Abstract
Cryptochromes are blue light receptors that regulate photomorphogenesis in plants and the circadian clock in animals and plants. Arabidopsis cryptochrome 2 (CRY2) mediates blue light inhibition of hypocotyl elongation and photoperiodic control of floral initiation. CRY2 undergoes blue light-induced phosphorylation, which was hypothesized to be associated with CRY2 photoactivation. To further investigate how light activates CRY2, we analyzed the physiological activities and phosphorylation of various CRY2 fusion proteins in transgenic plants. Our results showed that an 80-residue motif, referred to as NC80, was sufficient to confer the physiological function of CRY2. The GUS-NC80 fusion protein expressed in transgenic plants is constitutively active but unphosphorylated, suggesting that the blue light-induced CRY2 phosphorylation causes a conformational change to derepress the NC80 motif. Consistent with this hypothesis, the CRY2 C-terminal tail was found to be required for the blue light-induced CRY2 phosphorylation but not for the CRY2 activity. We propose that the PHR domain and the C-terminal tail of the unphosphorylated CRY2 form a “closed” conformation to suppress the NC80 motif in the absence of light. In response to blue light, the C-terminal tail of CRY2 is phosphorylated and electrostatically repelled from the surface of the PHR domain to form an “open” conformation, resulting in derepression of the NC80 motif and signal transduction to trigger photomorphogenic responses.
Keywords: blue light, cryptochrome
Cryptochromes are photolyase-like photoreceptors that mediate blue light regulation of development and the circadian clock (1–3). Arabidopsis cryptochrome 1 (CRY1) and cryptochrome 2 (CRY2) mediate primarily blue light inhibition of hypocotyl elongation and photoperiodic promotion of floral initiation, respectively (4, 5). CRY1 and CRY2 also have overlapping functions, because the cry1 mutant shows delayed flowering time under certain light or temperature conditions (6, 7), whereas the cry2 mutant exhibits reduced inhibition of hypocotyl elongation under low fluence rates of blue light (8). The molecular mechanisms underlying photoactivation of cryptochromes remain poorly understood, although it has been proposed that blue light activates cryptochromes in plants by changing their redox status, protein phosphorylation, and/or conformation to activate the photoreceptors (9–12).
Cryptochromes possess two domains, the N-terminal PHR (photolyase related) domain of ≈500 residues and a C-terminal extension of various lengths (2, 13). The PHR domain shares sequence similarity to photolyases, and it acts as the chromophore-binding domain that noncovalently binds flavin and pterin (14, 15). In addition, PHR is also involved in intra- and intermolecular protein–protein interactions (16–20). The C-terminal domain of cryptochrome is involved in functions such as nuclear localization, protein stability, posttranslational modification, and protein–protein interactions (1–3).
Photoreceptors are commonly known to undergo light-induced conformational changes (21), but how cryptochromes change their conformation in response to light remains unclear. Based on a study of GUS-CCT fusion proteins, which contain the reporter enzyme GUS (β-glucuronidase) and the C-terminal domain of Arabidopsis CRY1 or CRY2, it has been proposed that the C-terminal domain of Arabidopsis cryptochromes acts as the effector domain for the photoreceptor in mediating physiological responses (9). Transgenic plants expressing GUS-CCT1 or GUS-CCT2 showed constitutive photomorphogenic phenotypes such as suppressed hypocotyl elongation and expanded cotyledons, resembling the phenotype of the cop (constitutive photomorphogenic) or det (deetiolated) mutants (22, 23). It was proposed that the C-terminal domain of Arabidopsis cryptochromes interacts with the E3 ubiquitin ligase COP1 to trigger light signal transduction (24, 25). It has also been recently shown that the PHR domain of CRY1 can interact with not only the C-terminal domain of CRY1 intramolecularly but also the PHR domain of CRY1 intermolecularly (20, 26). Moreover, the intramolecular interaction between the two domains of CRY1 may be altered by a light-dependent conformation change (26). These results provide compelling evidence supporting a blue light-dependent conformation change in Arabidopsis CRY1.
Arabidopsis CRY1 and CRY2 undergo blue light-induced phosphorylation, which was proposed to be associated with photoactivation of the photoreceptors (10, 27, 28). Mammalian cryptochromes are also phosphoproteins, although it remains unclear whether light regulates phosphorylation of mammalian cryptochromes (29–31). In contrast to the plant cryptochromes, phosphorylation has been proposed to cause inactivation of animal cryptochromes (29). It was reported, based on a site-specific mutagenesis study, that phosphorylation of a single serine residue in the PHR domain was sufficient to inactivate mammalian cryptochromes (29). Because phosphorylation of Arabidopsis cryptochromes involves multiple serine residues, whereas a site-specific mutagenesis study has yielded no definitive conclusion concerning the role of phosphorylation (X.Y. and C.L., unpublished work), we sought a different approach to investigate the role of CRY2 phosphorylation.
We report here a study of the structure–function relationship of Arabidopsis CRY2. Based on the analyses of the linear structures, physiological activities, and phosphorylation of different fusion proteins expressed in transgenic plants, we propose that blue light-induced phosphorylation of CRY2 causes a conformational change to derepress an 80-residue region located between the N-terminal PHR domain and the C-terminal tail of CRY2 and activation of the photoreceptor.
Results
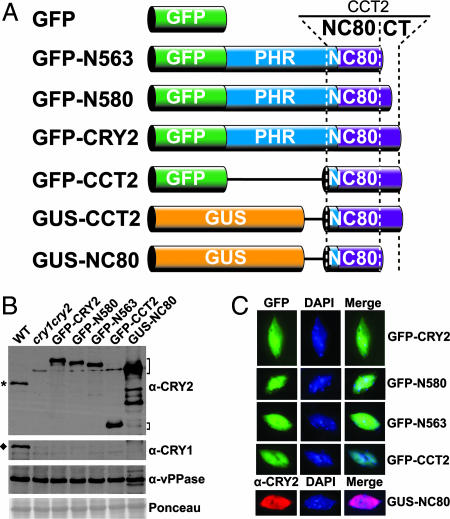
To systematically examine the relationship of the primary structure, physiological activity, and phosphorylation of CRY2, we prepared transgenic Arabidopsis lines expressing fusion proteins of GFP or GUS fused to various fragments of Arabidopsis CRY2 in the cryptochrome mutant background (32). The cry1cry2 mutant exhibits a long hypocotyl when grown in blue light and delayed flowering when grown in long-day photoperiods (32). A fusion protein that rescues either phenotypic defect of the cry1cry2 parent is regarded as physiologically active. Fig. 1 shows the linear structures (Fig. 1A), levels of protein expression (Fig. 1B), and subcellular localization (Fig. 1C) of the fusion proteins tested in this report. These CRY2 fusion proteins were all found in the nucleus, and most of them were expressed at comparable levels in the transgenic lines selected for analyses. The amino acid residues, physiological activities, and light regulation of phosphorylation of the CRY2 fusion proteins described in this report are summarized in Table 1.
Fig. 1.
The structure, expression, and nuclear localization of CRY2 fusion proteins. (A) Diagrams depicting the linear structures of CRY2 fusion proteins analyzed in this report. GUS, β-glucuronidase; PHR, photolyase-related domain; CCT2, CRY2 C-terminal domain; CT, C-terminal tail; NC80, 80 residues between the N- and C-terminal domains. (B) Immunoblot of protein extract prepared from indicated genotypes fractionated by a 10% SDS/PAGE gel and probed with anti-CRY2 (α-CRY2), anti-CRY1 (α-CRY1) antibodies. The blot was also probed with anti-vacuolar pyrophosphatase (α-vPPase) antibody and stained with Ponceau S as controls to show the relative loadings. Brackets indicate CRY2 fusion proteins, the asterisk indicates endogenous CRY2, and the diamond indicates endogenous CRY1. (C) Nuclear localization of CRY2 fusion proteins. Fluorescent microscopy images of GFP (green), nuclear stain by DAPI (blue), and immunofluorescence of GUS-NC80 (red) probed with anti-CRY2 antibody (α-CRY2) are shown.
Table 1.
A summary of the structure and activity of various CRY2 fusion proteins examined in this report
| Construct | CRY2 residues | MW, kDa | Floral promotion | Hypocotyl inhibition | Pi |
|---|---|---|---|---|---|
| GFP | 0 | 29.2 | − | − | N/A |
| GFP-CRY2 | 1–612 | 97.5 | + | + | + |
| GFP-N580 | 1–580 | 94.2 | + | + | + |
| GFP-N563 | 1–563 | 92.4 | + | + | + |
| GFP-CCT2 | 486–612 | 41.7 | − | − | “++” |
| GUS-CCT2 | 486–612 | 82.8 | ++ | ++ | ++ |
| GUS-NC80 | 486–565 | 77.6 | ++ | ++ | − |
Floral promotion: −, lack of apparent effect on flowering time; +, early flowering in LD; ++, early flowering in both LD and SD. Hypocotyl inhibition: −, long hypocotyl; +, short hypocotyl in blue light; ++, short hypocotyl in light and dark. Pi (phosphorylation of the fusion protein): −, no phosphorylation; +, blue light-induced phosphorylation; ++, constitutive phosphorylation; “++”, constitutive but low level phosphorylation. NA, not analyzed; MW, molecular mass.
The C-Terminal Tail of CRY2 Is Dispensable for the Two Major Physiological Activities of the Photoreceptor.
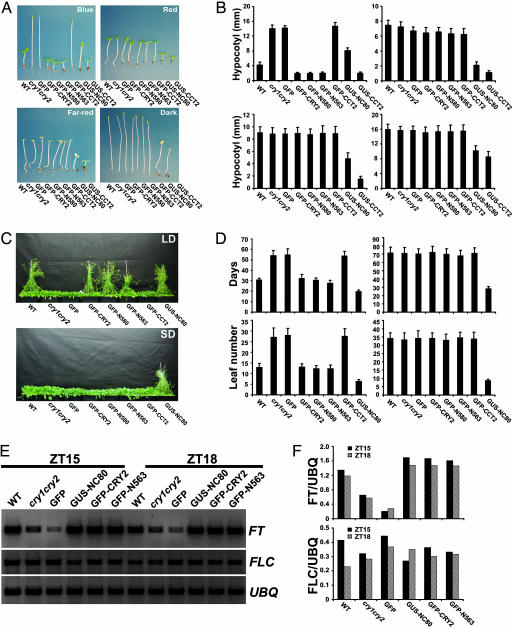
Promotion of floral initiation and inhibition of hypocotyl elongation are the two major physiological activities of CRY2. Fig. 2 shows these two phenotypes of all of the transgenic lines tested. GFP-CRY2 behaves like a wild-type CRY2; both act only in response to blue light. The transgenic seedlings expressing GFP-CRY2 completely rescued the long-hypocotyl phenotype of the cry1cry2 parent when grown in continuous blue light, and they are indistinguishable from the wild type or the cry1cry2 parent when grown in dark, red light, or far-red light (Fig. 2 A and B). As expected, the negative control of cry1cry2 expressing GFP showed no phenotypic alterations (Fig. 2 A and B), whereas the other control expressing GUS-CCT2 exhibited constitutive photomorphogenic responses (Fig. 2 A and B) (9). To our surprise, transgenic plants expressing GFP-N563 and GFP-N580 that contain different lengths of deletion of the CRY2 C-terminal tail showed similar blue light-dependent activity to that of GFP-CRY2; they exhibited short hypocotyls only in blue light (Fig. 2 A and B).
Fig. 2.
Phenotypes of transgenic plants expressing CRY2 fusion proteins. (A and B) Hypocotyl phenotype. (A) Representative 5-day-old seedlings of indicated genotypes were grown in continuous blue light (19 μmol m−2s−1), red light (10 μmol m−2s−1), far-red light (3 μmol m−2s−1), or in the dark. All transgenic lines, except GUS-CCT2, are in the cry1cry2 background. (B) The means of hypocotyl lengths of seedlings (representatives shown in A) and the standard deviations (n ≥ 20) are shown. Light conditions in B are in the same order as in A. (C and D) Flowering-time phenotype. (C) Indicated genotypes were grown in LD (16-h light/8-h dark) for 43 days or in SD (8-h light/16-h dark) for 55 days when the pictures were taken. (D) The flowering times measured as days to flower and the number of rosette leaves at the day floral buds became visible, and the standard deviations (n ≥ 20) are shown. LD (Left), SD (Right). (E and F) RT-PCR showing the level of mRNA expression of FLC, FT, and UBQ in plants of indicated genotypes grown in LD (16-h light/8-h dark). Samples were harvested at 15 (ZT15) and 18 (ZT18) hours after light was on. The signal of FT and FLC shown in E were digitized and normalized against UBQ (F).
The GFP-CRY2, GFP-N563, and GFP-N580 fusion proteins also rescued the late-flowering phenotype of the cry1cry2 parent grown in long-day photoperiods, and they do not seem to affect the flowering time in plants grown in short-day photoperiods (Fig. 2 C and D). As expected, the level of FT mRNA is noticeably lower in the cry1cry2 mutant than in the wild type (33, 34), but it returned to normal in the transgenic GFP-CRY2 or GFP-N563 plants (Fig. 2 E and F), suggesting that the GFP-CRY2 and GFP-N563 fusion proteins promote floral initiation by the same mechanism as that of the endogenous CRY2. Because the transgenic plants expressing similar levels of GFP-CRY2, GFP-N563, or GFP-N580 showed similar extents of phenotypic changes (Figs. 1B and 2 A–D), we concluded that the three CRY2 fusion proteins have comparable specific activities and that the CRY2 C-terminal tail of 49 residues (residues 564–612) is not required for the physiological activity of CRY2.
Homodimerization of CRY2 Is Important for CRY2 Function.
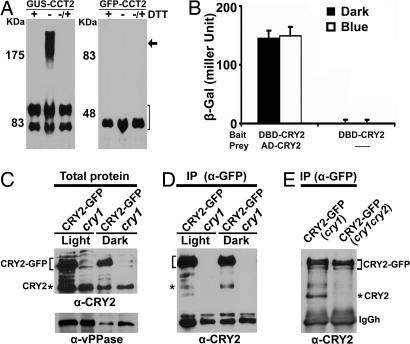
Another unexpected result of our experiment was that GFP-CCT2 is physiologically inactive, because it failed to rescue the cry1cry2 mutant (Fig. 2 A and B). GFP-CCT2 contains the identical CRY2 sequence (residues 486–612) as that of GUS-CCT2, which is known to be active or “more active” than the endogenous CRY2 (9) (Fig. 2 A and B). We reasoned that the strong activity of GUS-CCT2 might be associated with intermolecular GUS–CCT2 interactions, because GUS is known to form oligomers in vivo (35, 36). To test this possibility, we analyzed the GUS-CCT2 and GFP-CCT2 fusion proteins extracted from plants in either reduced or oxidized conditions. In the presence of the reducing agent DTT, the GUS-CCT2 signal was detected by the anti-CRY2 antibody as two fast-migrating bands at the position close to the 83-kDa marker (Fig. 3A Left and Table 1). These two bands apparently represent the GUS-CCT2 monomers, although the lower band of the two may result from partial proteolysis during sample preparation. In the absence of DTT, the GUS-CCT2 signal was detected in three bands, a slow-migrating broad band at the position above the 175-kDa marker in addition to the two fast-migrating monomeric bands. The 175-kDa band apparently contains GUS-CCT2 dimers associated by disulfide bonds, because it disappears completely in the presence of DTT (Fig. 3A Left). In contrast to GUS-CCT2, GFP-CCT2 migrates as a single band corresponding to the mass of GFP-CCT2 monomer, regardless of the presence of DTT (Fig. 3A Right). These results suggest that GUS-CCT2 and GFP-CCT2 may exist as homodimers and monomers in vivo, respectively, which explains why GUS-CCT2, but not GFP-CCT2, is physiologically active.
Fig. 3.
Homodimerization of GUS-CCT2 and CRY2. (A) Proteins of transgenic plants expressing GUS-CCT2 or GFP-CCT2 were extracted and boiled in SDS/PAGE sample buffer in the presence (+DTT) or absence (−DTT) of DTT (100 mM). To an aliquot of the samples extracted in the absence of DTT, DTT was added after boiling, and the samples were reboiled before analysis (−/+ DTT). Samples were fractionated in 8% (Left) or 10% (Right) SDS/PAGE gels, and the immunoblots were probed with anti-CRY2. The sizes (kDa) of molecular mass markers are shown to the left of each blot. The arrow indicates the GUS-CCT2 dimer. The bracket indicates GUS-CCT2 or GFP-CCT2 monomers. (B) Yeast two-hybrid assay showing intermolecular CRY2 interaction. The β-galactosidase activities in yeast grown in liquid medium in the dark or blue light (15 μmol m−2s−1) are shown. DNA binding domain (DBD) and activation domain (AD) are fused to CRY2 as indicated. (C) Immunoblots showing CRY2-GFP and endogenous CRY2 in transgenic seedlings expressing CRY2-GFP in the cry1 background and the cry1 control. Seedlings were grown in dark (Dark) or continuous white light (Light). The antibodies used in the immunoblots are indicated at the bottom of each blot. (D) Co-IP of CRY2-GFP and the endogenous CRY2. The cry1 mutant and cry1 transgenic line expressing CRY2-GFP were grown in dark or continuous white light, the immunoprecipitation reactions using the anti-GFP antibody were fractionated by SDS/PAGE, blotted, and probed with anti-CRY2 antibody. (E) Co-IP similar to D except that the samples were prepared from 6-day-old etiolated seedlings expressing CRY2-GFP in the cry1cry2 mutant background. The bracket and asterisk indicate CRY2 fusion proteins and endogenous CRY2, respectively; IgGh indicates the IgG heavy chain.
We next examined whether dimerization is an intrinsic property of the full-length CRY2, using the yeast two-hybrid assay and coimmunoprecipitation (co-IP) assays. The result of the yeast two-hybrid assay showed that CRY2 forms homodimers in yeast cells (Fig. 3B). In the co-IP experiment, CRY2-GFP was immunoprecipitated with the anti-GFP antibody to test whether it might also pull down the endogenous CRY2 (Fig. 3 C and D). Fig. 3D shows that the anti-GFP antibody immunoprecipitated not only CRY2-GFP (≈97 kDa) but also a ≈70-kDa band in samples prepared from plants grown in dark or light. This ≈70-kDa band is apparently the endogenous CRY2, because it has the molecular mass of CRY2, and it was recognized by the anti-CRY2 antibody but not by the anti-GFP antibody (Fig. 3E and data not shown). A possibility that this ≈70-kDa band might be a partial proteolytic product of CRY2-GFP was further ruled out because it was absent in the control co-IP reaction using the sample extracted from the cry1cry2 mutant that expresses only CRY2-GFP but not the endogenous CRY2 (Fig. 3E). These results are consistent with the notion that CRY2 homodimerizes in vivo.
The intermolecular interaction between CRY2 proteins appears relatively weak or transient, because no significant amount of the CRY2 dimer was detected by gel filtration, glycerol gradient centrifugation, or native gel electrophoresis experiments (H.Y. and C.L., unpublished work). CRY1 was also reported to form homodimers by weak interactions; the physical association of the endogenous CRY1 to the epitope-tagged Myc-CRY1 was detected only after formaldehyde cross-link treatment (20). The PHR domain of CRY2 is most likely the dimerization domain, because GFP-CCT2 showed no dimerization (Fig. 3A), which is also similar to that reported for CRY1 (20). Interestingly, although CRY2-GFP can pull down the endogenous CRY2 in a co-IP reaction (Fig. 3 C–E), GFP-CRY2 failed to show such activity in repeated tests (data not shown). It is conceivable that the attachment of GFP to the N terminus of CRY2 (in GFP-CRY2) disturbed the weak or transient interactions between the PHR domains of GFP-CRY2 and the PHR domain of the endogenous CRY2, whereas attachment of GFP to the C terminus of CRY2 did not affect such interactions.
The NC80 Motif of CRY2 Has Light-Independent Physiological Activities.
It is intriguing that GFP-N563, which contains a deletion of the C-terminal tail of CRY2, and GUS-CCT2, which contains a deletion of the N-terminal PHR domain of CRY2, are both physiologically active in vivo (Figs. 1 and 2 and Table 1). The simplest interpretation of these seemingly perplexing results would be that the two fusion proteins possess a common sequence required for function. Indeed, there is a 78-residue sequence overlap between GUS-CCT2 (486–612) and GFP-N563 (1–563) (Fig. 1A). The hypothesis that this small region of CRY2 contains the active site would predict that expression of this short sequence of CRY2 should complement the cry mutations but in a blue light-independent manner. To test this possibility, we prepared transgenic lines expressing GUS-NC80 in the cry1cry2 (Fig. 2) or cry1 mutants [supporting information (SI) Fig. 5]. GUS-NC80 is composed of the marker enzyme GUS, which provides the means of dimerization, and 80-aa residues of CRY2 (residues 486 to 565), which includes all of the 78 residues found in both GUS-CCT2 and GFP-N563 (Table 1). It is referred to as NC80 because this 80-aa sequence is located between the N-terminal PHR domain and the C-terminal tail of CRY2 (Fig. 1A). The NC80 sequence contains the bipartite nuclear localization signal of CRY2 (residue 541–557) (37), and it is located in the nucleus (Fig. 1C).
Transgenic plants expressing GUS-NC80 developed hypocotyls significantly shorter than those of the cry1cry2 parent grown in not only blue light but also red light, far-red light, and in the dark (Fig. 2 A and B). Therefore, GUS-NC80 is constitutively active in deetiolation. GUS-NC80 is also active in promoting floral initiation. Transgenic expression of GUS-NC80 rescued the late-flowering phenotype of the cry1cry2 mutant (Fig. 2 C and D) and the defective expression of the FT gene (Fig. 2 E and F). We conclude that NC80 contains a major active site of CRY2 responsible for the signal transduction processes regulating both hypocotyl inhibition and floral promotion.
Blue Light-Induced Phosphorylation of CRY2 Contributes to the Conformational Change Derepressing NC80.
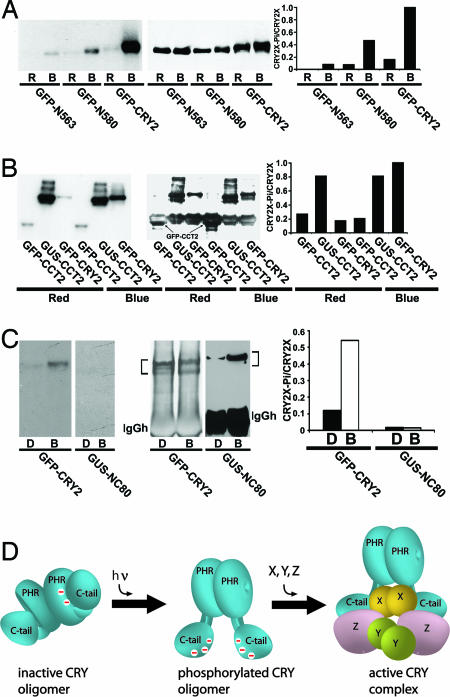
We have previously proposed that blue light-induced phosphorylation is associated with blue light-dependent activation of cryptochromes in plants (10, 27). However, cryptochrome phosphorylation might also cause desensitization or inactivation, as was reported for mammalian cryptochromes (29). To investigate this question, we analyzed the phosphorylation of CRY2 fusion proteins examined in this report (Fig. 4). Consistent with the observation that GFP-CRY2, GFP-N580, and GFP-N563 exhibited blue light-dependent physiological activity, all three fusion proteins showed blue light-induced phosphorylation (Fig. 4A). The extent of relative phosphorylation (normalized to the level of expression) of the three fusion proteins are in the order of GFP-CRY2 > GFP-N580 > GFP-N563 (Fig. 4A). Because 30% of the CRY2 C-terminal tail (14 of the 47 residues) are serines, and serines are the phosphorylated residues of Arabidopsis cryptochromes (28) (X.Y. and C.L., unpublished work), this result suggests that the CRY2 C-terminal tail is an important phosphorylation motif and that either deletion or phosphorylation of the CRY2 C-terminal tail derepress NC80. The observation that GFP-N563 showed relatively weaker phosphorylation but strong physiological activity may be explained by deletion of the CRY2 C-terminal tail reducing not only phosphorylatable residues but also the repressive effect of an unphosphorylated CRY2 C-terminal tail on NC80. On the other hand, the fact that GFP-N563 remains blue light-responsive confirms that, in addition to the C-terminal tail, the PHR domain is also involved in the suppression of NC80 (9). The constitutively active GUS-CCT2 is strongly phosphorylated; the inactive GFP-CCT2 is only weakly phosphorylated (Fig. 4B). Therefore, phosphorylation of the CRY2 C-terminal tail and resulting derepression of NC80 may both depend on CRY2 homodimerization.
Fig. 4.
In vivo phosphorylation of CRY2 fusion proteins. (A–C) Seedlings grown in fluorescent red light (A and B) or in the dark (C) for 5–6 days were excised, incubated with [32P]H3PO4 in either the initial growth condition or blue light (15 μmol m−2s−1 in A and B or 26 μmol m−2s−1 in C) for 30 (A and B) or 15 min (C). Fusion proteins were immunoprecipitated by using anti-GFP (A), anti-CRY2 or anti-GUS (B and C) antibodies, fractionated by SDS/PAGE, blotted to nitrocellulose membranes, exposed to x-ray film, and probed with the antibodies indicated. The relative phosphorylation (Right, CRY2-Pi/CRY2X) was calculated by dividing the radioactive signal of the autoradiograph (Left) with the level of respective fusion proteins detected by the immunoblot (Center). D, dark; B, blue light; R, red light. (D) A model depicting light-induced phosphorylation, conformational change, and activation of CRY2. Negative charges resulting from phosphorylation (−) and unidentified CRY2-interacting proteins (X, Y, Z) are indicated.
No phosphorylation was detected for the GUS-NC80 fusion protein (Fig. 4C). This result argues against the alternative hypothesis that phosphorylation of CRY2 might repress or inactivate NC80. It is interesting to compare the constitutively active but unphosphorylated GUS-NC80 with the constitutively active and constitutively phosphorylated GUS-CCT2 (Figs. 2 and 4B) (10). Because the only sequence difference between GUS-CCT2 and GUS-NC80 is the lack of the CRY2 C-terminal tail in GUS-NC80 (Fig. 1A), the fact that NC80 can be constitutively activated by either constitutive phosphorylation of the C-terminal tail (GUS-CCT2) or deletion of it (GUS-NC80) demonstrates that the functional significance of CRY2 phosphorylation lies primarily in the conformational changes it causes to derepress NC80 but not in the phosphorylation of NC80 per se.
Discussion
As summarized in Table 1, the CRY2 fusion proteins examined in this study can be grouped into three categories: (i) those that are phosphorylated and active only in response to blue light (GFP-CRY2, GFP-N563, and GFP-N580), (ii) constitutively phosphorylated and constitutively active (GUS-CCT2), and (iii) unphosphorylated but constitutively active (GUS-NC80). Taking into account the sequence differences among all of the CRY2 fusion proteins examined, these results can be collectively explained by the hypothesis that the blue light-dependent phosphorylation of the CRY2 holoprotein causes a conformational change to derepress the NC80 motif.
The crystal structure of the PHR domain of Arabidopsis CRY1 (CRY1-PHR) has been recently solved (38). The PHR domain of Arabidopsis CRY1 and Escherichia coli DNA photolyase (≈30% identical) have similar structural folds that are almost superimposable (38, 39). However, DNA photolyase and CRY1-PHR have quite different surface features despite their similar structural fold. For example, photolyases, but not CRY1-PHR, have a positively charged groove near the FAD-access cavity. This difference is consistent with the fact that DNA photolyase binds and repairs its DNA substrate in this groove (2, 39), whereas CRY1 lacks DNA-repairing activity (14, 15). More importantly, CRY1-PHR contains an overall negative electrostatic potential on the surface where the C-terminal domain is most likely to interface, whereas the corresponding surface areas of photolyase are either positively charged or uncharged (38). Computational modeling shows that CRY2-PHR and CRY1-PHR (≈60% identical) have very similar structures (13) (J.K. and C.L., unpublished work). Provided a similar negative surface potential of the CRY2-PHR domain, the phosphorylated C-terminal tail of CRY2 would be electrostatically repelled from its surface. The separation of the CRY2 C-terminal tail from the PHR domain would increase the solvent accessibility of the NC80 motif to cause derepression of the photoreceptor.
Taken together the above analysis, our results described in this report, and work from others support a hypothesis of how a cryptochrome may respond to light. According to this model (Fig. 4D), the PHR domain and the C-terminal tail of an unphosphorylated cryptochrome form a closed conformation in the dark to repress the NC80 motif from functioning. Upon absorption of photons, the C-terminal tail is phosphorylated and electrostatically repelled from the negatively charged PHR domain, resulting in an open conformation that derepresses the NC80 motif to trigger the physiological responses. The model shown in Fig. 4D is consistent with the involvement of proteins such as COP1 that interact with the C-terminal domain of CRY2 (24, 25). However, other proteins that interact with CRY2 at locations within or outside the NC80 motif or the C-terminal domain cannot be excluded at present. Further elucidation of the photoactivation mechanism of plant cryptochromes requires identification and study of all cryptochrome-interacting proteins.
Materials and Methods
Plant Materials.
All Arabidopsis lines studied in this report are derived from accession Col-4 (10, 32). Transgenic plants expressing CRY2 fusion proteins were prepared in the cry1, cry2, or cry1cry2 mutant background (5, 8, 32), which showed similar phenotypes, although only those in the cry1cry2 background are shown. The holocryptochrome and apocryptochrome have been referred to as cry and CRY, respectively (8), but we used CRY to represent both holocryptochrome and apocryptochrome in this report because of the difficulty to unambiguously distinguish the two in our discussion. Additional information for the preparation of transgenic lines and experimental light conditions can be found in SI Methods.
Protein Analyses.
Immunoblot, immunoprecipitation, and in planta32P labeling were as described (10, 27) with minor modifications. A commercial yeast two-hybrid (40, 41) system was used to test cryptochrome interactions. Additional details about protein analyses can be found in SI Methods.
Supplementary Material
Acknowledgments
We thank Yana Bernatavichute and Dr. Steve Jacobsen for assistance with the immunofluorescence microscopy study, Drs. D. Ehrhardt and Z. Wang (Carnegie Institute, Stanford, CA) for the pEGAD vector and anti-GFP antibody, respectively. This work is supported in part by National Institutes of Health Grant GM56265 (to C.L.), the Changjiang scholarship (to C.L.), and the 985 higher-education enhancement fund (to Hunan University). J.K., J.L., and K.T.B. were partially supported by predoctoral University of California, Los Angeles-National Science Foundation/Integrative Graduate Education and Research Traineeship Bioinformatics Training Award DGE-9987641, the University of California Mexico–U.S. (MEXUS)–El Consejo Nacional de Ciencia y TecnologÍa (CONACYT) Postdoctoral Fellowship, and the Better Opportunities for Young Scientists in Chosen Areas of Science and Technology (BOYSCAST) Award from India, respectively.
Abbreviations
- co-IP
coimmunoprecipitation
- CRY2
cryptochrome 2.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701912104/DC1.
References
- 1.Cashmore AR. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- 2.Sancar A. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 3.Lin C, Todo T. Genome Biol. 2005;6:220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad M, Cashmore AR. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Yang H, Mockler TC, Lin C. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 6.Blazquez MA, Ahn JH, Weigel D. Nat Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- 7.Bagnall DJ, King RW, Hangarter RP. Planta. 1996;200:278–280. doi: 10.1007/BF00208319. [DOI] [PubMed] [Google Scholar]
- 8.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 10.Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- 11.Giovani B, Byrdin M, Ahmad M, Brettel K. Nat Struct Biol. 2003;10:489–490. doi: 10.1038/nsb933. [DOI] [PubMed] [Google Scholar]
- 12.Partch CL, Sancar A. Photochem Photobiol. 2005;81:1291–1304. doi: 10.1562/2005-07-08-IR-607. [DOI] [PubMed] [Google Scholar]
- 13.Lin C, Shalitin D. Annu Rev Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- 14.Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR. Science. 1995;269:968–970. doi: 10.1126/science.7638620. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra K, Kim ST, Batschauer A, Dawut L, Sancar A. Biochemistry. 1995;34:6892–6899. doi: 10.1021/bi00020a037. [DOI] [PubMed] [Google Scholar]
- 16.Busza A, Emery-Le M, Rosbash M, Emery P. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- 17.Chaves I, Yagita K, Barnhoorn S, Okamura H, van der Horst GT, Tamanini F. Mol Cell Biol. 2006;26:1743–1753. doi: 10.1128/MCB.26.5.1743-1753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu H, Conte F, Green CB. Curr Biol. 2003;13:1653–1658. doi: 10.1016/j.cub.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 19.Hirayama J, Nakamura H, Ishikawa T, Kobayashi Y, Todo T. J Biol Chem. 2003;278:35620–35628. doi: 10.1074/jbc.M305028200. [DOI] [PubMed] [Google Scholar]
- 20.Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ. Plant Cell. 2005;17:1569–1584. doi: 10.1105/tpc.104.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes MG. In: Photoreceptor Evolution and Function. Holmes MG, editor. London: Academic; 1991. pp. 1–20. [Google Scholar]
- 22.Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- 23.Deng X-W, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 25.Yang HQ, Tang RH, Cashmore AR. Plant Cell. 2001;13:2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- 27.Shalitin D, Yu X, Maymon M, Mockler T, Lin C. Plant Cell. 2003;15:2421–2429. doi: 10.1105/tpc.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouly JP, Giovani B, Djamei A, Mueller M, Zeugner A, Dudkin EA, Batschauer A, Ahmad M. Eur J Biochem. 2003;270:2921–2928. doi: 10.1046/j.1432-1033.2003.03691.x. [DOI] [PubMed] [Google Scholar]
- 29.Sanada K, Harada Y, Sakai M, Todo T, Fukada Y. Genes Cells. 2004;9:697–708. doi: 10.1111/j.1356-9597.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- 30.Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. J Biol Chem. 2002;277:17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 32.Mockler TC, Guo H, Yang H, Duong H, Lin C. Development (Cambridge, UK) 1999;126:2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
- 33.Yanovsky MJ, Kay SA. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- 34.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 35.Kato A, Hayashi M, Nishimura M. Plant Cell Physiol. 1999;40:586–591. doi: 10.1093/oxfordjournals.pcp.a029581. [DOI] [PubMed] [Google Scholar]
- 36.Matsushita T, Mochizuki N, Nagatani A. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 37.Guo H, Duong H, Ma N, Lin C. Plant J. 1999;19:279–287. doi: 10.1046/j.1365-313x.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- 38.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Proc Natl Acad Sci USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park HW, Kim ST, Sancar A, Deisenhofer J. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 40.Chien CT, Bartel PL, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wittmann S, Chatel H, Fortin MG, Laliberte JF. Virology. 1997;234:84–92. doi: 10.1006/viro.1997.8634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.