Abstract
J proteins are obligate cochaperones of Hsp70s, stimulating their ATPase activity and thus allowing them to function in multiple cellular processes. In most cellular compartments, an Hsp70 works with multiple, structurally divergent J proteins. To better understand the functional specificity of J proteins and the complexity of the Hsp70:J protein network, we undertook a comprehensive analysis of 13 J proteins of the cytosol of the yeast Saccharomyces cerevisiae. Phenotypes caused by the absence of four proteins, Sis1, Jjj1, Jjj3, and Cwc23, could not be rescued by overexpression of any other cytosolic J protein, demonstrating the distinctive nature of J proteins. In one case, that of Zuo1, the phenotypic effects of the absence of a J protein could be rescued by overexpression of only one other J protein, Jjj1, which, like Zuo1, is ribosome-associated. In contrast, the severe growth phenotype caused by the absence of the cytosol's most abundant J protein, Ydj1, was substantially rescued by expression of J domain-containing fragments of many cytosolic J proteins. We conclude that many functions of Hsp70 chaperone machineries only require stimulation of Hsp70's ATPase activity by J protein partners. However, a subset of Hsp70 functions requires specific J protein partners, likely demanding either sublocalization within the compartment or binding to specific client proteins.
Keywords: Hsp40, Hsp70, molecular chaperone, multigene family
Through their action in protein folding, degradation, translocation across membranes, and disassembly of protein complexes, molecular chaperones are important participants in many crucial cellular processes (1, 2). Hsp70s and their J protein partners (at times referred to as Hsp40s) constitute an important component of the cellular “chaperone” in virtually all living systems (3). In most cellular compartments, an Hsp70 has multiple J protein partners. The cytosol, a hub of activity for a variety of crucial cellular processes, is no exception. With a goal of better understanding the degree of functional overlap among J proteins, as well as their individual specificities, we choose the cytosol of Saccharomyces cerevisiae as the focus of a comprehensive analysis of J protein function.
Although J proteins are obligate partners of Hsp70s, it is the Hsp70 that has long been considered the core of the Hsp70:J protein chaperone machine by virtue of their client protein interaction, which is modulated by nucleotide binding (4). ATP binding fosters rapid binding to client proteins, whereas nucleotide hydrolysis stabilizes the interaction. The cytosol of S. cerevisiae contains two predominant classes of Hsp70s, Ssa (SSA1–4) and Ssb (SSB1–2) (5). Yeast cells expressing only one representative of a class grow very similarly to wild-type cells under a variety of conditions. But the classes are functionally distinct, because a member of the Ssa class cannot substitute for an Ssb Hsp70 and vice versa (6).
A role of all J proteins is stimulation of the ATPase activity of their partner Hsp70s. Such stimulation fosters productive interaction of Hsp70s with their client proteins (4). The ≈65-aa J domain, the defining feature of all of the J proteins, is responsible for the stimulation. The J domain is characterized by a highly conserved histidine–proline–aspartic acid (HPD) tripeptide signature motif that is important for J domains' stimulatory activity. Despite the omnipresent J domain, J proteins, as a group, are strikingly dissimilar, varying significantly in their domain organization and localization within the cytosol (7). Other domains have been shown to associate directly with client proteins, thereby fostering client protein interaction with Hsp70 or promoting localization to a particular site within a cellular compartment.
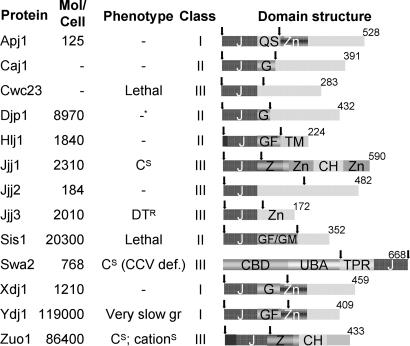
Historically, J proteins have been divided into three classes (I, II, and III). The class I designation is based on the motifs/domains present in the founding member of this group, DnaJ of Escherichia coli: an N-terminal J domain, followed by a glycine/phenylalanine (G/F)-rich region, four repeats of the CxxCxGxG-type zinc finger, and a C-terminal extension known to bind client proteins (8). The cytosol of S. cerevisiae has three J proteins that have been placed in class I: Apj1, Xdj1, and Ydj1 (7). Class II J proteins, by definition, have a similar structure, in that they have a J domain and a G/F region. However, class II J proteins lack the CxxCxGxG-type zinc finger domain. Cytosolic class II J proteins include Caj1, Djp1, Hlj1, and Sis1. All of the other J proteins have been arbitrarily placed in class III. A few S. cerevisiae class III J proteins contain zinc-binding domains, but these are not of the type found in class I proteins. Jjj1 contains C2H2--type zinc fingers, whereas Jjj3 has a CSL-type zinc finger (Fig. 1). Apart from structural differences among J proteins, the relative abundance of cytosolic J proteins also varies dramatically (Fig. 1). Thus, it is possible that potential functional overlap between J proteins is masked by differences in their expression levels.
Fig. 1.
Cytosolic J proteins of S. cerevisiae. Molecules per cell are as reported by Ghaemmaghami et al. (11). No data available is indicated by −. Phenotypes tested are as follows: cold sensitivity (CS), resistance to diphtheria toxin (DTR), very compromised for growth (very slow gr.), sensitive to cations (cationS), and defective in uncoating CCVs (CCV def.). Information was obtained from the Saccharomyces Genome database, www.yeastgenome.org. J protein classes were taken from Walsh et al. (7). In the domain structure, arrows indicate the J domain fragments used (Apj11–161, Caj11–139, Cwc231–139, Djp11–127, Hlj11–179, Jjj11–128, Jjj21–132, Jjj31–124, Sis11–167, Swa2362–668, Xdj11–146, Ydj11–134, and Zuo11–234). Numbers indicate amino acids in each full-length J protein. Regions in which these amino acids are highly represented include QS, G, GF, and GF/GM (listed by their standard single-letter amino acid code). J, J domain; Zn, zinc finger; CBD, clathrin-binding domain; UBA, ubiquitin association; TPR, tetratricopeptide repeat; Z, zuotin-like; CH, charged; TM, transmembrane.
To better understand the functional diversity displayed by J proteins, we undertook an analysis of cytosolic J proteins of S. cerevisae. Several J proteins of the cytosol appeared to be functionally unique, because overexpression of no other J protein was able to substitute for their functions. We also found that the function of the most abundant J protein of the cytosol, Ydj1, could be carried out by expression of the J domain from several diverse J proteins. Our analysis of a single cellular compartment likely provides a paradigm for understanding the general principles of diversity of function of J protein:Hsp70 machineries in other cellular compartments and in other organisms.
Results
Deletion Phenotypes of Genes Encoding Cytosolic/Nuclear J Proteins.
As a starting point for our analysis of cytosolic J protein function, we constructed or obtained strains having a deletion of each of the 10 genes that encode a predominantly cytosolic J protein, as defined in ref. 9: Apj1, Djp1, Jjj1, Jjj2, Jjj3, Sis1, Swa2, Xdj1, Ydj1, and Zuo1. We also analyzed three other J proteins: Cwc23, which, although predominantly nuclear, is in the cytosol as well; Hlj1, an endoplasmic reticulum (ER) membrane protein whose J domain faces the cytosol (10); and Caj1, whose localization has not been reported, but which lacks apparent sequences for targeting to any organelle. The phenotypes of the knockout strains, all in the W303 genetic background, were similar to those previously reported, with the exception of Δswa2, which grew slowly at low temperatures. Seven deletion strains, those lacking Cwc23, Jjj1, Jjj3, Sis1, Swa2, Ydj1, and Zuo1, had easily assayable phenotypes and became the focus of our studies. The reported abundance of J proteins within the cytosol ranges from >100,000 molecules per cell of Ydj1 to only 125 molecules per cell of Apj1 (11). Because of this large variation, we reasoned that functional overlap might exist among these proteins that is not evident from the phenotypes displayed by the single gene knockouts. Therefore, overexpression constructs were made for all 13 cytosolic J proteins by cloning the corresponding ORFs under the glyceraldehyde 3-phosphate dehydrogenase (GPD) promoter in a high-copy plasmid. Transformants harboring constructs containing Caj1, Djp1, Hlj1, and Xdj1 were either not obtained or grew very poorly, presumably because overexpression of these J proteins was deleterious. Therefore, these expression plasmids were not included in the experiments discussed below.
J Domain Fragments Are Sufficient for Robust Growth of Δydj1.
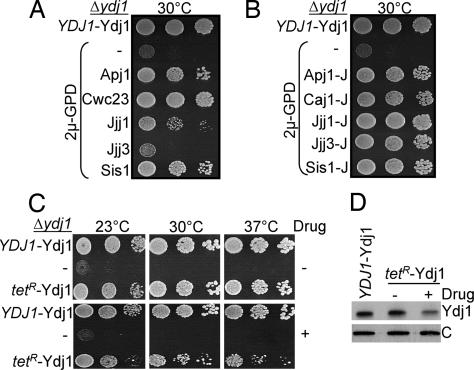
We began our analysis with Ydj1, because it is the most abundant cytosolic J protein. Ydj1 partners with Ssa Hsp70s in protein folding and translocation of proteins into endoplasmic reticulum (ER) and mitochondria (12–15). Δydj1 cells are viable but have a severe growth defect at all temperatures. As expected from earlier reports (12), increased expression of Sis1 improved growth of Δydj1 cells [Fig. 2A and supporting information (SI) Fig. 6A]. Overexpression of Apj1 also dramatically rescued the growth defect of Δydj1 cells (Fig. 2A). Such rescue was not particularly surprising, because Apj1, like Ydj1, is a class I J protein. However, overexpression of the class III J protein, Cwc23, also rescued Δydj1 cells significantly (Fig. 2A), even though it only has a J domain in common with Ydj1.
Fig. 2.
Ability of other J proteins and lower levels of Ydj1 to rescue Δydj1 growth phenotype. (A–C) Ten-fold serial dilution of Δydj1 cells expressing the indicated proteins were plated onto minimal medium and incubated for 3 days at the indicated temperatures. The following were used as controls: Ydj1 under control of its own promoter in a centromeric vector, pRS314 (YDJ1-Ydj1); and empty vector (−). (A and B) Overexpressing full-length (A) or J domain fragments of cytosolic J proteins, as indicated in the legend to Fig. 1 (B), driven by the GPD promoter from a 2μ plasmid. The complete set of plates is shown in SI Fig. 6A. (C) Ydj1 under control of the tetracycline-regulatable promoter (tetR-Ydj1) or empty vector (−) in the absence (−) or presence (+) of 0.5 μg/ml doxycycline (drug). (D) Total lysates prepared from cells grown in the absence (−) or presence (+) of 0.5 μg/ml doxycycline (drug) were resolved by SDS/PAGE, electro-blotted, and probed with anti-Ydj1 antibodies and, as loading control, anti-Ssc1 antibodies (indicated by “C”).
Because of rescue by divergent J proteins that had no obvious sequence similarity beyond their J domains, we reasoned that the J domain alone might be sufficient. Therefore, we designed constructs to express J domain-containing fragments at high levels. In contrast to the full-length proteins, J domain fragments of many cytosolic J proteins were able to substantially rescue Δydj1 (Fig. 2B and SI Fig. 6B). At a variety of temperatures, the J domain fragment of Jjj1 permitted the most robust growth of any fragment tested. This rescue depended on J domain function, because Jjj1-JH32Q, having an alteration in the histidine–proline–aspartic acid (HPD) motif, did not rescue (SI Fig. 6C). Even though expression of full-length Caj1 and Djp1 were deleterious (data not shown), their J domain fragments were able to rescue Δydj1 (SI Fig. 6B). In addition, J domain fragments from a number of class III J proteins, such as Jjj3-J, rescued, although the full-length construct did not (Fig. 2 A and B). We are unable to make a comprehensive summary regarding the ability of each full-length protein or J domain fragment to rescue because of the lack of tools to measure levels of expression of each polypeptide. However, we can draw the general conclusion that a variety of J domain-containing fragments are competent to carry out functions of Ydj1.
Normal Levels of a J Domain Fragment Are Sufficient for Rescue of Δydj1.
The rather efficient rescue of Δydj1 by J domain fragments raised two questions: (i) What level of full-length Ydj1 is required for wild-type growth? (ii) How robust is the growth of cells expressing such levels of a J domain? To answer the first question, the coding region of Ydj1 was placed under the control of the tetracycline repressible promoter (tetR). In the absence of drug, the level of Ydj1 expressed from tetR was indistinguishable from that expressed by the endogenous promoter (Fig. 2D). When Ydj1 expression was reduced to ≈40% of normal levels after addition of drug, significant growth defects were observed at all of the temperatures tested (Fig. 2 C and D), thereby indicating that a high level of Ydj1 protein is required for wild-type growth.
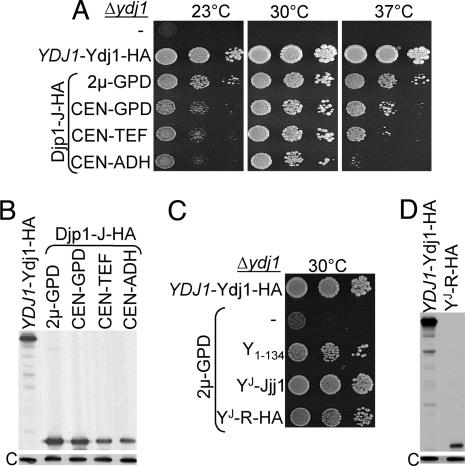
To answer the second question, we needed to quantitatively compare the level of expression of Ydj1 and J domain fragment. To accomplish this, several J domain fragments, as well as full-length Ydj1, were tagged with HA-epitopes. Ydj1-HA, which was expressed at levels very similar to that of untagged protein driven by the endogenous YDJ1 promoter, allowed robust growth of Δydj1 cells at all temperatures tested (SI Fig. 7 A and B). Several of the tagged J domain constructs rescued much more poorly than their untagged constructs, likely because of changes in expression levels or effects of the tag on function or both. However, the J domain fragment of Djp1 having the HA-epitope tag (Djp1-J HA) rescued the growth of Δydj1 cells as well as the untagged construct (SI Fig. 7C). This similarly robust rescue enabled us to use HA-specific antibody to compare the expression levels of a J domain and Ydj1-HA and to thus compare the ability of fragments to substitute for full-length Ydj1. Using promoters of variable strengths, Djp1-J HA was expressed in Δydj1 cells at levels ranging from ≈0.6- to 1.7-fold that of Ydj1 expressed from its own promoter (Fig. 3B). At all expression levels tested, significant rescue was observed (Fig. 3A). Plating efficiency and rate of colony formation of strains having different constructs were similar at 30°C. At 23°C and 37°C, more disparity among the strains was observed. Cells expressing the J domain fragment at 60% of the normal level of Ydj1 did not form colonies at 23°C or 37°C under the conditions tested. Thus, a J domain-containing fragment, when expressed at levels at which full-length Ydj1 protein is normally present, can support growth under conditions that cells lacking wild-type Ydj1 cannot, but it is unable to fully substitute for Ydj1.
Fig. 3.
A J domain fragment expressed at levels comparable to wild-type Ydj1 substantially rescues Δydj1. (A and C) Serial dilutions of Δydj1 cells expressing J domain fragments, only a vector (−), or HA-tagged Ydj1 under control of the YDJ1 promoter (YDJ1-Ydj1-HA) were spotted on minimal media and incubated at 30°C for 3 days. (A) HA-tagged Djp1 J domain fragment (Djp1-J HA) driven by promoters of different strengths from either high-copy (2μ) or centromeric (CEN) plasmids. (C) N-terminal 134 aa of Ydj1 (Ydj11–134) or the J domain of Ydj1 (amino acids 1–63) fused to either an additional 64 (64–128) amino acids of Jjj1 (YJ Jjj1) or a random sequence (YJ R-HA) driven by the GPD promoter in 2μ plasmids. (B and D) Total cell lysates of strains were subjected to SDS/PAGE, electro-blotted, and probed with anti-HA antibodies and, as loading control, anti-Ssc1 antibodies (indicated by “C”).
Importance of G/F for J Domain Function.
The results of the experiments described above suggest that a J domain by itself may be sufficient for substantial rescue of Δydj1. However, the J domain-containing fragments used in this study had amino acids in addition to the J domain, raising the question about what role, if any, these extra amino acids might play. It has been suggested that the G/F region, a defining feature of class I and II J proteins, may be critical for their function (16). However, the ability of Jjj1-J (Jjj11–128), which includes the J domain (amino acids 1–62) plus 66 additional amino acids, to rescue Δydj1 demonstrates that a G/F region is not required, because Jjj1 is a class III J protein and thus has no such region (Fig. 2B).
However, this result does not exclude the possibility that sequences normally adjacent to a J domain, even though they may not be similar in sequence, play important roles. To ask specifically whether the J domain of a class I protein requires a G/F region for rescue of Δydj1, we tested a chimera between amino acids 1–60 of Ydj1 and 61–128 of Jjj1 (YJ Jjj1). Overexpression of YJ Jjj1 rescued Δydj1 efficiently (Fig. 3C). To extend this analysis, we expressed a chimera encoding the J domain of Ydj1, followed by 32 aa that are not present in any J protein, plus a 32-aa 3xHA tag. This construct, YJ R-HA, rescued the growth defect of Δydj1 cells at 30°C, the physiologically optimum temperature for yeast (Fig. 3C), even though it is expressed at lower levels (compare Fig. 3B and Fig. 3C). Thus, we conclude that the rescue of Δydj1 by J domain-containing fragments is due solely to the function of the J domain.
Jjj3, a Specialized J Protein, Requires Its C-Terminal CSL Zinc Finger Domain.
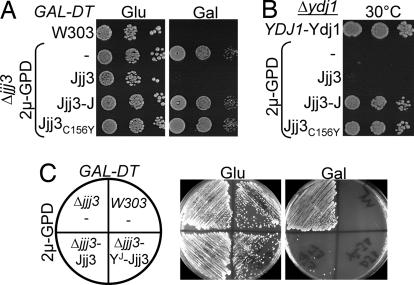
We continued our analysis of strains lacking other cytosolic J proteins. Jjj3 plays an essential role in the biosynthesis of diphthamide (DPH), an unusual amino acid formed by posttranslational modification of a conserved histidine found in the translation elongation factor, eEF2 (17). This modified amino acid is the target for ADP-ribosylating diphtheria toxin (DT) produced by Corynebacterium diphtheriae. As a result, cells lacking Jjj3 (Fig. 4A) or other proteins involved in this pathway are tolerant to DT. For testing Jjj3 function in vivo, the overexpression constructs were transformed into a Δjjj3 strain harboring a plasmid encoding the catalytic subunit of DT under the control of a galactose-inducible promoter. None of the J domain fragments, including that of Jjj3 itself, or full-length proteins could perform Jjj3's function in vivo, as evidenced by the ability of transformants to grow on galactose-based media (Fig. 4A and data not shown).
Fig. 4.
Effect of Jjj3's CSL zinc finger and J domain on function. All strains tested contained a plasmid-encoding diphtheria toxin under galactose regulation. Shown are wild-type control strain with empty vector (W303), Δjjj3 with empty vector (−) or 2μGPD vector expressing full-length Jjj3 (Jjj3), J domain fragment of Jjj3 (Jjj3-J), Jjj3C156Y or a chimera having the N-terminal 63 aa of Ydj1, and amino acids 70–172 of Jjj3 (YJ Jjj3). Transformants were plated on minimal media containing either 2% glucose (Glu) or galactose (Gal) and incubated for 3 days at 30°C.
The inability of the J domain fragment of Jjj3 to substitute for the full-length protein (Fig. 4A) suggested to us that sequences in addition to the J domain may be critical for Jjj3's role in DPH biosynthesis. Jjj3 is a small protein, having only 94 residues, encompassing a CSL zinc finger in addition to the J domain. To determine whether this motif is important for Jjj3 function, we tested a mutant protein, Jjj3C156Y, having a tyrosine substituted for the cysteine in the conserved CSL tripeptide. Jjj3C156Y cells were tolerant to DT (Fig. 4A), indicating that the CSL domain is critical for Jjj3's function. In addition, we tested whether chimera YJ Jjj3, containing the J domain of Ydj1 (amino acids 1–63) and C-terminal region of Jjj3 (amino acids 70–172), could substitute for Jjj3. This construct was functional in DPH biosynthesis, because Δjjj3 cells harboring this plasmid were sensitive to DT (Fig. 4C). Thus, although a J domain is required for Jjj3's function (SI Fig. 8), it is not a Jjj3 specificity determinant.
A Jjj3 J domain fragment, but not full-length protein, was competent to substitute for Ydj1 when overexpressed (Fig. 2 A and B). We asked whether alteration of the zinc finger would allow full-length Jjj3 to rescue Δydj1 cells. Jjj3C156Y rescued growth of Δydj1 cells as the Jjj3-J domain fragment (Fig. 4B).
Specialization of Essential J Proteins, Cwc23 and Sis1.
We also tested two essential J proteins: Cwc23, which is implicated in RNA splicing (7), and Sis1, which is required for the maintenance of the [RNQ+] prion and is thought to play an important role in translation initiation (18, 19). Δcwc23 and Δsis1 haploids carrying their respective wild-type genes on a URA3-based plasmid, as well as one of the expression plasmids, were plated on 5-fluoroorotic acid (5-FOA)-containing plates to select for cells having lost the wild-type gene. None of the heterologous full-length or J domain-containing fragments rescued either deletion strain (data not shown), suggesting that Sis1 and Cwc23 contain sequences specifically required for their essential functions.
Specialization of the Ribosome-Associated J Proteins, Zuo1 and Jjj1.
Zuo1 and Jjj1 are both ribosome-associated J proteins. Zuo1 is the J protein partner of Ssb, the specialized ribosome-associated Hsp70 that binds nascent chains exiting the ribosome (20). Jjj1 functions in 60S subunit biogenesis with Ssa (21). All constructs were tested for rescue of Δjjj1 and Δzuo1. None of our expression plasmids rescued the cold sensitivity of cells lacking Jjj1, indicating a specialized function of this J protein (data not shown). As reported recently (21), overexpression of full-length Jjj1 partially rescued the cation and cold sensitivity of Δzuo1 cells. However, no other construct had an effect on the growth of Δzuo1 cells, indicating that Jjj1 is unique in its ability to partially substitute for Zuo1.
Swa2, the Auxilin Homolog.
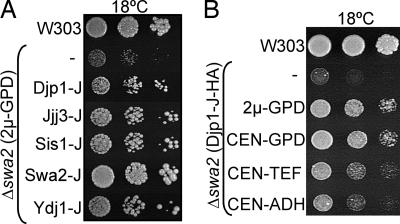
Swa2 is involved in the uncoating of clathrin-coated vesicles (CCVs) (22). Swa2 is a complex protein, having clathrin-binding domains (CBDs), a ubiquitin association domain (UBA), tetratricopeptide repeat (TPR) motifs, and a C-terminal J domain. All of the full-length and the J domain-containing constructs were transformed into Δswa2 cells. None of the full-length J protein constructs were able to rescue the cold sensitivity of Δswa2 (data not shown). However, several J domain-containing fragments, including at least one from each class, partially rescued the cold sensitivity of Δswa2 (Fig. 5A). As observed in the case of Δydj1, the level of rescue positively correlated with expression levels (Fig. 5B).
Fig. 5.
J domain-containing fragments of some cytosolic J proteins partially rescued the cold sensitivity of Δswa2. Serial dilutions of Δswa2 cells containing the indicated plasmids were spotted on minimal media, and plates were incubated at 18°C for 10 days. Shown are wild-type control strain (W303) and Δswa2 harboring empty vector (−). (A) 2μ-GPD plasmids expressing indicated J domain fragments. (B) Djp1-J HA fragment expressed from indicated promoters of variable strengths were transformed in Δswa2.
Discussion
We began a broad analysis of cytosolic J proteins to better understand the degree of functional diversity that exists within a single cellular compartment. As discussed below, a complex picture emerges, with the cytosolic compartment having a mixture of “general” and “specialty” J proteins.
The General J Protein, Ydj1.
Surprisingly, the severe growth defects caused by the absence of the most abundant cytosolic J protein, Ydj1, could be substantially alleviated by the expression of J domains from a variety of J proteins. This sufficiency of the J domains to substitute for Ydj1 implies that the core ability of a J protein, the ability to stimulate the ATPase activity of its partner Hsp70, is sufficient for many cellular processes. Such rescue required neither a J domain from a particular class of J proteins nor expression at levels much higher than normal Ydj1 levels. Thus, even though full-length Ydj1 is competent to bind and deliver client proteins to Ssa Hsp70 and is a requirement for in vitro refolding of luciferase (8), we propose that this activity is not required for many of its in vivo functions.
However, we do not mean to imply that client protein binding is never critical for the function of “general” J proteins. First, a J domain, even when expressed at the level at which Ydj1 is normally expressed, does not fully rescue the growth defect caused by the absence of Ydj1. Second, our laboratory previously reported that the C-terminal substrate-binding domain of either Ydj1 or Sis1 is required for robust growth of yeast cells (23). It is likely that certain client proteins require interaction with a J protein for efficient presentation to Ssa but that binding to either Ydj1 or Sis1 often suffices in these cases. It is likely that other cytosolic J proteins functionally overlap with Ydj1 in vivo, as well. For example, both Ydj1 and class II Hlj1 participate in the degradation in the cytosol of proteins extracted from the endoplasmic reticulum (ER) (10).
The G/F Region of Class I and II J Proteins.
The ability of the Ydj1 J domain, without any of the amino acids normally flanking it being present, to substantially rescue the growth defect of cells lacking Ydj1 established the sufficiency of the J domain itself. However, this rescue also raised a question concerning the importance of the G/F region, a defining feature of class I and II J proteins that lies adjacent to the J domain. Its functional significance has been an ongoing point of debate, in part due to the report that the growth defects of Δydj1 could be rescued by a fragment containing the J domain plus the G/F region, but not by a construct encoding only the J domain of Ydj1 (24). However, the results reported here indicate that the need for such sequences is nonspecific, but rather may be needed for structural stability.
Nevertheless, the sufficiency of the J domain does not mean that, in the context of full-length proteins, the region rich in glycines never serves an important purpose. In analyses of both DnaJ of E. coli and Sis1 of S. cerevisiae, the G/F region has been shown to be functionally important. However, in these two cases, the specific sequences found to be critical were neither glycines nor phenylalanines. For example, the defects caused by the deletion of the G/F region of DnaJ of E. coli were mimicked by alterations of the D or I/V of DI/VF repeats found at the end of the G/F region (16). In the case of Sis1, alteration of a single D or N residue in a small, 12-aa “insertion segment” that distinguishes its G/F region from that of Ydj1 abolished the ability of a J domain plus G/F fragment, Sis11–121, to carry out Sis1's essential functions (25).
Consistent with the idea that it is not the glycines and phenylalanines themselves in a G/F-rich region that are functionally important, a mutant YDJ1 protein lacking the entire G/F region rescued a Δydj1 strain and full-length Ydj1 (26). Furthermore, a closer look at the amino acid sequence of J proteins found in the yeast cytosol revealed that even the presence of a G/F-rich region as the distinguishing feature of class I and II J proteins is somewhat arbitrary. For example, in reviews (3, 7), Apj1, Caj1, Djp1, Hlj1, Sis1, Xdj1, and Ydj1 were classified as class I or II proteins, meaning that, by definition, a G/F region was present. The predominance of glycines and phenylalanines in the 33 aa C-terminal to the J domain of Ydj1 is obvious, with 15 glycines and seven phenylalanines. However, the presence of a G/F region adjacent to the J domain of Apj1 and Djp1 is not as clear. Apj1 has two glycines and three phenylalanines; Djp1 has six glycines and four phenylalanines.
Specialist J Proteins.
As a counterpoint to the generality of Ydj1 function, the degree of specificity among J proteins is also illuminated by the results of our study. Seven of the 13 J protein deletion strains analyzed had assayable phenotypes. Of these, the phenotypes of four: Δcwc23, Δsis1, Δjjj1, and Δjjj3 could only be rescued by expression of the deleted genes, suggesting a high degree of specificity. What is the basis of such specificity? Although there is still much to be learned, the data presented here and elsewhere provide some clues. Tethering to a particular location within a particular cellular compartment may be important. For example, Δzuo1 is not listed above, because its phenotype could be partially rescued by one other J protein, Jjj1. Both Jjj1 and Zuo1 stably associate with ribosomes (21, 27). In cases in which tethering is important, an extremely high local concentration of the J protein may be required to recruit an Hsp70 partner to a particular site of action, a criteria, which in the case of Zuo1, would only be met by the other ribosome-associated J protein, Jjj1.
Interestingly, although both Jjj1 and Zuo1 are ribosome-associated, and Jjj1 can partially substitute for Zuo1, these two J proteins partner with different Hsp70s, Zuo1 with Ssb (27) and Jjj1 with Ssa (21). This ability of Ssa to function with an alternative J protein and substitute for Ssb is reminiscent of the ability of the human Zuo1 ortholog, Mpp11, to substitute for Zuo1 (28). In doing so, Mpp11 partners with Ssa, consistent with Ssb being present only in fungi and with Ssa orthologs being found in all eukaryotes. This ability also underscores the specificity that lies within the J protein group of proteins that, at least in many cases, eclipses the specificity of Hsp70s. Indeed, in the case of the yeast cytosol, both comparison of J domain sequences and functional information is consistent with Zuo1 being the only J protein partner of Ssb, with the other 12 working with Ssa.
One effect of the sequestering of a J protein, either by localization to a particular site or by interaction with a particular client protein, may be its inability to substitute in vivo for another J protein, even if it is otherwise functionally competent. This idea is consistent with the observation that, unlike full-length Jjj3, a Jjj3C156Y construct, which is unable to perform Jjj3's specialized function in DPH biosynthesis, was able to rescue Δydj1 as the Jjj3 J domain construct. Although, the role of Jjj3's zinc finger in DPH synthesis is not known, one can speculate that it might either bind to a client protein or to a protein complex involved in DPH biosynthesis. Therefore, in cases such as Jjj3, the sequestration might prevent functional overlap.
However, increased expression of full-length J proteins such as Caj1, but not their J domains, is deleterious to cells. In fact, the J domain fragment of Caj1 rescued the growth defects caused by the absence of Ydj1 quite effectively. At this point, we can only speculate about the cause of the toxicity. Perhaps these J proteins are deleterious because they bind client proteins “inappropriately” or because they recruit Hsp70 to specific sites, depleting the pool available to function with specialized J proteins.
Swa2 presents an unusual case and, on the basis of our data, cannot be easily classified as a general or specialty J protein. It is structurally complex and highly specialized for uncoating of CCVs (29). However, the cold sensitivity of Δswa2 could be rescued by a number of J domain-containing constructs. Our result is consistent with the ability of a fragment containing the J domain and the adjacent tetratricopeptide repeat (TPR) motifs of Swa2 itself to partially complement the defect of Δswa2 in α-factor processing (30), even though it cannot bind clathrin. The simplest explanation is that such specialization, that is direct binding to clathrin, is not absolutely critical under the conditions tested. It should also be noted that Swa2 is present in <1,000 molecules per cell, whereas even ≈100,000 molecules per cell of a J domain does not allow wild-type growth of Δswa2 cells. Similarly, the apparent importance of the substrate-binding domain of Ydj1 at higher temperatures may be due to the higher demand for refolding of partially denatured proteins.
Conclusions
In summary, our results suggest that important general functions of J proteins can be carried out by J domains, indicating only a requirement for stimulation of Hsp70s' ATPase activity. The specificity of J proteins is largely governed by regions outside the J domain. Mechanistically understanding the basis of specificity of individual J protein in the yeast cytosol demands further study, but substrate specificity and sequestration to particular sites within a cellular compartment likely play significant roles. However, it is also possible that subtle but important alterations in the cycle of binding and release of Hsp70s from particular client proteins may be important as well.
Materials and Methods
Genetic Methods.
Knockout strains were constructed in the W303 genetic background by first swapping the KanMX cassette in the respective deletion strains from the knockout library collection (Open Biosystems, Huntsville, AL) (31) with LEU2 by using a linearized marker-swap plasmid (32). The disrupted gene:marker cassette was then PCR amplified by using specific flanking primers and then used for one-step disruption in W303. In the case of the DJP1 deletion, a disruption cassette plasmid was constructed that included the 5′ UTR-LEU2-3′ UTR in pBluescript (Stratagene, La Jolla, CA) by using standard protocols (33) and transformed directly into W303. Δjjj1 (21), Δsis1 (34), and Δzuo1 (20) strains were previously described. Δcwc23 and Δhlj1 strains [from P. Ahlquist (University of Wisconsin, Madison) and J. Brodsky (University of Pittsburgh, Pittsburgh, PA), respectively] were back-crossed six times with W303. In vivo diphthamide biosynthesis was monitored by scoring viability of yeast cells upon conditional expression of a galactose-inducible diphtheria toxin (GAL-DT) plasmid pLMY101 (35).
Construction of J Protein Overexpression Plasmids.
ORFs corresponding to the full-length and J domain-containing fragments (see legend to Fig. 1) of all of the cytosolic J proteins were PCR amplified by using appropriate gene-specific primers and were cloned into 2μ- or centromere (CEN)-based plasmids under different promoters (36). For regulated expression of Ydj1, the complete ORF was cloned in a tetracycline-repressible vector pCM184 (37). Selected J proteins were HA-tagged by in-frame cloning of DNA encoding a 3xHA tag after the insertion of a NotI site by QuikChange PCR (Stratagene) before the stop codon in each coding sequence. The 2μGPD-YJ Jjj1 chimera was constructed by in-frame fusion of the DNA encoding amino acids 1–60 of Ydj1 with amino acids 61–128 of Jjj1 protein. For making 2μGPD-YJ R-HA, a 32-aa sequence unrelated to the J proteins (LPPWWQQLALAASLVPLAWLSHQKHCPGLNLS), followed by a 3xHA tag, was inserted after codon 63 of Ydj1 by PCR sewing using overlapping primers. This resulted into a 128-aa protein fragment containing the J domain of Ydj1, with the other segment having no relationship to a J protein. The 2μGPD-YJ Jjj3 chimera was constructed by in-frame fusion of amino acids 1–63 of Ydj1 to amino acids 70–172 of Jjj3 by PCR sewing.
Other Methods.
Total proteins were isolated by treating cells with 0.1 N NaOH and resuspended in SDS sample buffer (62.5 mM Tris·HCl, pH 6.8, 5% glycerol, 2% SDS, 2% β-mercaptoethanol, and 0.01% bromophenol blue). Protein was detected as previously described by using anti-HA mouse 12CA5 (Roche Biochemicals, Indianapolis, IN) and anti-Ydj1 rabbit antibodies (34). Quantification was done with ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Supplementary Material
Acknowledgments
We thank J. Brodsky and P. Ahlquist for yeast strains; R. J. Collier (Harvard Medical School, Boston, MA) for plasmid pLMY101; and A. Meyer, R. Aron, P. Yang, A. Andrew, and J. Marszalek for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM31107 (to E.A.C.).
Abbreviations
- CCV
clathrin-coated vesicle
- DPH
diphthamide
- DT
diphtheria toxin
- G/F
glycine/phenylalanine
- GPD
glyceraldehyde 3-phosphate dehydrogenase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702357104/DC1.
References
- 1.Hartl F, Hayer-Hartl M. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Weissman JS, Horwich A. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Cheetham ME, Caplan AJ. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig EA, Huang P, Aron R, Andrew A. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 5.Craig EA, Huang P. In: Protein Folding Handbook. Buchner J, Kiefhaber T, editors. Vol 4. Germany: Wiley-VCH, Weinheim; 2005. pp. 490–515. [Google Scholar]
- 6.Craig EA, Ziegelhoffer T, Nelson J, Laloraya S, Halladay J. Cold Spring Harbor Symposia on Quantitative Biology: Protein Kinesis: The Dynamics of Protein Trafficking and Stability. Vol XV. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1995. pp. 441–449. [Google Scholar]
- 7.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z, Cyr DM. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 9.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 10.Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 12.Caplan AJ, Douglas MG. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caplan AJ, Cyr DM, Douglas MG. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- 14.Atencio DP, Yaffe MP. Mol Cell Biol. 1992;12:283–291. doi: 10.1128/mcb.12.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker J, Walter W, Yan W, Craig EA. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cajo GC, Horne BE, Kelley WL, Schwager F, Georgopoulos C, Genevaux P. J Biol Chem. 2006;281:12436–12444. doi: 10.1074/jbc.M511192200. [DOI] [PubMed] [Google Scholar]
- 17.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Mol Cell Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong T, Arndt KT. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- 19.Sondheimer N, Lopez N, Craig EA, Lindquist S. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA. Proc Natl Acad Sci USA. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pishvaee B, Costaguta G, Yeung BG, Ryazantsev S, Greener T, Greene LE, Eisenberg E, McCaffery JM, Payne GS. Nat Cell Biol. 2000;2:958–963. doi: 10.1038/35046619. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JL, Craig EA. J Cell Biol. 2001;152:851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JL, Craig EA. Mol Cell Biol. 2000;20:3027–3036. doi: 10.1128/mcb.20.9.3027-3036.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez N, Aron R, Craig EA. Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aron R, Lopez N, Walter W, Craig EA, Johnson J. Genetics. 2005;169:1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 28.Hundley HA, Walter W, Bairstow S, Craig EA. Science. 2005;308:1032–1034. doi: 10.1126/science.1109247. [DOI] [PubMed] [Google Scholar]
- 29.Lemmon SK. Curr Biol. 2001;11:R49–R52. doi: 10.1016/s0960-9822(01)00010-0. [DOI] [PubMed] [Google Scholar]
- 30.Xiao J, Kim LS, Graham TR. Mol Biol Cell. 2006;17:3281–3290. doi: 10.1091/mbc.E06-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 32.Voth WP, Jiang YW, Stillman DJ. Yeast. 2003;20:985–993. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook JE, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 34.Yan W, Craig EA. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattheakis LC, Shen WH, Collier RJ. Mol Cell Biol. 1992;12:4026–4037. doi: 10.1128/mcb.12.9.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 37.Gari E, Piedrafita L, Aldea M, Herrero E. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.