Abstract
The dominance of sexual reproduction is still an unresolved enigma in evolutionary biology. Strong advantages of sex have to exist, because only a few parthenogenetic taxa persist over evolutionary timescales. Oribatid mites (Acari) include outstanding exceptions to the rule that parthenogenetically reproducing taxa are of recent origin and doomed to extinction. In addition to the existence of large parthenogenetic clusters in oribatid mites, phylogenetic analyses of this study and model-based reconstruction of ancestral states of reproduction imply that Crotoniidae have reevolved sexuality from parthenogenetic ancestors within one of those clusters. This reversal in reproductive mode is unique in the animal kingdom and violates Dollo's law that complex ancestral states can never be reacquired. The reevolution of sexuality requires that ancestral genes for male production are maintained over evolutionary time. This maintenance likely is true for oribatid mites because spanandric males exist in various species, although mechanisms that enable the storage of genetically ancestral traits are unclear. Our findings present oribatid mites as a unique model system to explore the evolutionary significance of parthenogenetic and sexual reproduction.
Keywords: oribatid mites, parthenogenesis, spanandric males, automixis, ancient asexuals
The enigma of the evolution of sex comprises two processes, the origin and the maintenance of sex. Theories on the advantages of sex mainly refer to the improvement of the progeny's fitness in sexual populations despite reducing the overall number of offspring (1–2). Nevertheless, one of the enduring mysteries of biology is the prevalence of sexual reproduction in eukaryotes. Because parthenogenetic species do not waste resources in producing males (the now-classic “two-fold” advantage) and do not break up favorable gene combinations, they should rapidly out-compete sexual species in most environments (1–2). Why this is not true has been debated for decades, with so many answers having been proposed (3–5) that a second enigma has emerged: How could a few animal lineages have maintained parthenogenetic reproduction over considerable evolutionary time, avoiding extinction long enough to radiate and form monophyletic clades? The most studied examples of such “ancient asexual scandals” (1) are darwinulid ostracods (6), bdelloid rotifers (7), and several large clusters within oribatid mites (8–11).
Mites exhibit a bewildering array of genetic systems and reproductive modes (12, 13), and parthenogenetic reproduction has evolved numerous times. Parthenogenesis is most common in Oribatida, a widespread and abundant group of soil invertebrates. An estimated 9% of species are parthenogenetic, which is one to two orders of magnitude higher than in other animal groups (8). Most parthenogenetic oribatid mites are clustered in species-rich clades with no known sexual species, making each such clades an independent “asexual scandal” (8, 9, 11, 14).
The pattern of reproductive modes is most varied in Desmonomata, a speciose group with an age of at least 100 million years (11, 15), probably predating the break-up of Pangea (16). Although most families in this group are either entirely parthenogenetic or sexual, there is also one with mixed reproductive modes (Table 1) (17). All parthenogenetic species have a highly female-biased sex ratio, with most populations having >99% females, whereas sexual species comprise at least 30% males (14, 17). Evidence of these patterns comes from culturing and population studies of a wide range of species throughout the world, representing most known genera (14, 17). However, phylogenetic relationships among the sexual and parthenogenetic taxa have been addressed only superficially.
Table 1.
Name of sequenced individuals, fragment length, GenBank accession numbers, distribution, and mode of reproduction for all specimens analyzed in this study
| Taxa | Fragment length, bp |
GenBank accession nos. |
Distribution | Reproductive mode | Refs. | ||||
|---|---|---|---|---|---|---|---|---|---|
| 18S | hsp82 | ef1α | 18S | hsp82 | ef1α | ||||
| Enarthronota | |||||||||
| Hypochthoniidae | |||||||||
| Hypochthonius rufulus (Koch, 1835) | 1,782 | 531 | 543 | EF091427 | DQ090776 | AY632861 | Holarctic, Seychelles | Parthenogenetic | 51, 52, u.o. |
| Eniochthoniidae | |||||||||
| Eniochthonius minutissimus (Berlese, 1903) | 1,759 | 535 | 543 | EF091428 | DQ090773 | EF081329 | Cosmopolitan | Parthenogenetic | 51, u.o. |
| Lohmanniidae | |||||||||
| Lohmannia banksi (Norton et al., 1978) | 1,794 | 513 | 543 | AF022036 | DQ090777 | EF081330 | U.S.A. | Parthenogenetic | u.o. |
| Mixonomata | |||||||||
| Nehypochthoniidae | |||||||||
| Nehypochthonius porosus (Norton and Metz, 1980) | 1,741 | 535 | 543 | EF081308 | DQ090779 | EF081328 | U.S.A., Hawaii | Parthenogenetic | 53 |
| Phthiracaridae | |||||||||
| Steganacarus magnus (Nicolet, 1855) | 1,733 | 513 | 543 | AF022040 | DQ090781 | AY632837 | Holarctic, U.S.A. | Sexual | 51 |
| Atropacarus striculus (Koch, 1835) | 1,742 | 522 | 543 | EF091416 | DQ090782 | EF081309 | Holarctic, Oriental, Australian | Parthenogenetic | 32 |
| Euphthiracaroidea | |||||||||
| Rhysotritia duplicata (Grandjean, 1953) | 1,741 | 513 | 543 | EF091417 | DQ090780 | EF081310 | Palearctic | Parthenogenetic | 54, u.o. |
| Desmonomata | |||||||||
| Camisiidae | |||||||||
| Heminothrus paolianus (Berlese, 1913) | 1,741 | 528 | 543 | EF091423 | DQ090794 | EF081316 | Holarctic | Parthenogenetic | 17, 51, 55 |
| Platynothrus peltifer (Koch, 1839) | 1,741 | 525 | 543 | EF091422 | DQ090793 | AY632851 | Holarctic, Oriental, New Zealand, Neotropic | Parthenogenetic | 17, 40, 51, 56 |
| Camisia biurus (Koch, 1839) | 1,741 | 522 | 543 | EF081302 | EF081331 | EF081312 | Holarctic | Parthenogenetic | 17 |
| Camisia spinifer (Koch, 1835) | 1,741 | 522 | 543 | EF091420 | EF081332 | EF081313 | Holarctic, Oriental, South America | Parthenogenetic | 17, 51 |
| Crotoniidae | |||||||||
| Crotonia brachyrostrum (Hammer, 1966) | 1,741 | 522 | 543 | EF081303 | DQ090796 | EF081314 | Gondwanan | Sexual | 17 |
| Crotonia cf caudalis (Hammer, 1966) | 1,741 | 519 | 543 | EF081304 | DQ090795 | EF081315 | Gondwanan | Sexual | 17 |
| Hermanniidae | |||||||||
| Hermannia gibba (Koch, 1839) | 1,739 | 510 | 543 | EF091426 | DQ090800 | EF081327 | Holarctic, Seychelles | Sexual | 17, 51 |
| Nanhermanniidae | |||||||||
| Nanhermannia coronata (Berlese, 1913) | 1,741 | 535 | 543 | EF091421 | DQ090799 | AY632825 | Holarctic, Neotropic | Parthenogenetic | 17, 51, u.o. |
| Malaconothridae | |||||||||
| Malaconothrus gracilis (van der Hammen, 1952) | 1,741 | 528 | 543 | EF091424 | EF081339 | EF081311 | Holarctic, Neotropic | Parthenogenetic | 17, 51 |
| Nothridae | |||||||||
| Nothrus silvestris (Nicolet, 1855) | 1,741 | 535 | 543 | EF091425 | DQ090802 | AY573591 | Holarctic, Australian | Parthenogenetic | 17, 51, u.o. |
| Nothrus silvestris bistilus (Jacot, 1937) | 1,741 | 535 | 543 | EF081305 | EF081333 | EF081323 | East of U.S.A. | Parthenogenetic | 17 |
| Nothrus truncatus (Banks, 1895) | 1,742 | 535 | 543 | EF081306 | EF081334 | EF081322 | U.S.A. | Parthenogenetic | 17 |
| Novonothrus flagellatus (Hammer, 1966) | 1,741 | 531 | 543 | EF081307 | DQ090801 | EF081324 | New Zealand, Australia, Gondwanan | Sexual | 17 |
| Trhypochthoniidae | |||||||||
| Archegozetes longisetosus (Aoki, 1965) | 1,748 | 510 | 543 | AF022027 | DQ090798 | EF081321 | Pantropical | Parthenogenetic | 17, 51, 55, u.o. |
| Mainothrus badius (Berlese, 1905) | 1,741 | 519 | 543 | EF081301 | EF081338 | EF081318 | Holarctic, Neotropic | Parthenogenetic | 17 |
| Mucronothrus nasalis (Willmann, 1929) | 1,741 | 528 | 543 | EF081299 | DQ090797 | EF081319 | Boreal, Australian, Neotropic | Parthenogenetic | 17, 55, 57 |
| Trhypochthonius americanus (Ewing, 1908) | 1,741 | 528 | 543 | EF081298 | EF081337 | EF081317 | U.S.A. | Parthenogenetic | 17, 51, 55 |
| Trhypochthoniellus crassus (Berlese, 1904) | 1,741 | 525 | 543 | EF081300 | EF081336 | EF081320 | Holarctic, Australian, Neotropic, Ethiopic | Parthenogenetic | 17, 55 |
| Brachypylina | |||||||||
| Achipteriidae | |||||||||
| Achipteria coleoptrata (Linnaeus, 1758) | 1,741 | 510 | 543 | EF091418 | EF081335 | AY632776 | Holarctic, Neotropic, Oriental | Sexual | u.o. |
| Carabodidae | |||||||||
| Carabodes femoralis* (Nicolet, 1855) | 1,740 | 510 | 543 | EF091429 | DQ090786 | EF081325 | Holarctic, Pantropic | Sexual | u.o. |
| Eutegaeidae | |||||||||
| Eutegaeus curviseta (Hammer, 1966) | 1,741 | 510 | 543 | EF081297 | DQ090789 | EF081326 | Gondwanan | Sexual | u.o. |
| Phenopelopidae | |||||||||
| Eupelops plicatus (Koch, 1835) | 1,740 | 522 | 543 | EF091419 | DQ090783 | AY632797 | Holarctic, Nearctic | Sexual | 51, u.o. |
| Tectocepheidae | |||||||||
| Tectocepheus velatus (Michael, 1880) | 1,746 | 516 | 543 | EF093781 | EF093770 | EF093763 | Cosmopolitan | Parthenogenetic | 51, u.o. |
Of the sexual taxa, Crotoniidae are most puzzling (17). The sexuality of these soil- and tree-dwelling mites may simply reflect the ancestral reproductive mode of Desmonomata, but unlike the other taxa, they are not globally distributed; their range is essentially Gondwanan (Table 1). Also, they are morphologically similar to Camisia, a widespread and rather derived genus of the parthenogenetic Camisiidae. The Gondwana distribution and the morphological similarity suggest that Crotoniidae may have evolved from within Camisiidae and thereby reevolved sexuality. The regain of sex would contrast Dollo's law, which states that complex characters never reevolve once they are lost (18). If true, the reevolution of sexuality in oribatid mites would be the first such reversal known in the animal kingdom and would add to the mystique of sex as “the queen of problems in evolutionary biology” (2).
We tested the hypothesis that sexuality reevolved in Crotoniidae by investigating its phylogenetic position among a wide range of sexual and parthenogenetic oribatid mites by using a combined data set of partial sequences of the ribosomal 18S region (18S), the heat shock protein 82 gene (hsp82), and the elongation factor 1 alpha gene (ef1α).
Results
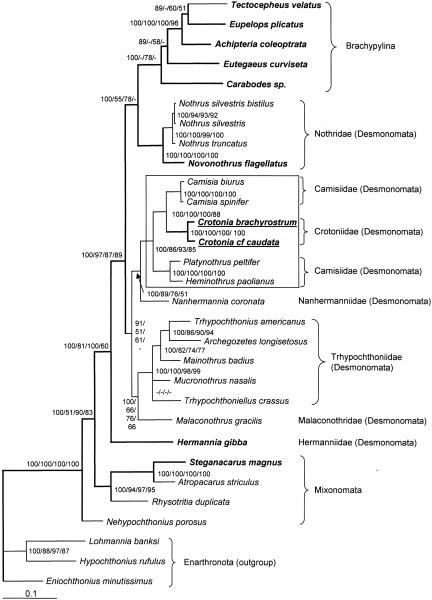
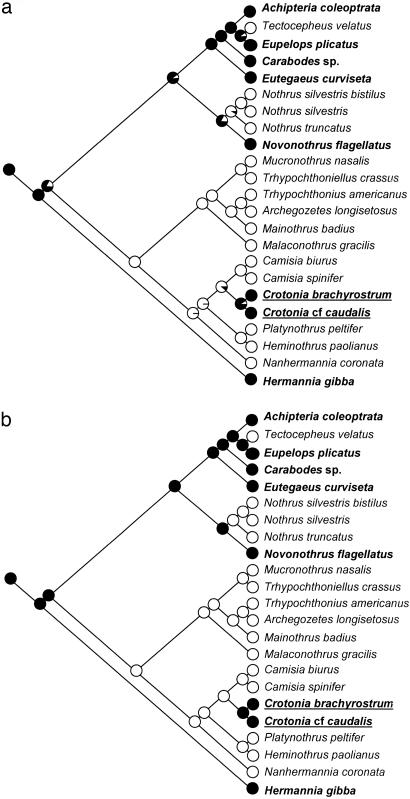
Phylogenetic analyses with neighbor joining (NJ), maximum likelihood (ML), maximum parsimony (MP), and Bayesian algorithms were based on a supermatrix with 2,897 base pairs and 30 taxa. All algorithms gave nearly identical tree topologies, which largely agree with those based on morphological data and earlier molecular studies (Fig. 1) (9, 19, 20). Although Desmonomata as a whole were paraphyletic, all internal taxa except Camisiidae were monophyletic. The sexual genus Novonothrus was basal in Nothridae, supported by high bootstrap and posterior probability values. ML and MP analyses of character evolution consistently assigned sexuality as the ancestral state of Nothridae (Fig. 2).
Fig. 1.
Bayesian tree of combined sequences of the ribosomal 18S region, the heat-shock protein 82, and the elongation factor 1 alpha of 30 oribatid mite taxa. Enarthronota are used as outgroup. Numbers at nodes represent posterior probabilities for Bayesian analyses and bootstrap support values for NJ, ML, and MP analyses. Sexual lineages are indicated by boldfaced lines and font; species that likely reevolved sexual reproduction are both boldfaced and underlined.
Fig. 2.
Cladogram of the Desmonomata on the basis of ML. Ancestral state of nodes is analyzed by ML on the basis of a symmetrical model with equal rates for the loss and regain of sex (a) and MP (b). Filled circles indicate sexual reproduction; open circles indicate parthenogenetic reproduction. Sexual species are in boldface; species that likely reevolved sexual reproduction are both boldfaced and underlined.
By contrast, the sexual genus Crotonia clustered within Camisiidae, a large parthenogenetic family of ≈80 species, with Camisia being its sister-taxon. In this topology, four successive outgroups of Crotonia (two inside and two outside Camisiidae) are entirely parthenogenetic. Monophyly of Camisiidae/Crotoniidae was supported by high bootstrap and posterior probability values (Fig. 1). ML and MP analyses of character evolution assigned parthenogenesis as the ancestral reproductive mode of the Camisiidae/Crotoniidae clade (Fig. 2). ML analysis estimated the rates of loss and regaining of sex to be 0.12 under a symmetrical model of character evolution; under the asymmetrical model, the rate of loss was three times that of regaining sex (0.18 and 0.06, respectively). More biased assumptions for the loss of sex (5:1, 10:1) gave similar results (data not shown).
Results from phylogenetic analyses and the reconstruction of the ancestral states of reproduction support the hypothesis that Crotoniidae reevolved sexual reproduction from parthenogenetic ancestors and contradicts Dollo's law. Therefore, the loss of the complex process of sexuality likely is not irreversible in evolution.
Discussion
The atavistic resurrection of complex ancestral traits, contrary to Dollo's law, appears to be more frequent than commonly thought (21–28). Morphological examples include the reevolution of shell coiling in Gastropoda after 10 million years of absence (22, 23), the reappearance of wings in several lineages of stick insects (24), and the regaining of ancestral muscles in bowerbirds (25). Life history examples include the reevolution of feeding larvae within a group of direct-developing species in the gastropod Crepipatella (26) and reversal to a free-living state in several parasites (27). Atavisms are also present in humans (28). Another example relates to reproductive biology; the plant Hieracium pilosella (29) reevolved sexuality but from a recent and narrow parthenogenetic lineage. The reevolution of sexuality in ancient parthenogenetic clusters of oribatid mites as suggested by this study is, to our knowledge, previously unrecognized in the animal kingdom.
Much of what has been written about large parthenogenetic clusters in oribatid mites has focused on Desmonomata (9, 10, 14), especially Camisiidae, Malaconothridae, and Trhypochthoniidae. Our data support monophyly of species-rich parthenogenetic taxa within Desmonomata and therefore that parthenogenetic lineages of oribatid mites are not evolutionary “dead-ends”; they have persisted and radiated to form clusters, e.g., the parthenogenetic genus Nothrus with 67 species. These lineages of oribatid mites join bdelloid rotifers and darwinulid ostracods (6, 7, 30) as “ancient asexual” groups, which challenge the view that sexual reproduction is indispensable for long-term survival and radiation of lineages.
Adding to the “scandal” of ancient asexuals (1), the results of our study suggest that Crotoniidae have reevolved sex from parthenogenetic ancestors. Reevolution of sex likely resulted from changes in evolutionary forces. In contrast to Camisiidae which typically colonize soil organic layers, Crotoniidae species frequently colonize trees; soil collections may prove to be accidental for many species (31). Generally, parthenogenesis predominates in oribatid mite communities in soil, whereas the bark of trees and mosses are colonized almost exclusively by sexual species (32, 33). This dominance of sexuality suggests that sexual reproduction is necessary for coping with the more heterogeneous environment (tangled bank hypothesis; see ref. 34) or increased exposure to antagonists (red queen hypothesis; see ref. 35) in aboveground habitats. Furthermore, the reproductive mode is affected by the availability of resources. In soil, the permanent availability of resources (litter material and detritus) may explain the widespread occurrence of parthenogenesis (36). Although Crotonia has changed to a tree-dwelling life cycle, the mode of reproduction may have changed accordingly.
How reversion from parthenogenetic to sexual reproduction occurred remains unclear. In animal taxa with cyclical parthenogenesis (intermittent mixis), pure bisexual reproduction can reevolve by the abandonment of the parthenogenetic part of the life cycle, but cyclical parthenogenesis is unknown in oribatid mites (14). Reversion to sexual reproduction may be facilitated because transitions between different modes of reproduction exist in higher mite taxa (12). The studied parthenogenetic oribatid mite species reproduce by automictic thelytoky in which the meiotic maturation division is followed by fusion of haploid nuclei to restore diploidy (37). In automictic species, the reversion to sexual reproduction requires that the ability to produce males has been maintained during long evolutionary periods of parthenogenesis. Many parthenogenetic species of oribatid mites are known to produce rare nonfunctional (spanandric) males (17), as is common for parthenogenetic animals in general (38), including the “ancient asexual” darwinulid ostracods (39). In parthenogenetic oribatids, nonfunctionality of males is caused by incomplete spermatogenesis, and females ignore spermatophores if they are formed (40). Nonfunctionality has also been indicated by population genetic studies because populations are unaffected by male presence (41). Why spanandric males persist despite the costs to produce them is unclear. Presumably, such males form as developmental “accidents,” as in other parthenogenetic species (42) and, being rare events, the costs of nonfunctional males may be negligible. Irrespective of the mechanisms involved, the occasional formation of spanandric males in parthenogenetic Camisiidae presumably facilitated the capture of functionality of ancestral genes for male production over long evolutionary timescales and therefore the reevolution of sex in Crotoniidae. Knowledge of the genetic and epigenetic mechanisms controlling developmental cascades that lead to male production will answer these questions.
Conclusion
In summary, parthenogenetic radiations are infrequent events in evolution, and the previously unrecognized reevolution of sexual reproduction from parthenogenetic ancestors is even more rare. In general, most parthenogenetic taxa have close sexual relatives and contain few species; in Desmonomata, especially in Camisiidae/Crotoniidae, the pattern is reversed with few sexual taxa within a large cluster of parthenogenetic species. Results of the present study suggest that Crotoniidae indeed reevolved sex, which is a spectacular case of breaking Dollo's law, implying that parthenogenesis is not necessarily an evolutionary dead end. The reevolution of sexual reproduction in Crotoniidae within the ancient clade of parthenogenetic Camisiidae suggests that sexual reproduction is indispensable at certain environmental conditions. Oribatids are an ideal model group to explore these conditions and therefore unravel the enigma of the evolution of sexual reproduction and the conditions under which these reproductive modes prevail.
Materials and Methods
Taxon Sampling.
In total, 30 oribatid mite species were sampled. Oribatid mites are commonly ascribed to six major groups, Palaeosomata, Enarthronota, Parhyposomata, Mixonomata, Desmonomata, and Brachypylina (19, 40). Parthenogenetic clusters are most common in Enarthronota and Desmonomata, which are early- and middle-derivative groups, respectively. We focused on Desmonomata, comprising seven families with 36 genera and ≈500 described species (17, 19, 43). In addition to having the large parthenogenetic families Trhypochthoniidae (68 spp.), Malaconothridae (104 spp.), Camisiidae (92 spp.), and Nanhermanniidae (56 spp.), Desmonomata include two families, Crotoniidae (45 spp.) and Hermanniidae (80 spp.), that reproduce only sexually and one family, Nothridae (54 spp.), that has both sexual and parthenogenetic genera. Representatives of all seven families of Desmonomata were included to ascertain whether sexuality in these families appeared to be ancestral or derived with respect to other Desmonomata (Table 1). Camisiidae were most heavily sampled, because a close relationship to Crotoniidae was hypothesized. Other desmonomatan families were represented by a single genus, because their reproductive modes were internally constant.
Several species of Brachypylina, the “higher” oribatid mites, were sampled to ascertain monophyly or paraphyly of Desmonomata. Members of Enarthronota and Mixonomata were sequenced for use as respective outgroups and were selected on the basis of earlier phylogenetic studies (9, 19, 20). Parhyposomata and Palaeosomata were not included, because they are small taxa having no apparent bearing on our objectives.
Oribatid mites were collected from litter and soil at different localities in Germany, Poland, the U.S., New Zealand, and Russia. We complemented the data set with sequences available at GenBank (Table 1).
Sample Preparation, PCR, and Sequencing.
Total DNA was extracted from 1 to 10 individuals by using Qiagen (Hilden, Germany) DNeasy kit for animal tissues following the manufacturer's protocol (but elution in 30 μl instead of 400 μl; Qiagen). Amplifications for the18S region, hsp82, and ef1α were performed either in 50-μl volumes containing 1 μl of each primer (100 pmol/μl), 4–8 μl of DNA, and 25 μl of HotStarTaq Mastermix (2.5 units of HotStarTaq polymerase, and 200 μM each dNTP and 15 mM MgCl2 buffer solution; Qiagen) or in 25-μl volumes by using half the amount of reagents. The primers used and the PCR programs are given in supporting information (SI) Tables 2 and 3. PCR products were visualized on 1% agarose gels and purified by using QIAquick PCR Purification kit (Qiagen). PCR products were either prepared for direct sequencing or cloned by using Qiagen PCR Cloning kit and transformed into Escherichia coli Nova Blue Singles competent cells (Novagen, Darmstadt, Germany) by heat shock by using the manufacturer's protocol. The plasmids were purified by using FastPlasmid mini kit (Eppendorf, Hamburg, Germany). DNA was sequenced by Scientific Research and Development (Oberursel, Germany), Qiagen Genomic Services, (Hilden, Germany), or Macrogen (Seoul, Korea). All sequences are available at GenBank (for accession numbers, see Table 1).
Alignment and Phylogenetic Analysis.
Because the parameters of the evolutionary models of the three data sets were very similar, DNA sequences of 18S, hsp82, and ef1α of 30 oribatid mite taxa were combined in a supermatrix and aligned by using the default settings in ClustalX (44); the alignment was modified by eye. The evolutionary model parameters were determined with Modeltest 3.7 (45) by using a hierarchical likelihood ratio test. The model of evolution was TrN+I+G (46) with base frequencies A = 0.3082, C = 0.2238, G = 0.2484, gamma distribution shape parameter α = 0.5819 for four categories of among-site variation, and fraction of invariant sites I = 0.5915. The substitution rates were estimated as A-C, A-T, C-G, and G-T = 1.0, A-G = 2.7550, and C-T = 4.8958. Phylogenetic trees were constructed by using NJ, MP, and ML algorithms as implemented in PAUP* 4b10 (47). MP and ML trees were constructed with a heuristic search of 100 random additions, and the tree-bisection reconnection (TBR) branch-swapping algorithm with the option to collapse zero branch length. A strict consensus tree was constructed for both. Reliability of the branches was ascertained by bootstrap analyses for NJ (100,000 replicates), ML (100 replicates, heuristic search), and MP (10,000 replicates, heuristic search) in PAUP*. Bayesian phylogenetic analysis was performed with MrBayes version 3.1.2 (48) by using the settings for GTR+I+G with three independent runs of 3 million generations and four chains each; rate matrix and base frequencies were estimated, and trees were sampled every 300 generations. A majority consensus tree was generated by using a burn-in of 2,000.
Ancestral states and the history of character evolution were investigated with parsimony and likelihood algorithms by using the StochChar package in Mesquite (49, 50). Likelihood analyses were calculated under a symmetrical model with equal rates for the loss and regaining of sex and an asymmetrical model with independent rates estimated by ML algorithm. Asymmetrical models with higher rates for the loss of sex (5:1, 10:1) were also tested. Probabilities were calculated assuming equal length for all branches on the basis of the topology of the ML and Bayesian tree.
Separate analyses (NJ, MP, ML, and Bayesian) of the three data sets gave slightly different topologies among desmonomatan families, but internal topologies were identical (data not shown). The Camisiidae/Crotoniidae group was always supported by high support values, and Novonothrus occupied a basal position within Nothridae.
Supplementary Material
Acknowledgments
We thank J. M. Cianciolo (University of Indiana, Bloomington, IN), N. Lindberg (Uppsala University, Uppsala Sweden), J. Illig (Technische Universität Darmstadt), and D. Sandmann (Technische Universität Darmstadt) for collecting and providing specimens; M. Laumann (Universität Tübingen, Tübingen, Germany) for providing hsp82 and ef1α sequence data; R. H. Cruickshank, M. Heethoff, and I. Schaefer for supporting the data analysis; and U. Brose, D. Mark Welch, W. C. Birky, and two anonymous reviewers for commenting on earlier drafts of the manuscript. This study was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft).
Abbreviations
- ML
maximum likelihood
- MP
maximum parsimony
- NJ
neighbor joining.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition footnote: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information databank, www.ncbi.nlm.nih.gov (accession nos. AF022027, AF022036, AF022040, AY573591, AY632776, AY632825, AY632837, AY632851, AY632861, DQ090773, DQ090776, DQ090777, DQ090779–83, DQ090786, DQ090789, DQ090793–DQ090802, EF081297–EF081339, EF091416–EF091429, EF093763, EF093770, and EF093781).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700034104/DC1.
References
- 1.Maynard Smith J. The Evolution of Sex. Cambridge: Cambridge Univ Press; 1978. [Google Scholar]
- 2.Bell G. The Masterpiece of Nature: The Evolution and Genetics of Sexuality. Los Angeles: Univ of California Press; 1982. [Google Scholar]
- 3.Birky CW. Genetics. 1996;144:427–437. doi: 10.1093/genetics/144.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vrijenhoek RC. In: Advances in Molecular Ecology. Carvalho G, editor. Amsterdam: IOS Press; 1998. pp. 151–172. [Google Scholar]
- 5.Barraclough TG, Birky CW, Burt A. Evolution (Lawrence, Kans) 2003;57:2166–2172. doi: 10.1111/j.0014-3820.2003.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 6.Martens K, Horne DJ, Griffiths HI. In: Sex and Parthenogenesis: Evolutionary Ecology of Reproductive Modes in Non-Marine Ostracods. Martens K, editor. The Netherlands: Backhuys, Leiden; 1998. pp. 37–55. [Google Scholar]
- 7.Mark Welch D, Meselson M. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 8.Norton RA, Palmer SC. In: The Acari: Reproduction, Development, and Life-History Strategies. Schuster R, Murphy PW, editors. London: Chapman & Hall; 1991. pp. 107–136. [Google Scholar]
- 9.Maraun M, Heethoff M, Schneider K, Scheu S, Weigmann G, Cianciolo J, Thomas RH, Norton RA. Exp Appl Acarol. 2004;33:183–201. doi: 10.1023/b:appa.0000032956.60108.6d. [DOI] [PubMed] [Google Scholar]
- 10.Schaefer I, Domes K, Heethoff M, Schneider K, Schoen I, Norton RA, Scheu S, Maraun M. J Evol Biol. 2006;18:184–193. doi: 10.1111/j.1420-9101.2005.00975.x. [DOI] [PubMed] [Google Scholar]
- 11.Heethoff M, Domes K, Laumann M, Maraun M, Norton RA, Scheu S. J. Evol. Biol. 2007;20:392–402. doi: 10.1111/j.1420-9101.2006.01183.x. [DOI] [PubMed] [Google Scholar]
- 12.Cruickshank RH, Thomas RH. Evolution (Lawrence, Kans) 1999;53:1796–1803. doi: 10.1111/j.1558-5646.1999.tb04563.x. [DOI] [PubMed] [Google Scholar]
- 13.Weeks AR, Marec F, Breeuwer JAJ. Science. 2001;292:2479–2482. doi: 10.1126/science.1060411. [DOI] [PubMed] [Google Scholar]
- 14.Norton RA, Kethley JB, Johnston DE, O'Connor BM. In: Evolution and Diversity of Sex Ratios. Wrensch DL, Ebbert MA, editors. New York: Chapman & Hall; 1993. pp. 8–99. [Google Scholar]
- 15.Norton RA, Bonamo PM, Grierson JD, Shear WA. J Paleont. 1998;62:259–269. [Google Scholar]
- 16.Hammer M, Wallwork JA. Biol Skr Dan Vid Selsk. 1979;22:1–31. [Google Scholar]
- 17.Palmer SC, Norton RA. Exp Appl Acarol. 1991;12:67–81. [Google Scholar]
- 18.Gould SJ. J Hist Biol. 1970;3:189–212. doi: 10.1007/BF00137351. [DOI] [PubMed] [Google Scholar]
- 19.Grandjean F. Acarologia. 1969;11:127–153. [Google Scholar]
- 20.Haumann G. Zur Phylogenie Primitiver Oribatiden (Acari: Oribatida) Germany: Verlag Technische Universität, Graz; 1991. [Google Scholar]
- 21.Marshall CR, Raff EC, Raff RA. Proc Natl Acad Sci USA. 1994;91:12283–12287. doi: 10.1073/pnas.91.25.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collin R, Cipriani R. Proc R Soc Lond B. 2003;270:2551–2555. doi: 10.1098/rspb.2003.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagel M. Trends Ecol Evol. 2004;19:278–280. doi: 10.1016/j.tree.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Whiting MF, Bradler S, Maxwell T. Nature. 2003;421:264–267. doi: 10.1038/nature01313. [DOI] [PubMed] [Google Scholar]
- 25.Rainkov RJ, Borecky SR, Berman SL. Condor. 1979;81:203–206. [Google Scholar]
- 26.Collin R, Chaparro OR, Winkler F, Véliz D. Biol Bull. 2007;212:83–92. doi: 10.2307/25066586. [DOI] [PubMed] [Google Scholar]
- 27.Cruickshank RH, Paterson AM. Trends Parasit. 2006;22:509–515. doi: 10.1016/j.pt.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Verhulst J. Acta Biotheoretica. 1996;44:59–73. doi: 10.1007/BF00046435. [DOI] [PubMed] [Google Scholar]
- 29.Chapman H, Houliston GJ, Robson B, Iline IA. Int J Plant Sci. 2003;164:719–728. [Google Scholar]
- 30.Butlin R, Schön I, Griffiths HI. In: Sex and Parthenogenesis: Evolutionary Ecology of Reproductive Modes in Non-Marine Ostracods. Martens K, editor. The Netherlands: Backhuys, Leiden; 1998. pp. 1–24. [Google Scholar]
- 31.Olszanowski Z. J Nat Hist. 1999;33:233–253. [Google Scholar]
- 32.Cianciolo J, Norton RA. Exp Appl Acarol. 2006;40:1–25. doi: 10.1007/s10493-006-9016-3. [DOI] [PubMed] [Google Scholar]
- 33.Erdmann G, Floren A, Lisenmair KE, Scheu S, Maraun M. Pedobiologia. 2006;50:433–441. [Google Scholar]
- 34.Ghiselin MT. The Economy of Nature and the Evolution of Sex. Los Angeles: Univ of California Press; 1974. [Google Scholar]
- 35.Van Valen LM. Evol Theory. 1973;1:1–30. [Google Scholar]
- 36.Scheu S, Drossel B. Proc R Soc Lond B. 2007;274:1225–1231. doi: 10.1098/rspb.2007.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrensch DL, Kethley JB, Norton RA. In: Mites: Ecological and Evolutionary Analysis of Life-History Patterns. Houck MA, editor. New York: Chapman & Hall; 1994. pp. 282–343. [Google Scholar]
- 38.Lynch M. Q Rev Biol. 1984;59:257–290. [Google Scholar]
- 39.Smith JS, Kamiya T, Horne DJ. Proc R Soc Lond B. 2006;273:1569–1578. doi: 10.1098/rspb.2005.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taberly G. Acarologia. 1988;29:95–107. [Google Scholar]
- 41.Palmer SC, Norton RA. Biochem Syst Ecol. 1992;20:219–231. [Google Scholar]
- 42.Groot TVM, Breeuwer JAJ. Exp Appl Acarol. 2006;39:257–271. doi: 10.1007/s10493-006-9019-0. [DOI] [PubMed] [Google Scholar]
- 43.Subias LS. Graellsia. 2004;60(Suppl):1–305. [Google Scholar]
- 44.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucl Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 47.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (And Other Methods) Version 4.0. Massachusetts: Sinauer Associates, Sunderland; 1999. [Google Scholar]
- 48.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 49.Maddison WP, Maddison DR. Mesquite: A Modular System for Evolutionary Analysis. 2003 ( www.mesquiteproject.org.), Version 1.0.
- 50.Maddison WP, Maddison DR. StochChar: A Package of Mesquite Modules for Stochastic Models of Character Evolution. 2005 ( www.mesquiteproject.org), Version 1.06.
- 51.Grandjean F. C R Séanc Acad Sci. 1941;212:463–467. [Google Scholar]
- 52.Luxton M. Pedobiologia. 1981;21:312–340. [Google Scholar]
- 53.Norton RA, Metz L. Ann Entomol Soc Amer. 1980;73:54–62. [Google Scholar]
- 54.Harding DJL. In: Evans GO, editor. Proceedings of the 2nd International Congress of Acarology; Budapest: Akademiai Kiadio; 1969. [Google Scholar]
- 55.Palmer SC, Norton RA. Exp Appl Acarol. 1990;8:445–456. [Google Scholar]
- 56.Taberly G. C R Séanc Acad Sci. 1958;246:1655–1657. [PubMed] [Google Scholar]
- 57.Travé J. Acarologia. 1973;15:522–533. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.