Abstract
Helicobacter pylori is a gastric-dwelling pathogen responsible, with acid secretion, for peptic ulcer and a 20-fold increase in the risk of gastric cancer. Several transcriptomes have been described after short-term exposure to acidity in vitro, but there are no data identifying the effects of chronic gastric exposure on bacterial gene expression. Comparison of the in vivo to the in vitro transcriptome at pH 7.4 identified several groups of genes of known function that increased expression >2-fold, and three of these respond both to acidity in vitro and to gastric infection. Almost all known acid acclimation genes are highly up-regulated. These include ureA, ureB, and rocF and the pH-gated urea channel, ureI. There is also up-regulation of two groups of motility and chemotaxis genes and for pathogenicity island genes, especially cagA, a predictor for pathogenicity. Most of these genes interact with HP0166, the response element of the pH-sensing two-component histidine kinase, HP0165/HP0166, ArsRS. Based on the pH profile of survival of ureI deletion mutants in vitro and their inability to survive in gastric acidity, the habitat of the organism at the gastric surface is acidic with a pH ≤ 4.0. Hence, the pH of the habitat of H. pylori on the surface of the stomach largely determines the regulation of these specific groups of genes.
Keywords: acid acclimation, gastric pH, global transcriptome, microarray
The acidity of the mammalian stomach is important for preventing the transit of many potentially injurious bacteria to the intestine. Although some pathogens, such as Vibrio cholerae or pathogenic Escherichia coli, are able to transit the highly acidic gastric environment because of expression of acid resistance or tolerance genes (1), only Helicobacter pylori, a Gram-negative, microaerophilic neutralophile, is able to colonize the human stomach. Studies have been performed in vitro to examine the effect of short term exposure (30–120 min) to acidic pH on H. pylori gene regulation (2–5). Under these conditions, the organism up-regulates acid acclimation and other groups of genes (2). Only analysis of gene expression in the stomach can evaluate the effect of long-term exposure to the gastric environment and identify whether it is acidic or neutral and provide information on the bacterial response to gastric habitation.
An important component of the response of H. pylori to acid is acid acclimation (6–8). Acid acclimation is the ability of H. pylori to maintain periplasmic pH at 6.1 in the face of an external pH at least as low as pH 2.5 (6, 9, 10). Hence, by maintaining a near neutral periplasmic pH, the cytoplasmic pH experiences relatively small excursions from its optimal pH in the presence of acid, allowing the organism to grow in gastric acidity as if it were in a neutral environment. Many of the important components of the acid acclimation system have been identified in vitro by biochemical, physiological, and transcriptomal analysis (5, 6, 10–12).
The importance of some of the individual components of the acid acclimation system has been confirmed by gene deletion correlating the survival of H. pylori exposed to an acid pH in vitro with the ability of the mutants to infect the stomach (6, 13, 14). Other genes have been identified that contribute to or are essential for gastric infection, such as genes involved in motility and chemotaxis (15–17). Some of these studies have relied on examination of the effects of in vitro culture under different conditions on gene expression or bacterial survival to identify genes of interest for subsequent knockout studies. Others have relied on the loss of infective ability of deletion mutants (2–5, 13, 18). It has not been determined whether any of these genes change expression when the organism colonizes the stomach.
The median pH of the gastric lumen is 1.4 (19). This has resulted in the suggestion that there is a barrier to back diffusion of protons across the mucus layer and neutralization by bicarbonate secretion by the gastric surface cells (20). The perceived need for a barrier derives from the original finding that artificial phospholipid bilayers were proton permeable (21). More recent work showed that this result was due to contamination of the phospholipid and that bilayer proton permeability is similar to that of small cations. Hence, in the absence of proton conductance on the apical membrane of gastric epithelial cells, no proton barrier is required (22, 23).
Nevertheless, the pH at the gastric surface remains important and controversial. Studies using glass tip pH microelectrodes suggested the presence of a gastric barrier comprised of mucus and HCO3− secretion maintaining the surface pH at ≈6.0 even with a luminal pH of 1.0 to 2.0 (24). In contrast, studies using pH sensitive fluorimetric dyes concluded that the pH at the epithelial surface was maintained at pH 4 independent of luminal pH (25). Recent work using open tip electrodes shows that there is loss of the proton barrier in mice infected with H. pylori (26). Examination of the changes and the magnitude of expression of the acid acclimation genes in vivo can determine the degree of acidity at the gastric surface. Similarly, expression levels of motility and pathogenicity island (PAI) genes that are regulated by acid in vitro will provide additional evidence as to the pH of the gastric environment of H. pylori.
To determine the nature of the genes expressed by H. pylori in vivo, we isolated the bacterial RNA from infected gerbil stomachs and performed transcriptomal analysis. Because the luminal pH and the pathological consequences of H. pylori infection of the gerbil stomach is similar to that of the human, it is likely that the gene expression profile will resemble that occurring during colonization of man (19, 27). The animals were infected with H. pylori expressing Green Fluorescent Protein (GFP) to allow localization of the infection using confocal microscopy. The organisms were found adherent to the surface epithelial cells and in the overlying mucus layer (data not shown).
Here, we present the first set of data on the transcriptome of H. pylori inhabiting the stomach. A previous publication (28) used DNA capture to identify only those genes that were unique to the gastric environment but did not survey global changes as compared with in vitro culture conditions. Several groups of genes markedly increased expression in vivo compared with that of the bacterium in vitro. These data show the differences between gastric dwelling bacteria and bacteria cultured in vitro, reflecting the response to gastric habitation. These differences indicate that the gastric environment at the site of infection is acidic. There is good agreement between the data presented here and PCR studies on a few selected genes from human or monkey biopsies (29, 30).
Results
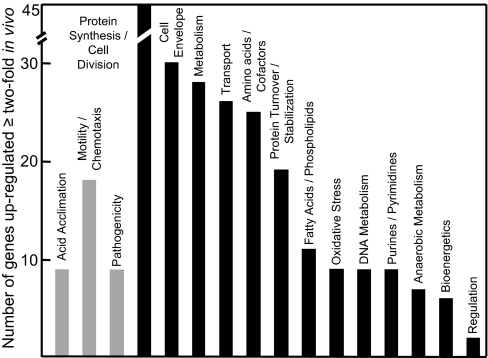
Microarray analysis of gene expression of H. pylori isolated from the gerbil stomach 10 days after inoculation identified 16 groups of genes of known function that increased expression >2-fold compared with H. pylori grown on TSA plates in a microaerobic environment at pH 7.4. There were also ≈100 genes of unknown function that increased by the same amount. Fig. 1 shows the number of genes in the functional groups that were up-regulated twofold or greater in the in vivo transcriptome. Members of two of these groups have been identified as required for infection of animal models. These are genes encoding proteins involved in acid acclimation and in motility and chemotaxis. The motility and chemotactic proteins allow the organism to penetrate the gastric mucus layer to find its site of colonization, probably a specific niche in the gastric epithelium (31). A third group of genes was also up-regulated in vivo encoding for proteins of the Cag PAI, one of which, CagA, has often been considered as a marker for induction of gastric pathology.
Fig. 1.
Genes of known function in the annotated H. pylori genome up-regulated after gastric infection. On the left are three groups of genes discussed in the text, acid acclimation, motility/chemotaxis, and pathogenicity (gray bars). On the right, in descending order, are groups of genes of known function that are up-regulated >2-fold after gastric infection compared with in vitro culture.
qPCR.
Two genes that were up-regulated in gerbil infection were selected for confirmation by real time PCR. One gene, HP0166, which was unchanged on the microarray in vivo, was chosen as a control. This gene did not show a change in the gastric environment but was up-regulated in acid in vitro. The results are in Table 1. This experiment confirmed the up-regulation of ureA and ureI in the gerbil infection model. There was no change in HP0166 (ompR).
Table 1.
The difference in cycle number between the in vivo and in vitro conditions as determined by qPCR
| Gene | ureA | ureI | ompR |
|---|---|---|---|
| ΔC(T) | 7.03 | 2.04 | 0.03 |
ΔC(T) = C(T)in vivo − C(T)in vitro, where C(T) is the cycle greater than one standard deviation above background.
Acid Acclimation.
A comparison of the in vivo transcriptome to the in vitro transcriptome (Table 2) shows that most of the known acid acclimation genes are up-regulated in the gerbil stomach. There was a relatively larger increase in vivo of the expressions of ureA, ureB, hypB, and rocF as compared with the others of this group. The genes encoding the structural subunits of urease, ureA and ureB, showed a very large increase in the gastric transcriptome, indicating an increased requirement for urease in the stomach, likely due to prolonged exposure to high acidity and variable urea and maybe peptic modification of the outer membrane. Accompanying this increased transcription, the genes encoding accessory subunits involved in Ni2+ insertion that converts the apoenzyme to active urease, ureE and ureG, and the Ni2+ donating hydrogenase accessory proteins hypA and hypB were also up-regulated. The absence of up-regulation of ureF and ureH may be due to variable stability of these mRNAs that has been described (32). The genes encoding the proton-gated urea channel, UreI and α-carbonic anhydrase, which are essential for acid acclimation, were also up-regulated in the stomach (6, 10). There was a large increase of expression of rocF, the gene encoding arginase. There was also increased expression of the three genes encoding enzymes able to generate intrabacterial ammonia, namely amidases amiE and amiF and aspartase, aspA. Also in the table are the in vitro pH 4.5 induced transcriptomal studies that showed up-regulation of the acid acclimation genes. Except for hypB and aspA, all of the acid acclimation genes of this group are members of the acid sensing ArsRS regulon (7).
Table 2.
Expression of acid acclimation genes up-regulated ≥2-fold in vivo compared with pH 7.4 in vitro and comparison with previously published in vitro acid-induced global transcriptome analyses
| Accession no. | Increase | Description | In vitro increased transcription to low pH by ref. |
|---|---|---|---|
| HP0072 | 29.90 ± 8.23 | Urease beta subunit (urea amidohydrolase) (ureB)* | 4 |
| HP0073 | 21.96 ± 4.38 | Urease alpha subunit (urea amidohydrolase) (ureA)* | 3, 4 |
| HP0071 | 5.04 ± 2.06 | Urease accessory protein (ureI)*† | 4, 5 |
| HP0070 | 1.79 ± 0.53 | Urease accessory protein (ureE)* | 5 |
| HP0068 | 3.64 ± 1.22 | Urease accessory protein (ureG)* | 4, 5 |
| HP0900 | 10.16 ± 1.39 | Hydrogenase expression/formation protein (hypB) | 5 |
| HP0869 | 2.57 ± 0.19 | Hydrogenase expression/formation protein (hypA)† | 3, 5 |
| HP1186 | 2.68 ± 0.98 | Carbonic anhydrase † | 5 |
| HP0294 | 5.31 ± 1.78 | Aliphatic amidase (amiE)* | 3–5 |
| HP1238 | 1.97 ± 0.25 | Formamidase (amiF)* | 3, 4 |
| HP0649 | 2.42 ± 0.60 | Aspartase (aspA) | |
| HP1399 | 15.94 ± 2.90 | Arginase (rocF)*† | 3, 5 |
Motility and Chemotaxis.
Expression of 18 genes involved in motility and chemotaxis increased in the gastric gene profile (Table 3). Some members of this group are essential for infection, such as flaA and flaB, encoding for the structural subunits of the flagellum. There was increased in vivo expression of the chemotaxis gene cheY (HP1067) and the two cheV genes (HP0393, HP0019). There was also increased expression of two genes encoding for transmembrane receptors, the methyl-accepting chemotaxis proteins, tlpB (HP0103), and HP0599.
Table 3.
Expression of motility and chemotactic genes up-regulated ≥2-fold in vivo compared with pH 7.4 in vitro and comparison with previously published in vitro acid-induced global transcriptome analyses
| Accession no. | Mean ± SEM | Description | In vitro increased transcription to low pH, by ref. |
|---|---|---|---|
| HP0601 | 20.69 ± 14.06 | Flagellin A (flaA) | 5 |
| HP0115 | 2.19 ± 1.51 | Flagellin B (flaB) | 4, 5 |
| HP0870 | 4.53 ± 0.79 | Flagellar hook (flgE) | |
| HP0908 | 3.07 ± 0.39 | Flagellar hook (flgE) | 4 |
| HP1585 | 3.93 ± 0.44 | Flagellar basal-body rod protein (flgG)* | 5 |
| HP1559 | 2.98 ± 0.77 | Flagellar basal-body rod protein (flgB) | 4, 5 |
| HP1557 | 4.45 ± 0.95 | Flagellar basal-body protein (fliE) | 5 |
| HP0325 | 2.27 ± 0.50 | Flagellar basal-body L-ring protein (flgH) | |
| HP0907 | 3.56 ± 0.68 | Hook assembly protein, flagella (flgD) | |
| HP1233 | 2.50 ± 0.54 | H. pylori predicted coding region HP1233flgJ-like | |
| HP1192 | 5.38 ± 1.66 | Secreted protein involved in flagellar motility* | 3, 5 |
| HP0232 | 4.17 ± 1.37 | Secreted protein involved in flagellar motility | 2 |
| HP1462 | 2.22 ± 0.61 | Secreted protein involved in flagellar motility | |
| HP1067 | 2.34 ± 0.66 | Chemotaxis protein (cheY) | 5 |
| HP0393 | 4.38 ± 0.72 | Chemotaxis protein (cheV) | 5 |
| HP0019 | 2.79 ± 0.75 | Chemotaxis protein (cheV) | 5 |
| HP0103 | 2.87 ± 1.76 | Methyl-accepting chemotaxis protein (tlpB) | 2 |
| HP0599 | 10.17 ± 0.24 | Methyl-accepting chemotaxis protein |
Genes identified as members of the ArsRS regulon are in bold.
*See ref. 7.
Twelve of this group of genes were up-regulated in the acidic in vitro transcriptome, hence again showing acidity of the gastric environment of H. pylori. Only three genes in this group, flgG, HP1192 (encoding a motility related secreted protein) and cheY (encoding a chemotaxis protein) are members of the ArsRS regulon and therefore the pH-dependent regulation of expression of the remaining nine genes does not depend on ArsRS but on unknown regulators.
PAI.
Many H. pylori strains contain the Cag PAI, whose genes encode for many components of a type IV secretory system (T4SS) (15, 62). These genes are not essential for infection, because many strains isolated from the human stomach lack this PAI. However, because of their up-regulation at pH 4.5 in vitro, interaction with HP0166 and their likely clinical importance, this group is included here (33). As shown in Table 4, nine genes of the Cag PAI were up-regulated in the stomach, five of which respond to acid in vitro and eight of which interact with the response element of the ArsRS, HP0166.
Table 4.
Expression of Cag PAI genes up-regulated ≥2-fold in vivo compared with pH 7.4 in vitro and comparison with previously published in vitro acid-induced global transcriptome analyses
| Accession no. | Mean ± SEM | Description | In vitro increased transcription to low pH, by ref. |
|---|---|---|---|
| HP0547 | 21.10 ± 5.06 | Cag PIA protein (cag26) cagA* | |
| HP0546 | 5.90 ± 2.71 | Cag PIA protein (cag25)* | |
| HP0543 | 3.06 ± 0.15 | Cag PIA protein (cag22)* | 5 |
| HP0542 | 2.66 ± 0.49 | Cag PIA protein (cag21)* | 5 |
| HP0541 | 4.22 ± 0.59 | Cag PIA protein (cag20)* | 5 |
| HP0532 | 3.46 ± 1.37 | Cag PIA protein (cag12) | 5 |
| HP0531 | 3.04 ± 0.86 | Cag PIA protein (cag11)* | |
| HP0520 | 2.26 ± 0.51 | Cag PIA protein (cag1)* | 5 |
| HP0525 | 2.47 ± 0.50 | virb11/ptlH homolog |
Genes identified as members of the ArsRS regulon are in bold.
*See ref. 7
cag26 encodes the protein, CagA, which has been associated with virulence of infection and is exported and injected into the host cell (34) and this gene increases expression about 20-fold probably because of adhesion of the organism to the gastric surface and CagA secretion into the host cell. Without secretion as is the case in vitro, there is no change in expression of this gene. Because seven of these genes belong to the ArsRS regulon, they probably respond to the acidity of the gastric environment of H. pylori.
Discussion
Analysis of gene expression by the organism dwelling in the stomach is required to explain the unique ability of H. pylori to colonize the human stomach. H. pylori global transcriptomal analysis has been limited to studies of gene regulation in vitro although many of these in vitro studies have measured expression in acidic compared with neutral media but do not fully represent the gastric conditions encountered by the organism (2–5). Only one study included urea to more closely mimic the conditions of the stomach (5). The pH at the site of colonization is controversial and other factors may play a role in adaptation of the organism to its gastric environment. The data on acid acclimation genes in vivo show that H. pylori experiences and resides in acid and also expresses many motility and chemotaxis genes to allow the bacteria to move on the gastric surface or the adjacent mucus layer. PAI genes thought to play an important role in the gastric mucosal response to infection are also increased in the in vivo transcriptome (35).
Acid Acclimation.
Acid acclimation is the ability of H. pylori to maintain periplasmic pH close to neutrality in the presence of extra bacterial acidity to maintain cytoplasmic pH at physiological levels. Several acid acclimation genes are required for Helicobacter colonization of animal models. These genes include urease, those involved in its biosynthesis, and UreI, the proton gated urea channel essential for adequate access of medium urea to intrabacterial urease and periplasmic α-carbonic anhydrase (6, 10, 13). Intrabacterial urease along with UreI and the periplasmic α-carbonic anhydrase act in concert to buffer the periplasm to pH 6.1 even at a pHout 2.5. These in vitro data were obtained by measurement of inner membrane potential, cytoplasmic pH, and the use of a pH sensing dye to directly visualize periplasmic alkalization using ureI and HP1186 (α-carbonic anhydrase) deletion mutants or acetazolamide to inhibit α-carbonic anhydrase (6, 9, 36).
The in vivo H. pylori transcriptome showed an increase in the level of message for the acid acclimation genes ureA, ureB, several urease accessory genes required for Ni2+ insertion, ureI and α-carbonic anhydrase when compared with bacteria grown under optimal laboratory conditions. There was also an increased transcription of the hydrogenase accessory genes, hypA and hypB. These genes encode for proteins involved in nickel regulation and are implicated in the activation of the urease apoenzyme. Deletion of either hypA or hypB results in significant reduction in urease activity by 40- and 200-fold, respectively (37). Therefore, along with the increase in expression of ureA and ureB, a concomitant increase would occur in Ni2+ sequestering proteins to provide this essential cation to the apoenzyme (38, 39). The increase in expression of these genes emphasizes the need for high urease content for colonization of the niche occupied by H. pylori in the gerbil stomach (31).
There was also increased expression of three genes encoding enzymes able to generate intrabacterial ammonia, the amidases (amiE and amiF), and aspartase, aspA. These enzymes may contribute to neutralization of entering protons under acidic conditions by production of ammonia. These enzymes may also contribute to periplasmic buffering and thus acid acclimation. None of these genes have been shown to be essential either for colonizing the mouse stomach or for acid survival in vitro. However, the intragastric pH of the mouse stomach is two units higher than that of the gerbil and may account for the up-regulation of these genes in the gerbil in vivo transcriptome (13, 40).
There was a large increase of expression of the gene encoding arginase (rocF) by about 15-fold in vivo, possibly to provide an alternate source for intracellular urea. Arginase generates intrabacterial urea, and although not required for infection of the mouse stomach, it is required for acid survival in vitro in the absence of urea (41–43). Urea concentrations at the site of colonization may not be maintained at adequate levels, which may account for the up-regulation of arginase seen in vivo.
Except for aspA, all of the genes identified in this group showed similar changes in transcription in in vitro transcriptomal analyses when the bacteria were exposed to various levels of acidity (3–5). The above data make it likely that the site of infection of the gerbil stomach by H. pylori is acidic requiring the increased production of proteins encoded by the acid acclimation genes in this group.
It is thought that the gastric mucus is a barrier to luminal acid, protecting the gastric epithelium from damage. Recent data have challenged this supposition. Early work using glass microelectrodes suggested the presence of a pH gradient in the gastric mucus with the luminal face being acidic and the epithelial surface being neutral. This pH gradient through the mucus was maintained until luminal pH fell to <2.0, whereupon surface pH equaled luminal pH (44). More recent work using open tip ionophore based electrodes indicated the presence of a gastric surface pH barrier at a luminal pH of 2.0 or even 1.0 (45). However, using similar electrodes, it appears that infection with H. pylori of the mouse stomach disrupts this putative gastric barrier, allowing acidification of the gastric surface (26). An alternative approach to determine the surface pH gradient, using pH sensitive fluorescent probes, shows that, in agreement with the earlier microelectrode data, the gradient disappeared when the lumen was maintained at pH 2.0 (25). These studies show that the pH at the gastric epithelial surface can be maintained at pH 4.0 independent of luminal pH until the luminal pH falls to <3.0, and then surface pH equals luminal pH. The luminal pH after the gerbils were killed was found to be pH 3.0 (data not shown), hence the surface pH is at least 3.0. The median pH of the stomach of this species is similar to that of man, pH 1.4 (13, 19, 27).
The pH at the gastric surface can be deduced from gastric physiology and from physiological studies of H. pylori itself. The antrum of the stomach is an endocrine organ and is also the major site of H. pylori infection in the human (46). Feeding stimulates the release of gastrin from the antral G-cell because of muscarinic and gastrin-releasing peptide (GRP) stimulation and by luminal aromatic amino acids (47). Gastrin stimulates the release of histamine from the enterochromaffin-like cell, which activates the H2 receptor of the parietal cell and is the major peripheral stimulant of acid secretion. The buffering action of food maintains luminal pH to between pH 4.0 and 5.0. After digestion, the intragastric pH falls to 3.0 or less. D-cells are located at the base of antral glands and, because their apical surface is exposed to the lumen, they sense surface pH and release somatostatin when the luminal pH falls to 3.0 (48). Somatostatin inhibits gastrin release. The antral D cells thus respond to acidic pH at a location well below the surface of the mucosa. This inhibition of gastrin release by luminal pH at the D cell suggests that gastric mucus and HCO3− secretion do not provide a barrier to protons at a luminal pH ≤ 3.0 (25, 49).
Other evidence supporting an acidic environment at the gastric surface is the finding that Hp ureI deletion mutants are unable to colonize either the mouse or gerbil and also do not survive at a pH <4.0 in vitro even in the presence of physiological urea concentrations (13). Expression of the UreI component of the acid acclimation system is essential for infection or colonization of the gerbil unless acid secretion is inhibited by administration of H, K ATPase inhibitors. H. pylori isogenic ureI deletion mutants infected the nonacid secreting gerbil stomach as effectively as the wild-type organism (13). However, restoration of acid secretion by discontinuing administration of the ATPase inhibitors completely eradicated the deletion mutant but not the wild-type organism. Therefore, the bacteria require expression of UreI to survive acidity at the site of colonization on the gastric surface. It is likely that this pH ≤ 4.0, the lowest pH at which H. pylori survives in the absence of either urea or UreI (9).
Except for the hypB and aspA, all of the acid acclimation genes in table 1 are members of the ArsRS regulon. These data provide further evidence for exposure of the periplasm in vivo to a pH that activates HP0165 at a pH ≈5.9, the pH for half maximal activation of UreI and close to the pKa of histidine (50, 51). This is close to the calculated periplasmic pH found in vitro when the pHout was pH 3.0 (9). It would seem that the regulation of these genes in vivo when the organism is already on the surface of the gastric epithelium is also a compelling argument for their requirement for colonization, not just transit to the gastric surface. Hence, the in vivo transcriptome and the ureI deletion infection experiments indicate that H. pylori has to resist acid both in transit from the gastric lumen to the gastric surface and at its site of colonization. A model of the in vivo acid acclimation mechanism and gene regulation by the ArsRS two-component system is shown in Fig. 2.
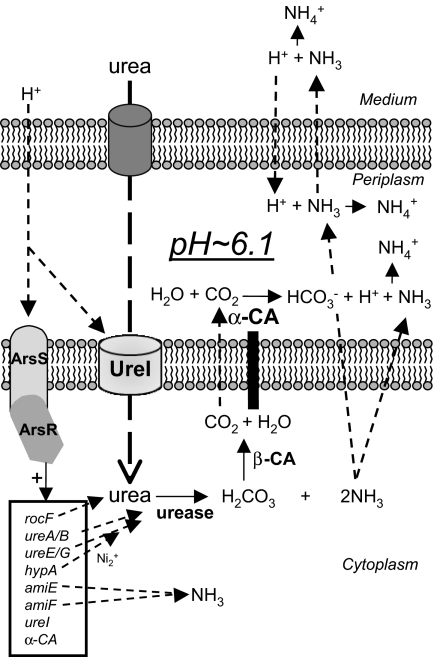
Fig. 2.
The mechanism of acid acclimation by H. pylori. Urea and protons enter the periplasm through outer membrane porins. Acid activation of UreI results in 300-fold acceleration of urea entry into the bacterial cytoplasm. At pH 6.1, the ArsRS two-component system is also activated and results in the up-regulation of the Ni2+ insertion genes, ureE and ureG, that contribute to increase of urease activity by converting the apoenzyme into active urease. The potential ammonia producing enzymes, amiE and amiF, and the urea producing rocF (arginase) are also up-regulated by this two-component system, as is α-carbonic anhydrase (α-CA) and ureI. Hydrolysis of urea results in the production of H2CO3 and 2NH3. The former is converted into CO2 and H2O by the cytoplasmic β-carbonic anhydrase (β-CA), and both the CO2 and NH3 diffuse rapidly through the inner membrane into the periplasm. There, the CO2 is converted by α-CA into HCO3− and H+. The bicarbonate buffers the periplasm, maintaining periplasmic pH at 6.1, and the NH3 neutralizes this proton and also protons entering the periplasm. Ammonia is also able to diffuse through the outer membrane to neutralize medium acidity. Cytoplasmic NH3 can also neutralize protons entering the cytoplasm. The acid acclimation genes that are up-regulated by ArsR (HP0166) are shown in the box.
Motility and Chemotaxis.
To infect the stomach, H. pylori must transit the mucus layer from the gastric lumen to access the epithelial surface, its site of infection. This movement is directed as H. pylori colonize a particular gastric niche, the transitional zone at the interface of the antral and fundic mucosa of the human stomach (31). It must continually seek out this niche as conditions vary between feeding and fasting. This directed movement requires motility and chemotaxis. H. pylori is spiral shaped with 2–6 unipolar sheathed flagella. Together, these attributes enable the organism to move in the gastric mucus (52).
Inoculation of gnotobiotic pigs with H. pylori strains of different motilities as determined by in vitro methods demonstrated that the rates of infection were proportional to the degree of motility of the organism and established the requirement for motility as a pathogenic factor (53). The flagellae are composed of two subunits, FlaA and FlaB. Mutation of either subunit alone or a double mutation abolished colonization in the gnotobiotic pig (54). Mutagenesis of a number of motility genes in H. pylori also abolished colonization and/or persistence in various animal infection models (16). Of the 13 genes encoding flagellar structural and biosynthetic proteins that were up-regulated in the gerbil stomach, flaA, flgD, flgE, and flgH, have been shown to be essential for gerbil infection. Eight of the 13 in vivo up-regulated genes were also up-regulated in in vitro transcriptomal comparison studies of H. pylori exposed to acid versus neutral pH (2–5). This commonality of increased expression of these flagellar genes between the in vivo transcriptome and acid-induced in vitro transcriptomes suggests that low pH at the site of colonization is one of the triggers for transcription of these genes. Also, there was an increase in expression of the chemotactic genes cheY, and the two cheV genes in both the in vivo and in vitro acid-induced transcriptomes suggesting that the trigger for the sigma factor σ80 is low pH (2, 5).
H. pylori has a positive chemotactic response to urea, sodium bicarbonate and the urease inhibitor, flurofamide (55). Because urea is an absolute requirement for acid survival by this gastric pathogen, it has been suggested that the chemotactic response to urea may be a requirement for colonization (55). Urease is not required for the positive chemotactic response to urea in a low viscosity medium but is required at higher viscosities (5.6–16.7 cP) (56). In H. pylori, chemo-attractants/repellents bind to inner membrane methyl-accepting chemoreceptor proteins (MCPs) (TlpA, TlpB and TlpC). Ligand binding to the chemoreceptor activates the histidine kinase, CheA, through the coupling protein CheW, phosphorylating the response regulator CheY that then binds to MotB the flagellar motor switch, affecting both flagellar direction and rotational velocity. cheY mutants of H. pylori strain N6 are nonmotile and unable to infect the gnotobiotic pig (57). Likewise, disruption of cheW and cheY in the SS1 strain renders the organism nonmotile and unable to infect the HSD/ICR mouse. In contrast, cheA, cheY, and cheW mutants of the SS1 strain were able to infect FVB/N mice but with an attenuated phenotype after 2 weeks of infection that returned to control levels after 6 months of infection (58). Disruption of the motB gene results in a nonmotile bacterium with intact flagella that is severely attenuated in its ability to infect the FVB/N mouse and incapable of infecting the HSD/ICR mouse (57, 59). Disruption of the putative MCP genes, tlpA and tlpC, had no effect on motility, but these mutants were also attenuated in their ability to infect mice. There was increased in vivo expression of the chemotaxis gene cheY (HP1067) and the two cheV genes (HP0393, HP0019) as in expression of these genes in the in vitro acid-exposed transcriptome (2, 5). Increased expression of the gene encoding the transmembrane receptor, methyl-accepting chemotaxis proteins, tlpB (HP0103) and HP0559 in the gerbil stomach but not under acidic conditions in vitro suggests that regulation of these genes, unlike that of cheV and cheY, is independent of environmental pH. Further, only three of these genes are part of the ArsRS regulon, suggesting regulation by factors other than environmental pH.
Pathogenicity.
It is thought that expression of the genes in the Cag PAI is related to generation of gastric pathology (35). Several genes of the Cag PAI are up-regulated by acidity in vitro (cag22, cag21, cag20, cag12, and cag1) and, along with cagA, are also part of the ArsRS regulon (5, 7). None of the in vitro acid-induced transcriptomes detected up-regulation of cagA, although it increased 21-fold in vivo. These findings suggest that the T4SS machinery is up-regulated when the organism senses acid so as to be primed to inject CagA. cagA is up-regulated following adhesion and secretion into the gastric cell due to the requirement for increased CagA synthesis. Therefore, cagA as a member of the ArsRS regulon requires not only acid for increased transcription but also adhesion to the host cell and increased protein turnover due to secretion into the host cell.
Thus, analysis of the in vivo transcriptome has shown up-regulation of several groups of genes, three of which are discussed here. The majority of the acid acclimation group of genes was up-regulated, and these are members of the pH sensitive ArsRS two-component system, indicating that this group is regulated by gastric acidity. Only three of the motility and chemotaxis group of genes are members of this regulon, but 12 of the 18 genes in this group were increased in vitro at pH 4.5, suggesting an additional acidity-dependent regulation. However, seven of nine PAI genes up-regulated in the stomach are members of this two-component system and hence respond to gastric acidity. Acidity is therefore a major factor in regulation of many of the genes, but other as-yet-undiscovered means of regulation must also be present.
Materials and Methods
Briefly, Mongolian gerbils were inoculated by gavage with H. pylori strain 69a expressing green fluorescent protein (GFP-Hp). Ten days after inoculation, the animals were killed, and their stomachs were rapidly removed. H. pylori RNA from the fundic and antral mucosae was isolated and enriched by using MICROBEnrich and MICROBExpress Bacterial mRNA enrichment kits (Ambion, Austin, TX). cDNA was synthesized by reverse transcription from in vivo H. pylori RNA in the presence of Cy5-dCTP and from in vitro cultured H. pylori RNA with Cy3-dCTP and hybridized to glass slides containing the 1,534 predicted ORFs of H. pylori strain 26695 (5). Microarray analysis was performed by using Phoretix Array software (Nonlinear Dynamic, Durham, NC). For details, see supporting information (SI) Materials and Methods.
Supplementary Material
Abbreviation
- PAI
pathogenicity island.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702300104/DC1.
References
- 1.Young GM, Amid D, Miller VL. J Bacteriol. 1996;178:6487–6495. doi: 10.1128/jb.178.22.6487-6495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang S, Lee CZ, Peck K, Sindici M, Matrubutham U, Gleeson MA, Wang JT. Infect Immun. 2001;69:1679–1686. doi: 10.1128/IAI.69.3.1679-1686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bury-Mone S, Thiberge JM, Contreras M, Maitournam A, Labigne A, De Reuse H. Mol Microbiol. 2004;53:623–638. doi: 10.1111/j.1365-2958.2004.04137.x. [DOI] [PubMed] [Google Scholar]
- 4.Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. Infect Immun. 2003;71:3529–3539. doi: 10.1128/IAI.71.6.3529-3539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Infect Immun. 2003;71:5921–5939. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcus EA, Moshfegh AP, Sachs G, Scott DR. J Bacteriol. 2005;187:729–738. doi: 10.1128/JB.187.2.729-738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pflock M, Finsterer N, Joseph B, Mollenkopf H, Meyer TF, Beier D. J Bacteriol. 2006;188:3449–3462. doi: 10.1128/JB.188.10.3449-3462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachs G, Weeks DL, Wen Y, Marcus EA, Scott DR, Melchers K. Physiology. 2005;20:429–438. doi: 10.1152/physiol.00032.2005. [DOI] [PubMed] [Google Scholar]
- 9.Scott DR, Weeks D, Hong C, Postius S, Melchers K, Sachs G. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 10.Weeks DL, Eskandari S, Scott DR, Sachs G. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 11.Scott DR, Marcus EA, Weeks DL, Lee A, Melchers K, Sachs G. Infect Immun. 2000;68:470–477. doi: 10.1128/iai.68.2.470-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott DR, Marcus EA, Weeks DL, Sachs G. Gastroenterology. 2002;123:187–195. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 13.Mollenhauer-Rektorschek M, Hanauer G, Sachs G, Melchers K. Res Microbiol. 2002;153:659–666. doi: 10.1016/s0923-2508(02)01380-3. [DOI] [PubMed] [Google Scholar]
- 14.Skouloubris S, Thiberge JM, Labigne A, De Reuse H. Infect Immun. 1998;66:4517–4521. doi: 10.1128/iai.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odenbreit S. Int J Med Microbiol. 2005;295:317–324. doi: 10.1016/j.ijmm.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 16.O'Toole PW, Lane MC, Porwollik S. Microb Infect Institut Pasteur. 2000;2:1207–1214. doi: 10.1016/s1286-4579(00)01274-0. [DOI] [PubMed] [Google Scholar]
- 17.van Vliet AH, Stoof J, Vlasblom R, Wainwright SA, Hughes NJ, Kelly DJ, Bereswill S, Bijlsma JJ, Hoogenboezem T, Vandenbroucke-Grauls CM, et al. Helicobacter. 2002;7:237–244. doi: 10.1046/j.1523-5378.2002.00088.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim N, Marcus EA, Wen Y, Weeks DL, Scott DR, Jung HC, Song IS, Sachs G. Infect Immun. 2004;72:2358–2368. doi: 10.1128/IAI.72.4.2358-2368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teyssen S, Chari ST, Scheid J, Singer MV. Dig Dis Sci. 1995;40:247–255. doi: 10.1007/BF02065405. [DOI] [PubMed] [Google Scholar]
- 20.Code CF. Scand J Gastroenterol. 1981;67:201–204. [PubMed] [Google Scholar]
- 21.Deamer DW. J Bioenerg Biomembr. 1987;19:457–479. doi: 10.1007/BF00770030. [DOI] [PubMed] [Google Scholar]
- 22.Gutknecht J. J Bioenerg Biomembr. 1987;19:427–442. doi: 10.1007/BF00770028. [DOI] [PubMed] [Google Scholar]
- 23.Priver NA, Rabon EC, Zeidel ML. Biochemistry. 1993;32:2459–2468. doi: 10.1021/bi00061a002. [DOI] [PubMed] [Google Scholar]
- 24.Schade C, Flemstrom G, Holm L. Gastroenterology. 1994;107:180–188. doi: 10.1016/0016-5085(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 25.Baumgartner HK, Montrose MH. Gastroenterology. 2004;126:774–783. doi: 10.1053/j.gastro.2003.11.059. [DOI] [PubMed] [Google Scholar]
- 26.Henriksnas J, Phillipson M, Storm M, Engstrand L, Soleimani M, Holm L. A J Physiol. 2006;291:G396–403. doi: 10.1152/ajpgi.00017.2006. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki SM, Matsuda MG, Misumi A, Honmyo U, Murakami A, Murata H, Sagara K, Kurano R, Okabe H. Dig Endosc. 2001;10:195–201. [Google Scholar]
- 28.Graham JE, Peek RM, Jr, Krishna U, Cover TL. Gastroenterology. 2002;123:1637–1648. doi: 10.1053/gast.2002.36589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boonjakuakul JK, Canfield DR, Solnick JV. Infect Immun. 2005;73:4895–4904. doi: 10.1128/IAI.73.8.4895-4904.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boonjakuakul JK, Syvanen M, Suryaprasad A, Bowlus CL, Solnick JV. J Infect Dis. 2004;190:946–956. doi: 10.1086/423142. [DOI] [PubMed] [Google Scholar]
- 31.Van Zanten SJ, Dixon MF, Lee A. Gastroenterology. 1999;116:1217–1229. doi: 10.1016/s0016-5085(99)70025-9. [DOI] [PubMed] [Google Scholar]
- 32.Akada JK, Shirai M, Takeuchi H, Tsuda M, Nakazawa T. Mol Microbiol. 2000;36:1071–1084. doi: 10.1046/j.1365-2958.2000.01918.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamaoka Y, Souchek J, Odenbreit S, Haas R, Arnqvist A, Boren T, Kodama T, Osato MS, Gutierrez O, Kim JG, et al. J Clin Microbiol. 2002;40:2244–2246. doi: 10.1128/JCM.40.6.2244-2246.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 35.Hatakeyama M. Int J Cancer. 2006;119:1217–1223. doi: 10.1002/ijc.21831. [DOI] [PubMed] [Google Scholar]
- 36.Athmann C, Zeng N, Kang T, Marcus EA, Scott DR, Rektorschek M, Buhmann A, Melchers K, Sachs G. J Clin Invest. 2000;106:339–347. doi: 10.1172/JCI9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson JW, Mehta NS, Maier RJ. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 38.Benoit S, Maier RJ. J Bacteriol. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta N, Olson JW, Maier RJ. J Bacteriol. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez V, Curi AP, Torkian B, Schaeffer JM, Wilkinson HA, Walsh JH, Tache Y. Gastroenterology. 1998;114:1125–1132. doi: 10.1016/s0016-5085(98)70417-2. [DOI] [PubMed] [Google Scholar]
- 41.McGee DJ, Radcliff FJ, Mendz GL, Ferrero RL, Mobley HLT. J Bacteriol. 1999;181:7314–7322. doi: 10.1128/jb.181.23.7314-7322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langford ML, Zabaleta J, Ochoa AC, Testerman TL, McGee DJ. Helicobacter. 2006;11:477–493. doi: 10.1111/j.1523-5378.2006.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGee DJ, Zabaleta J, Viator RJ, Testerman TL, Ochoa AC, Mendz GL. Eur J Biochem. 2004;271:1952–1962. doi: 10.1111/j.1432-1033.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- 44.Williams SE, Turnberg LA. Gut. 1981;22:94–96. doi: 10.1136/gut.22.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillipson M, Atuma C, Henriksnas J, Holm L. Am J Physiol. 2002;282:G211–G219. doi: 10.1152/ajpgi.00223.2001. [DOI] [PubMed] [Google Scholar]
- 46.Genta RM, Graham DY. Gastrointest Endosc. 1994;40:342–345. doi: 10.1016/s0016-5107(94)70067-2. [DOI] [PubMed] [Google Scholar]
- 47.Walsh JH. In: Physiology of the Gastrointestinal Tract. Johnson LR, editor. New York: Raven; 1994. pp. 1–129. [Google Scholar]
- 48.Schubert ML, Edwards NF, Makhlouf GM. Gastroenterology. 1988;94:317–322. doi: 10.1016/0016-5085(88)90418-0. [DOI] [PubMed] [Google Scholar]
- 49.Feldman M, Walsh JH. Gastroenterology. 1980;78:772–776. [PubMed] [Google Scholar]
- 50.Bury-Mone S, Skouloubris S, Labigne A, De Reuse H. Mol Microbiol. 2001;42:1021–1034. doi: 10.1046/j.1365-2958.2001.02689.x. [DOI] [PubMed] [Google Scholar]
- 51.Weeks DL, Gushansky G, Scott DR, Sachs G. J Biol Chem. 2004;279:9944–9950. doi: 10.1074/jbc.M312680200. [DOI] [PubMed] [Google Scholar]
- 52.Hazell SL, Lee A, Brady L, Hennessy W. J Infect Dis. 1986;153:658–663. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- 53.Eaton KA, Morgan DR, Krakowka S. Infect Immun. 1989;57:1119–1125. doi: 10.1128/iai.57.4.1119-1125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizote T, Yoshiyama H, Nakazawa T. Infect Immun. 1997;65:1519–1521. doi: 10.1128/iai.65.4.1519-1521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakamura H, Yoshiyama H, Takeuchi H, Mizote T, Okita K, Nakazawa T. Infect Immun. 1998;66:4832–4837. doi: 10.1128/iai.66.10.4832-4837.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foynes S, Dorrell N, Ward SJ, Stabler RA, McColm AA, Rycroft AN, Wren BW. Infect Immun. 2000;68:2016–2023. doi: 10.1128/iai.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terry K, Williams SM, Connolly L, Ottemann KM. Infect Immun. 2005;73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ottemann KM, Lowenthal AC. Infect Immun. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. J Bacteriol. 2006;188:1750–1761. doi: 10.1128/JB.188.5.1750-1761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heuermann D, Haas R. Mol Gen Genet. 1998;257:519–528. doi: 10.1007/s004380050677. [DOI] [PubMed] [Google Scholar]
- 62.Rozen S, Skaletsky H. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.