region. The highly conserved Trp residue in the tail of TRAP-like molecules from different Apicomplexan species has been experimentally associated with gliding and host-cell invasion (5, 17, 18, 38, 39).
Perspectives for Drug Design.
To evaluate opportunities for selective inhibition of the TRAP-binding site, our PfAldo:TRAP-tail complex structure was compared with the human aldolase isozymes. Human brain Aldolase C (HsC), which is the closest structural homolog to PfAldo according to the EBI SSM server (40), superimposed with PfAldo with an rms deviation of 0.90 Å for 329 residues and an overall sequence identity of 56% [1XFB, (32)]. The three human isoforms differ mutually in sequence by 28.7% (HsA vs. HsB), 16.7% (HsA vs. HsC), and 27.8% (HsB vs. HsC), and differ by 42.5–47.3% when compared with PfAldo. The nine active-site residues are invariant in all four enzymes. The four residues (E40, R48, R309, and Q312) defining the TRAP-W605 pocket are conserved in all three human aldolases, as are eight residues of the 25 helix–loop–helix residues undergoing the rigid body shift upon TRAP-tail binding by PfAldo (Fig. 3). Nevertheless, selective inhibitors of protozoan aldolases, including that from Plasmodium, have been reported (41). The observed flexibility in the active-site region of PfAldo suggests that small molecules might be discovered which selectively inhibit TRAP-binding to PfAldo without impairing the functioning of human aldolases by exploring differences between PfAldo and human aldolase residues at some distance from the active site, e.g., by contacting second and third shell residues of the binding site, or by preventing the shift of the helix–loop–helix unit which is required for creating the TRAP-binding site.
Simultaneous TRAP and Actin Binding.
The motion of the helix–loop–helix T42-D66 region identified in our structure is of particular interest as it partially corresponds to the biochemically identified actin-binding region of rabbit aldolase (42) equivalent to D39-N51 of PfAldo. Also, the four side chains of rabbit aldolase implicated in actin binding by mutagenesis studies (43), corresponding to R48, K112, R153, and K236 in PfAldo, are in close proximity to the TRAP-binding region revealed by our studies (Fig. 1A). Likely overlap of the TRAP and actin-binding sites of aldolase is also suggested by enzymatic activity studies of PfAldo indicating that TRAP-binding as well as actin binding is competitively inhibited by F1,6P (19). At the same time, Buscaglia and coworkers (19) have shown that PfAldo, TRAP-tail and actin can form a ternary complex, whereas Jewett and Sibley (18) provided evidence for the same complex in T. gondii. Aldolases in Apicomplexa therefore seem to have the remarkable property of binding both TRAP and actin with the overlapping and therefore mutually exclusive binding sites for these partner proteins, when considering a single aldolase subunit. However, taking into account that aldolase is tetrameric (29, 44), this apparent contradiction is resolved: different subunits of the same aldolase tetramer can bind different invasion machinery proteins. Interestingly, our hypothesis is in agreement with several studies reporting that different subunits of the same aldolase tetramer display different affinities for small molecule ligands (33, 34, 45–47) (see SI Materials and Methods). These studies and our results suggest that F1,6P aldolases are sophisticated allosteric assemblies where protein binding near the active site of one subunit affects the affinities for proteins near the active sites of other subunits. It might even be possible that a single PfAldo tetramer recruited for the invasion machinery interacts not only with two different proteins but also uses one or two of its subunits to catalyze the conversion of F1,6P.
Materials and Methods
Expression and purification of recombinant PfAldo in E. coli was performed according to a previously reported procedure (29) (see SI Materials and Methods). In vapor diffusion experiments, 500 nl of 10 mg/ml PfAldo were mixed with 500 nl of the reservoir solution in a 96-well CompactClover plate (Emerald Biostructures, Bainbridge Island, WA) and incubated at 25°C. The final optimized crystal growth conditions were as follows: 2.5 mM TRAP-tail, 0.5% n-dodecyl-β-d-maltoside, 50 mM cacodylic acid (pH 6.0), and 20% PEG 2000. No crystals were obtained without peptide present in the crystallization solution. Crystals could be obtained with the P. falciparum TRAP-tail hexapeptide, but they diffracted poorly. Crystals of the PfAldo-P. berghei TRAP-tail complex were allowed to equilibrate at room temperature for 2 min in a cryo-protecting solution consisting of 1 μl of 50% glycerol, 0.5% n-dodecyl-β-d-maltoside, 50 mM cacodylic acid (pH 6.0), 20% PEG 2000, and 5 mM P. berghei TRAP-tail. Crystals were then flash frozen in liquid nitrogen for further examination at synchrotron facilities.
Data were collected at the Advanced Light Source 8.2.2 and Stanford Synchrotron Radiation Laboratory 9-2 beamlines at λ = 0.9796 Å with 0.75°–1° rotation angles per image and 10- to 60-s exposure time. The structures were solved by molecular replacement techniques and refined with good statistics (see SI Materials and Methods and SI Table 1).
Supplementary Material
Acknowledgments
We thank Mark Robien, Tracy Arakaki, Ethan Merritt, Martin Criminale, Frank Zucker, and other members of the SGPP Consortium (www.sgpp.org) for assistance and stimulating discussions. We thank the staff at the Howard Hughes Medical Institute (HHMI) 8.2.2 beamline at the Advanced Light Source (ALS) and Stanford Synchrotron Radiation Laboratory (SSRL) 9-2 beamline for their assistance. This project was supported by National Institutes of Health National Institute of General Medical Sciences Grant 1P50 GM64655-01, for the SGPP Consortium, and by HHMI (to W.G.J.H.). This work was also supported in part by a grant from the NIH (to V.N.).
Abbreviations
- PfAldo
Plasmodium falciparum aldolase
- TRAP
thrombospondin-related anonymous protein
- F1,6P
fructose 1,6-bisphoshate
- HsA
human aldolase A (muscle)
- HsB
human aldolase B (liver)
- HsC
human aldolase C (brain).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2PC4 and ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0605301104/DC1.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korenromp EL, Williams BG, Gouws E, Dye C, Snow RW. Lancet Infect Dis. 2003;3:349–358. doi: 10.1016/s1473-3099(03)00657-1. [DOI] [PubMed] [Google Scholar]
- 3.Baird JK. N Engl J Med. 2005;352:1565–1577. doi: 10.1056/NEJMra043207. [DOI] [PubMed] [Google Scholar]
- 4.Kappe SHI, Buscaglia CA, Bergman LW, Coppens I, Nussenzweig V. Trends Parasitol. 2004;20:13–16. doi: 10.1016/j.pt.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Sibley LD. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- 6.Schmitz S, Grainger M, Howell S, Calder LJ, Gaeb M, Pinder JC, Holder AA, Veigel C. J Mol Biol. 2005;349:113–125. doi: 10.1016/j.jmb.2005.03.056. [DOI] [PubMed] [Google Scholar]
- 7.Schuler H, Mueller AK, Matuschewski K. Mol Biol Cell. 2005;16:4013–4023. doi: 10.1091/mbc.E05-02-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahoo N, Beatty W, Heuser J, Sept D, Sibley LD. Mol Biol Cell. 2006;17:895–906. doi: 10.1091/mbc.E05-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meissner M, Schluter D, Soldati D. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- 10.Herm-Gotz A, Weiss S, Stratmann R, Fujita-Becker S, Ruff C, Meyhofer E, Soldati T, Manstein DJ, Geeves MA, Soldati D. EMBO J. 2002;21:2149–2158. doi: 10.1093/emboj/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergman LW, Kaiser K, Fujioka H, Coppens I, Daly TM, Fox S, Matuschewski K, Nussenzweig V, Kappe SHI. J Cell Sci. 2003;116:39–49. doi: 10.1242/jcs.00194. [DOI] [PubMed] [Google Scholar]
- 12.Green JL, Martin SR, Fielden J, Ksagoni A, Grainger M, Lim B, Molloy JE, Holder AA. J Mol Biol. 2006;355:933–941. doi: 10.1016/j.jmb.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 13.Gaskins E, Gilk S, DeVore N, Mann T, Ward G, Beckers C. J Cell Biol. 2004;165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rees-Channer RR, Martin SR, Green JL, Bowyer PW, Grainger M, Molloy JE, Holder AA. Mol Biochem Parasitol. 2006;149:113–116. doi: 10.1016/j.molbiopara.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Bosch J, Turley S, Daly TM, Bogh SM, Villasmil ML, Roach C, Zhou N, Morrisey JM, Vaidya AB, Bergman LW, Hol WGJ. Proc Natl Acad Sci USA. 2006;103:4852–4857. doi: 10.1073/pnas.0510907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sultan AA, Thathy V, Frevert U, Robson KJH, Crisanti A, Nussenzweig V, Nussenzweig RS, Menard R. Cell. 1997;90:511–522. doi: 10.1016/s0092-8674(00)80511-5. [DOI] [PubMed] [Google Scholar]
- 17.Kappe S, Bruderer T, Gantt S, Fujioka H, Nussenzweig V, Menard R. J Cell Biol. 1999;147:937–943. doi: 10.1083/jcb.147.5.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jewett TJ, Sibley LD. Mol Cell. 2003;11:885–894. doi: 10.1016/s1097-2765(03)00113-8. [DOI] [PubMed] [Google Scholar]
- 19.Buscaglia CA, Coppens I, Hol WGJ, Nussenzweig V. Mol Biol Cell. 2003;14:4947–4957. doi: 10.1091/mbc.E03-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menard R. Cell Microbiol. 2001;3:63–73. doi: 10.1046/j.1462-5822.2001.00097.x. [DOI] [PubMed] [Google Scholar]
- 21.Buscaglia CA, Penesetti D, Tao MY, Nussenzweig V. J Biol Chem. 2006;281:1324–1331. doi: 10.1074/jbc.M506346200. [DOI] [PubMed] [Google Scholar]
- 22.Soldati D, Foth BJ, Cowman AF. Trends Parasitol. 2004;20:567–574. doi: 10.1016/j.pt.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 23.Miller LH, McAuliffe FM, Johnson JG. Prog Clin Biol Res. 1979;30:497–502. [PubMed] [Google Scholar]
- 24.Gaffar FR, Yatsuda AP, Franssen FFJ, de Vries E. Mol Biochem Parasitol. 2004;136:25–34. doi: 10.1016/j.molbiopara.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Baum J, Richard D, Healer J, Rug M, Krnajski Z, Gilberger TW, Green JL, Holder AA, Cowman AF. J Biol Chem. 2006;281:5197–5208. doi: 10.1074/jbc.M509807200. [DOI] [PubMed] [Google Scholar]
- 26.Jones ML, Kitson EL, Rayner JC. Mol Biochem Parasitol. 2006;147:74–84. doi: 10.1016/j.molbiopara.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Cowman AF, Crabb BS. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Gefflaut T, Blonski C, Perie J, Willson M. Prog Biophys Mol Biol. 1995;63:301–340. doi: 10.1016/0079-6107(95)00008-9. [DOI] [PubMed] [Google Scholar]
- 29.Kim H, Certa U, Dobeli H, Jakob P, Hol WGJ. Biochemistry. 1998;37:4388–4396. doi: 10.1021/bi972233h. [DOI] [PubMed] [Google Scholar]
- 30.Dalby A, Dauter Z, Littlechild JA. Protein Sci. 1999;8:291–297. doi: 10.1110/ps.8.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalby AR, Tolan DR, Littlechild JA. Acta Crystallogr D. 2001;57:1526–1533. doi: 10.1107/s0907444901012719. [DOI] [PubMed] [Google Scholar]
- 32.Arakaki TL, Pezza JA, Cronin MA, Hopkins CE, Zimmer DB, Tolan DR, Allen KN. Protein Sci. 2004;13:3077–3084. doi: 10.1110/ps.04915904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blom N, Sygusch J. Nat Struct Biol. 1997;4:36–39. doi: 10.1038/nsb0197-36. [DOI] [PubMed] [Google Scholar]
- 34.Choi KH, Mazurkie AS, Morris AJ, Utheza D, Tolan DR, Allen KN. Biochemistry. 1999;38:12655–12664. doi: 10.1021/bi9828371. [DOI] [PubMed] [Google Scholar]
- 35.Choi KH, Shi J, Hopkins CE, Tolan DR, Allen KN. Biochemistry. 2001;40:13868–13875. doi: 10.1021/bi0114877. [DOI] [PubMed] [Google Scholar]
- 36.St-Jean M, Lafrance-Vanasse J, Liotard B, Sygusch J. J Biol Chem. 2005;280:27262–27270. doi: 10.1074/jbc.M502413200. [DOI] [PubMed] [Google Scholar]
- 37.Buscaglia CA, Hol WG, Nussenzweig V, Cardozo T. Proteins. 2007;66:528–537. doi: 10.1002/prot.21266. [DOI] [PubMed] [Google Scholar]
- 38.Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos FC, Sinden RE. EMBO J. 1999;18:6221–6227. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuda M, Sakaida H, Chinzei Y. J Exp Med. 1999;190:1711–1716. doi: 10.1084/jem.190.11.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krissinel E, Henrick K. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 41.Dax C, Duffieux F, Chabot N, Coincon M, Sygusch J, Michels PAM, Blonski C. J Med Chem. 2006;49:1499–1502. doi: 10.1021/jm050237b. [DOI] [PubMed] [Google Scholar]
- 42.O'Reilly G, Clarke F. FEBS Lett. 1993;321:69–72. doi: 10.1016/0014-5793(93)80623-3. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Morris AJ, Tolan DR, Pagliaro L. J Biol Chem. 1996;271:6861–6865. [PubMed] [Google Scholar]
- 44.Millar JR, Shaw PJ, Stammers DK, Watson HC. Philos Trans R Soc London Ser B. 1981;293:209–214. doi: 10.1098/rstb.1981.0074. [DOI] [PubMed] [Google Scholar]
- 45.Grazi E, Trombetta G. Int J Biochem. 1987;19:197–200. doi: 10.1016/0020-711x(87)90332-6. [DOI] [PubMed] [Google Scholar]
- 46.Grazi E, Trombetta G. Arch Biochem Biophys. 1984;233:595–602. doi: 10.1016/0003-9861(84)90484-3. [DOI] [PubMed] [Google Scholar]
- 47.Grazi E, Trombetta G, Lanzara V. Biochem Biophys Res Commun. 1983;110:578–583. doi: 10.1016/0006-291x(83)91189-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.