Abstract
The alternative sigma factor σ54 has been implicated in diverse functions within the cells. In this study, we have constructed an rpoN mutant of Pseudomonas aeruginosa and investigated its importance as a target for antimicrobial agents, such as quinolones and carbapenems. The stationary-phase cells of the rpoN mutant displayed a survival rate approximately 15 times higher than that of the wild-type cells in the presence of quinolones and carbapenems. The stationary phase led to substantial production of pyoverdine by the P. aeruginosa rpoN mutant. Pyoverdine synthesis correlated with decreased susceptibility to antimicrobial agents. Quantitative real-time PCR revealed that stationary-phase cells of the rpoN mutant grown without an antimicrobial agent had approximately 4- to 140- and 2- to 14-fold-higher levels of transcripts of the pvdS and vqsR genes, respectively, than the wild-type strain. In the presence of an antimicrobial agent, levels of pvdS and vqsR transcripts were elevated 400- and 5-fold, respectively, in comparison to the wild-type levels. Flow cytometry assays using a green fluorescent protein reporter demonstrated increased expression of the vqsR gene in the rpoN mutant throughout growth. A pvdS mutant of P. aeruginosa, deficient in pyoverdine production, was shown to be susceptible to biapenem. These findings suggest that rpoN is involved in tolerance to antimicrobial agents in P. aeruginosa and that its tolerant effect is partly dependent on increased pyoverdine production and vqsR gene expression.
Pseudomonas aeruginosa is an opportunistic pathogen that infects immunocompromised hosts, causing infections that are especially difficult to eradicate. P. aeruginosa has evolved a mechanism to partly escape from the effects of antimicrobial agents without necessarily expressing a resistance mechanism. This mechanism has been introduced in the literature as antimicrobial tolerance. Antimicrobial tolerance can be defined as the intrinsic ability of bacteria to survive the killing effects of antimicrobial agents (23). The molecular basis of the tolerance is virtually unexplored. Under certain environmental conditions, such as an alteration in the nutritional supply, entry into the stationary phase, or high cell density, temperature, pH, or osmolarity, planktonic cells can turn on stress response genes and switch to a more tolerant phenotype (12). Stress response genes are regulated by different linked signals, such as quorum sensing, ppGpp, and poly(P) kinase. We have recently reported that increased basal levels of ppGpp under nongrowing conditions might be a signal leading to tolerance to quinolones in P. aeruginosa (25). Transcriptional regulators such as sigma factors are key elements in the bacterial adaptive responses needed for pathogenesis. For example, it has been shown that RpoS, a central regulator of the stress response, also plays a role in tolerance to quinolones and carbapenems in P. aeruginosa (11). RpoN is another important sigma factor that also appears to regulate virulence in P. aeruginosa. Work on RpoN has revealed that this sigma factor not only is important for the expression of flagella and pili (24, 26) but also has been recognized to govern a number of distinct functions, all of which seem to be important for adaptation and survival under unfavorable environmental conditions (1). Moreover, the role of RpoN in the regulation of virulence factors and its global negative control on the quorum-sensing system in P. aeruginosa have been demonstrated (2, 22). Studies performed with Escherichia coli have shown the relationship between a mutation in the rpoN gene and resistance to novobiocin, the coumarin antibiotic that inhibits DNA supercoiling by blocking the B subunit of DNA gyrase (4). The role of RpoN in the development and maintenance of tolerance to antimicrobial agents has not yet been defined. Taking into account the fact that different metabolic activities within the cells have been correlated with RpoN, we further wondered if RpoN could play a role as a novel target for antimicrobial agents in P. aeruginosa.
The data presented here demonstrate that an rpoN mutant during the stationary phase of growth encounters an iron-limited condition characterized by the secretion of the siderophore pyoverdine. We suggest that the tolerance of the rpoN mutant to antimicrobials may be connected with increased pyoverdine synthesis and with vqsR gene expression.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids that were used and constructed in this study are described in Table 1. Bacteria were grown at 37°C in Luria-Bertani (LB) broth (Difco Laboratories) unless otherwise indicated. Antibiotics were added as required at the following concentrations: for E. coli, ampicillin at 50 μg/ml, kanamycin at 25 μg/ml, gentamicin at 15 μg/ml, and tetracycline at 10 μg/ml; for P. aeruginosa, carbenicillin at 400 μg/ml, gentamicin at 200 μg/ml, tetracycline at 100 μg/ml, and sucrose at 5%. l-Glutamine at 1 mM was included in the medium for the overnight growth of PAO1 and the rpoN mutant. Where indicated, the medium was also supplemented with FeCl3 at a concentration of 100 μM.
TABLE 1.
Strains and plasmids used and constructed in this study
| Strain or plasmid | Relevant featurea | Source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Prototroph | N. Gotoh |
| DVR | rpoN::Tcr | This study |
| DVR1 | rpoN::Tcr carrying pDVR | This study |
| PGV | PAO1 carrying pGV | This study |
| RGV | rpoN::Tcr carrying pGV | This study |
| PVD | pvdS::Gmr | This study |
| E. coli | ||
| XL1-Blue | General cloning host | Stratagene, Inc. |
| S17-1 | thi endA recA hsdR with RP4-2Tc::Mu-Km::Tn7 integrated into the chromosome | 19 |
| SSC110 | rpsL (Strr) thr leu endA thi-1 lacY galK galT ara tonA tsx dam dcm supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] | Stratagene, Inc. |
| Plasmids | ||
| pGEM-T | TA cloning vector, high copy number; Apr | Promega |
| pCR2.1-TOPO | TA cloning vector, high copy number; Apr Kmr | Invitrogen |
| pACΩGm | pACYC184 derivative carrying Ω fragment; Gmr | 17 |
| pMOB3 | Kmr Cmr; 5% sucrose sensitive | 18 |
| pVqsR | pCR2.1-vqsR | This study |
| pRN | rpoN in pGEM-T | This study |
| pRN1 | rpoN::Tcr in pGEM-T | This study |
| pPVD | pvdS in pGEM-T | This study |
| pPVD1 | pvdS::Gmr in pGEM-T | This study |
| pPVD2 | pvdS::Gmr and mob in pGEM-T | This study |
| pMMB67EH | IncQ broad-host-range cloning vector, Apr Cbr | M. Tsuda |
| pDVR | pMMB67EH containing a functional rpoN gene in the opposite orientation to the tac promoter, Apr Cbr | This study |
| p67GFP | Broad-host-range vector carrying GFP; Apr Cbr | This laboratory |
| pGV | p67GFP carrying the vqsR promoter region | This study |
Apr, Gmr, Cbr, Kmr, and Tcr, resistance to ampicillin, gentamicin, carbenicillin, kanamycin, and tetracycline, respectively.
DNA analysis and manipulation.
Restriction enzymes and DNA polymerases were purchased from New England Biolabs (Beverly, MA), Toyobo (Osaka, Japan), and TaKaRa Shuzo (Kyoto, Japan) and were used under the conditions recommended by the manufacturers. Plasmid DNA was isolated using a QIAfilter plasmid maxi kit (QIAGEN, MD) or a plasmid miniprep kit according to the protocol provided by the manufacturer (Bio-Rad, CA). Chromosomal DNA was purified from P. aeruginosa PAO1 using a bacterial DNA kit (Omega Bio-Tek Inc., Doraville, GA). Treatment of DNA with enzymes, subcloning of DNA, and transformation of plasmids into E. coli and P. aeruginosa were carried out using standard methods (16). Where required, DNA fragments were isolated from agarose gels using a QIAGEN (Valencia, CA) gel extraction kit. Standard methods were used for the preparation of competent cells and for plasmid electroporation into E. coli (16). P. aeruginosa electrocompetent cells were prepared as described elsewhere (20).
Construction of the rpoN mutant.
To construct an rpoN knockout mutant, PCR amplification was used to obtain a 1.5-kb fragment of rpoN using primers 5′-ACCCGTAGTAGTGGATGGTGC-3′ and 5′-CAACGTCACACCAGTCGCTTG-3′. The amplified fragment was cloned into the pGEM-T vector, creating pRN. Next, the BssHII fragment was deleted from the rpoN gene and replaced with a 1.3-kb Tcr gene by using BssHII linkers to form pRN1. The constructed plasmid was transformed into PAO1 by electroporation, and the presence of the rpoN::Tcr allele at the proper location in the P. aeruginosa chromosome was confirmed by PCR with primers that hybridize outside and inside the rpoN gene and by DNA sequencing using the BigDye Terminator cycle sequencing ready reaction kit and an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Inc.).
Complementation of the rpoN mutant.
For the complementation experiments, a 2.5-kb EcoRI-HindIII fragment encompassing the rpoN gene was amplified and digested with EcoRI and HindIII. The generated fragment was subsequently ligated into an EcoRI-HindIII-digested broad-host-range vector, pMMB67EH, to yield pDVR. In this construct, the rpoN gene conserved its own promoter and Shine-Dalgarno sequence and was inserted in the opposite orientation with respect to the tac promoter.
Construction of the pvdS mutant.
For construction of the insertion within pvdS, primers pvdS1s (5′-TCTCCCTCCATCATTCGCAG-3′) and pvdS1a (5′-AGGACAACGCTGGGAAAGGAG-3′) were used to PCR amplify a 0.9-kb fragment encompassing the pvdS gene. The amplified fragment was inserted into the pGEM-T vector to yield pPVD. A gentamicin cassette was amplified from pACΩGm (17) as a StuI restriction fragment and cloned into the unique StuI site of the pvdS fragment, creating pPVD1. Plasmid pPVD2 was constructed by insertion of the MOB cassette from pMOB3 (18) as a NotI fragment into NotI-digested pPVD2. Biparental mating between E. coli S17-1(pPVD2) (19) and P. aeruginosa PAO1 was used to replace the wild-type gene with the mutant pvdS::Gmr allele. Double crossovers were selected on plates containing 5% sucrose and gentamicin (200 μg/ml). The resultant pvdS mutant was confirmed by PCR.
Construction of the vqsR-gfp transcriptional fusion.
For construction of the vqsR-gfp gene fusion, a 240-bp vqsR promoter region was PCR amplified from the genomic DNA of P. aeruginosa PAO1 by using primers vqsR1s (5′-CGGAATTCCGTCGAATAAACGCCAGTGCACAA-3′) and vqsR1a (5′-CGGGATCCCGCGCTTCGAGCAACTTTCCCA-3′) (underlined nucleotides represent engineered restriction enzyme sites EcoRI and BamHI, respectively). The amplified fragment was digested with EcoRI and BamHI and then ligated into the EcoRI-BamHI-digested green fluorescent protein (GFP) reporter vector p67GFP (K. Murakami, unpublished data).
Antimicrobial agents.
Ofloxacin (Sigma, St. Louis, MO), ciprofloxacin (Bayer Pharma, Germany), biapenem, and imipenem (Meiji Seika Co., Ltd., Tokyo, Japan) were used in the study.
Susceptibility testing.
The MIC and the minimal bactericidal concentration (MBC) of each agent were determined using the broth microdilution method, as previously described (10), with the following modification: the bacterial suspensions at a density of 1 × 106 cells/ml were incubated in LB broth. MICs were determined after 24 h of incubation at 37°C. The MIC was defined as the lowest concentration of the antimicrobial agent that completely inhibited the growth of the organism, as detected by the unaided eye. MBCs were measured by removing 10 μl from all wells containing no visible growth and plating the samples onto LB agar plates for further incubation at 37°C for 24 h.
Time-kill study.
For the time-kill studies, stationary-phase cells grown for 12 to 16 h and logarithmic-phase cells were used. Cells were harvested by centrifugation and resuspended in fresh LB broth before incubation with antimicrobial agents. Aliquots (0.1 ml) were taken after 0 to 12 h of incubation and plated in duplicate onto agar plates after serial dilutions to enumerate the surviving bacteria after 24 to 48 h of incubation at 37°C. Cell viability at each time point was expressed as the percentage of viable cells (CFU per milliliter) at time zero.
RNA isolation, RT-PCR, and qRT-PCR analysis.
Overnight LB broth-grown cultures were washed once and resuspended in fresh LB medium before the start of the experiment. Cells were sampled at time zero and at 1, 3, and 5 h after the resuspension in fresh medium with or without 8 μg/ml of ofloxacin, and their RNA was immediately stabilized with RNAprotect bacterial reagent (QIAGEN) and stored at −80°C. Total RNA was isolated with the RNeasy spin column (including an on-column DNase digestion step) according to the manufacturer's instructions (QIAGEN), treated with RQ1 DNase I (Promega) for 1 h at 37°C, and repurified through an RNeasy column. Approximately 650 ng of RNA was converted to cDNA. cDNA was synthesized using the SuperScript first-strand synthesis system (Invitrogen, Groningen, The Netherlands). The oligonucleotide probes for reverse transcription-PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR) were synthesized by Hokkaido System Science Co., Ltd. (Sapporo, Japan). RT-PCR and qRT-PCR were performed using primers pvdS2s (5′-AGATGTGGTCCAGGATGCGT-3′) and pvdS2a (5′-GTGTTCGAGGGTCGCGTAGT-3′), vqsR3s (5′-TTGCGGATATCGTCTCCGAA-3′) and vqsR3a (5′-TTTTCATCAGCGCGATGACC-3′), and rpsLs (5′-CGAACTATCAACCAGCTGGTG-3′) and rpsLa (5′-GCTGTGCTCTTGCAGGTTGTG-3′). As a control for RNA contamination by DNA, the PCR was performed on the same samples without first-strand synthesis.
A LightCycler (Roche Molecular Biochemicals) real-time PCR machine (software version 3.5) was used for the quantification of cDNA. For quantitative analysis of the pvdS, vqsR, or rpsL transcript by qRT-PCR, PCRs were performed using a LightCycler FastStart DNA MasterPLUS SYBR green I kit (Roche Applied Science, Mannheim, Germany) according to the specifications of the supplier.
qRT-PCRs were performed in 10-μl mixtures containing 2 μl of Master Mix, 1 μl of cDNA, and a 0.4 μM (each) forward and reverse primers. For quantitation, the pvdS and vqsR amplicons were first cloned into the pGEM-T and pCR2.1-TOPO cloning vectors, respectively; then purified recombinant plasmid DNAs containing the amplicon of interest were 10-fold serially diluted and used to generate external standard curves according to the manufacturer's instructions. For construction of the rpsL external standard curve, PAO1 genomic DNA was used. PCRs were performed in triplicate for each gene and sample. The 230-bp PAO1 rpsL and pvdS genes and a 210-bp fragment of the PAO1 vqsR gene were amplified using the following cycles: 95°C for 10 min; 35 cycles of 95°C, 54°C (rpsL and pvdS), and 52°C (vqsR) for 10 s; and 72°C for 30 s. To correct for differences in the amount of starting material, the ribosomal gene rpsL was chosen as a reference gene. Results were read with the “second derivative maximum”' algorithm of the software provided. The LightCycler software generated a standard curve by plotting “crossing cycle number” versus logarithms of the given concentrations for each control. The software calculated the concentrations of the genes studied with the aid of the standard curve.
Flow cytometric analysis.
P. aeruginosa cultures were maintained in LB broth supplemented with carbenicillin (400 μg/ml). To study growth phase-dependent vqsR expression by flow cytometry, the p67GFP shuttle vector, containing the GFP reporter under the control of the P. aeruginosa vqsR promoter region, was electroporated into P. aeruginosa PAO1 and DVR, and the transformants were designated PGV and RGV, respectively. Overnight cultures of PGV and RGV were diluted to an optical density at 595 nm (OD595) of 0.01 in LB broth supplemented with 400 μg of carbenicillin/ml and were incubated at 37°C with shaking. For each assay, the OD595 was determined at hourly intervals and the experiment was continued for 24 h. In another experiment, overnight cultures of PGV and RGV were washed, resuspended in fresh LB broth, and then supplemented with ofloxacin at a concentration of 8 μg/ml. Samples (0.5 ml) were taken after 0 to 5 h of incubation and assayed for flow cytometry. Prior to the measurement, bacterial cells were washed once in phosphate-buffered saline, resuspended in 0.5 ml of phosphate-buffered saline, and then serially diluted. A Coulter Epics XL flow cytometer (Beckman Coulter, Inc.) was used to measure the intensity of fluorescence of vqsR-gfp-producing bacteria. Fluorescence and scatter data were collected for 20,000 events, and mean fluorescence intensity was calculated. The relative fluorescent units represent the fluorescence values corrected for the background (PAO1 without gfp).
RESULTS
Susceptibility testing.
To address whether rpoN serves as a possible target for antimicrobial agents, insertional inactivation of the rpoN gene was performed in P. aeruginosa PAO1; the resulting mutant was designated DVR. The MICs and MBCs of antimicrobial agents for the study strains are summarized in Table 2. No significant difference was observed in the MICs between the wild-type strain and the rpoN mutant. The MBCs of quinolones for the rpoN mutant were slightly higher than those for the wild type. In contrast, the MBCs of carbapenems for the rpoN mutant were 8 to 16 times higher than those for the wild-type strain. The MICs and the MBCs for the pvdS mutant and the wild type were found to be almost the same, except for the MBC of imipenem, which was 4 times higher for the pvdS mutant than for the wild-type cells. The MBC is not a suitable or reliable measure of tolerance, because it is defined as the end point killing of more than 99.9% of cells; therefore, the assessment of both the rate and the extent of killing was performed using time-kill assays.
TABLE 2.
Susceptibilities of the wild type, the rpoN mutant, and the pvdS mutant to antimicrobial agents
| Antimicrobial agent | Wild type (PAO1)
|
rpoN mutant
|
pvdS mutant
|
|||
|---|---|---|---|---|---|---|
| MICa | MBCa | MIC | MBC | MIC | MBC | |
| Biapenem | 0.5 | 2 | 0.5 | 16 | 0.5 | 1 |
| Imipenem | 1 | 1 | 1 | 16 | 1 | 4 |
| Ofloxacin | 1 | 1 | 1 | 2 | 1 | 2 |
| Ciprofloxacin | 0.125 | 0.5 | 0.25 | 2 | 0.125 | 0.25 |
Expressed in micrograms per milliliter.
Impact of rpoN inactivation on tolerance to quinolones and carbapenems.
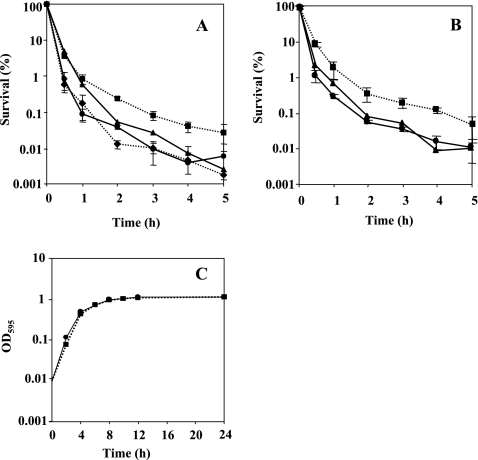
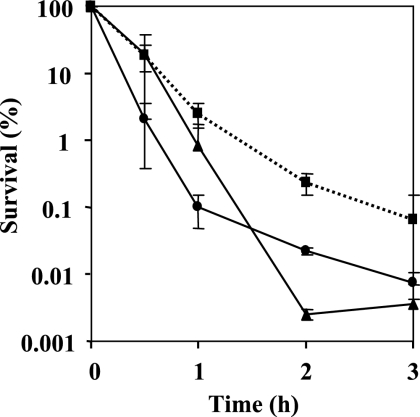
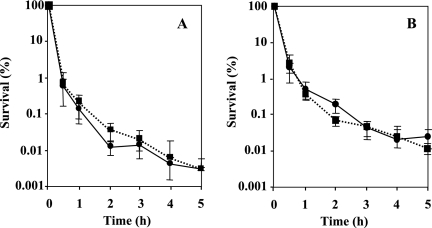
Examination of growth kinetics in LB medium supplemented with 1 mM glutamine showed that the growth rate of the mutant did not differ from the wild-type rate, indicating that the rpoN gene mutation has no effect on the growth rate under the conditions tested (Fig. 1C). For overnight growth, the wild-type and rpoN mutant cells were supplemented with 1 mM glutamine. The addition of glutamine had no effect on the susceptibility to antimicrobial agents following resuspension; therefore, glutamine was omitted from the resuspension. The results of time-kill studies in the presence of ofloxacin (8 μg/ml) and ciprofloxacin (2 μg/ml) are shown in Fig. 1A and B, respectively. The survival rate of the rpoN mutant in the presence of quinolones was 15 times higher than that of the wild type. To confirm that this phenotype was due to the loss of rpoN and not to a secondary mutation, we constructed a complementation plasmid encoding a wild-type copy of rpoN under the control of its native promoter contained on pDVR1, a pMMB67-based shuttle vector. The survival rate of the complemented strain DVR1, harboring pDVR1 (Fig. 1A and B), was as low as that of the wild type. The killing curves for biapenem (32 μg/ml) and imipenem (16 μg/ml) are shown in Fig. 2A and B, respectively. The stationary-phase cells of the rpoN mutant displayed reduced susceptibility to carbapenems; they were approximately 15 times less sensitive to biapenem and imipenem addition after 12 h than the wild-type strain. The complemented strain DVR1 restored the parental phenotype in the presence of carbapenems (Fig. 2A and B). It was apparent that the effect of killing was due to antibiotics, not to natural dying (data not shown). For growth of the log-phase culture, overnight cultures were diluted 1,000-fold in fresh medium and bacteria were cultured for ∼3 h to an OD600 of 0.25. Assessment of the sensitivity of logarithmic-phase cells of the rpoN mutant strain grown in LB to the addition of biapenem by comparing the survival rate with that of the wild-type strain also showed a clear difference. The logarithmic-phase cells were treated with biapenem at 32 μg/ml, and the killing curve is presented in Fig. 3. We observed that exposing logarithmic-phase cells of the rpoN mutant to biapenem, without glutamine supplementation, produced tolerance of biapenem addition. In contrast, when the rpoN mutant was supplemented with glutamine, tolerance was abolished. For the wild-type strain, glutamine addition had no effect on susceptibility to biapenem during the logarithmic phase. From these data, we initially concluded that the mechanism underlying tolerance in the logarithmic-phase cells of the rpoN mutant differs from that for the stationary-phase cells.
FIG. 1.
(A and B) Time-dependent killing study in the presence of 8 μg/ml of ofloxacin (A) and 2 μg/ml of ciprofloxacin (B) for stationary-grown cells of PAO1, DVR, and PVD. By taking the survival at time zero as 100%, the number of CFU was changed to a percentage. Circles on solid line, PAO1 (wild type); squares on dotted line, DVR (rpoN mutant); diamonds on dotted line, PVD (pvdS mutant). Complementation studies were performed by introducing the rpoN gene on pMMB67EH into DVR (triangles on solid line). (C) Growth curves showing culture absorbance at 595 nm plotted against time. Symbols are as explained for panels A and B. The experiment was performed in triplicate. Error bars, standard deviations. Where error bars are not shown, the standard deviation was within the size of the symbol.
FIG. 2.
Time-dependent killing study in the presence of biapenem at a concentration of 32 μg/ml (A) and imipenem at 16 μg/ml (B) for stationary-phase cells of PAO1, DVR, and PVD. Circles on solid line, PAO1 (wild type); squares on dotted line, DVR (rpoN mutant); diamonds on dotted line, PVD (pvdS mutant). Complementation studies were performed by introducing the rpoN gene on pMMB67EH into DVR (triangles on solid line). The survival of DVR supplemented with FeCl3 in the presence of carbapenems (squares on solid line) was also studied. By taking the survival at time zero as 100%, the number of CFU was changed to a percentage. Error bars, standard deviations for three determinations.
FIG. 3.
Time-dependent killing study in the presence of biapenem at 32 μg/ml for logarithmic-phase cells of PAO1 and DVR (with or without addition of 1 mM glutamine). By taking the survival at time zero as 100%, the number of CFU was changed to a percentage. Circles on solid line, PAO1 (wild type); squares on dotted line, DVR (rpoN mutant) with no glutamine addition; triangles on solid line, DVR with glutamine added. Error bars, standard deviations for three determinations.
The stationary-phase cells of the rpoN mutant trigger pyoverdine production.
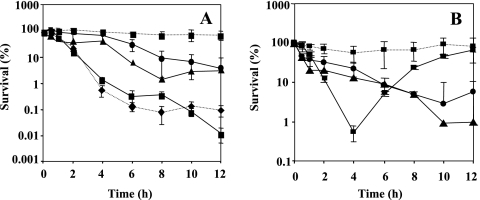
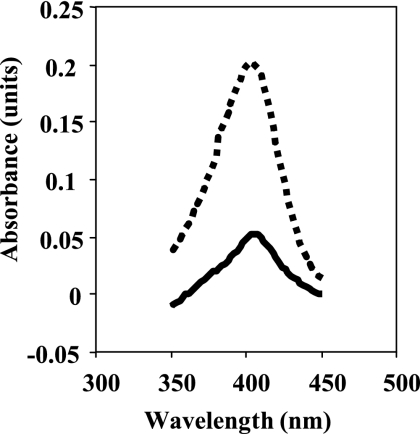
During the course of this study, we have observed that upon transition in the stationary phase and following resuspension in the fresh medium, the rpoN mutant significantly synthesized pigment. The observations raised the question of whether the pigment produced by the rpoN mutant could be a siderophore, pyoverdine, and would in a way represent iron limitation. In order to approach this problem, we allowed for two possibilities: (i) we postulated that parental-level susceptibility for the rpoN mutant could be restored in the presence of excess iron throughout growth, thus proving that iron limitation in the rpoN mutant led to increased tolerance to antimicrobials; and (ii) using a pvdS mutant, deficient in pyoverdine synthesis, we asked whether pvdS gene disruption affects susceptibility to antimicrobials. To prove the hypothesis mentioned above, we grew the rpoN mutant and the wild-type strain overnight in the presence of 100 μM FeCl3 and 1 mM glutamine. Before the start of the experiment, the cells were washed once and resuspended in fresh medium supplemented with quinolones and 100 μM FeCl3 (Fig. 4A and B). The addition of FeCl3 suppressed pyoverdine production and restored the parental phenotype in the presence of quinolones. The susceptibility of the rpoN mutant in the presence of carbapenems and FeCl3 is demonstrated in Fig. 2A and B. The addition of FeCl3 during the biapenem killing assay eliminated the tolerant effect in the rpoN mutant, and in the case of imipenem, regrowth occurred after 6 h. To further investigate the importance of pyoverdine production for the survival of the rpoN mutant in the presence of antimicrobial agents, we constructed a pvdS mutant by insertional inactivation and determined its susceptibilities to antimicrobials by killing curve assays. A pvdS mutant exhibited no change in antibiotic susceptibility relative to that of the wild type in the presence of ofloxacin (Fig. 1A). However, the survival of the pvdS mutant was affected by the presence of biapenem at 32 μg/ml (Fig. 2A). These data initially suggested that the pvdS gene is probably implicated in tolerance to biapenems but not to quinolones. To gain more insight into the pigment produced, we measured the absorbance of the filtered supernatant of the wild-type strain and the rpoN mutant after overnight growth. The absorption spectrum is shown in Fig. 5. We observed a peak at 403 nm, which is characteristic of pyoverdine (3). To determine whether siderophore production was related to the growth medium, we grew the rpoN mutant in NYB (2) and PTSB (13) media, commonly used for the growth of P. aeruginosa. We observed that in both media, the rpoN mutant showed the same growth kinetics as when it was grown in LB medium; however, we did not observe siderophore production, and spectrometric observations did not reveal an increase in the absorbance between A350 and A450 (data not shown). This observation implies that the increased pyoverdine production seen in the rpoN mutant might be only LB medium restricted or dependent. Taken together, we propose that the rpoN mutant of P. aeruginosa PAO1 secreted pyoverdine under conditions that repressed the production of pyoverdine in the wild-type strain.
FIG. 4.
Kinetics of killing by ofloxacin and ciprofloxacin in the presence of FeCl3. Numbers of CFU were determined at different time points after incubation with 100 μM FeCl3 in LB medium supplemented with ofloxacin at 8 μg/ml (A) or ciprofloxacin at 2 μg/ml (B). Circles on solid line, PAO1 (wild type); squares on dotted line, DVR (rpoN mutant). Error bars, standard deviations for three determinations.
FIG. 5.
Absorption spectrum of culture supernatants of the wild type (PAO1) (solid line) and the rpoN mutant (DVR) (dotted line) grown to the stationary phase in LB medium supplemented with 1 mM glutamine.
Transcriptional expression of the pvdS and vqsR genes.
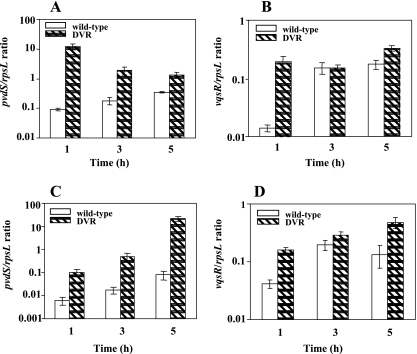
Previous studies with Vibrio harveyi (8) have demonstrated that rpoN together with luxO is implicated in siderophore production. PvdS is an alternative sigma factor controlling the expression of the genes required for synthesis of pyoverdine, an iron-chelating compound secreted by P. aeruginosa (7). Since no link between rpoN and siderophore production has been demonstrated in P. aeruginosa, our unexpected finding of increased production of pyoverdine in the stationary-phase cells of the rpoN mutant led us to begin our studies by assessing the transcript levels of pvdS following resuspension of the stationary-phase cells of the rpoN mutant and the wild type in fresh medium without or with antibiotic addition. RT-PCR was performed on total RNA (∼650 ng). qRT-PCR was used to confirm the results obtained using RT-PCR. The rpsL gene was used as an internal control to ensure that the same amount of total RNA from the wild type and the mutant strain was used. The starting quantity of cDNA from the wild type and the mutant was normalized using the rpsL gene. The results showed that without quinolone addition, at the 1-h time point, a 140-fold increase in the level of pvdS transcripts for the rpoN mutant in comparison to that for the wild type was observed. At the 3-h and 5-h time points, pvdS transcript levels remained approximately 10- and 5-fold higher, respectively, than those for the wild-type strain (Fig. 6A). To examine the contribution of antibiotic addition to the pvdS expression pattern, we performed qRT-PCR on the cDNAs obtained from the wild type and the rpoN mutant after the addition of 8 μg/ml ofloxacin. The results demonstrated a substantial increase in the level of pvdS transcripts for the rpoN mutant; at the 3-h time point, the rpoN mutant showed a 30-fold increase in pvdS transcripts compared to the wild type. Moreover, at the 5-h time point, a 400-fold increase in the pvdS/rpsL ratio was observed for the rpoN mutant (Fig. 6C). These results together indicate that antibiotic addition actually increased pvdS transcript levels for the rpoN mutant. This observation prompted us to investigate if some other genes might be upregulated by this significant increase in pvdS transcripts in the rpoN mutant. Since it was previously reported (5, 6) that VqsR controls the expression of the genes required for siderophore biosynthesis, we determined the levels of transcripts of the vqsR gene for both the wild type and the rpoN mutant. Interestingly, when we assayed vqsR transcripts in the rpoN mutant without ofloxacin addition, at the 1-h time point, we observed 15-fold-higher expression of vqsR in the rpoN mutant, which clearly correlated with increased levels of pvdS transcripts at the 1-h time point. At the 3-h time point, vqsR transcript levels decreased, reaching wild-type levels, but further increased about twofold in the rpoN mutant at the 5-h time point (Fig. 6B). We further analyzed the vqsR transcripts after the addition of ofloxacin, and the results clearly demonstrated that the transcription of the vqsR gene in the rpoN mutant was increased ∼2.5- to 5-fold in comparison to that for the wild-type strain (Fig. 6D).
FIG. 6.
Transcriptional expression of the pvdS and vqsR genes. Stationary-phase cells of the wild type (PAO1) and the rpoN mutant (DVR) were grown in the presence or absence of ofloxacin at a concentration of 8 μg/ml, and total RNA was isolated at time zero and at 1, 3, and 5 h. Shown are pvdS (A) and vqsR (B) transcripts in the absence of ofloxacin and pvdS (C) and vqsR (D) transcripts in the presence of ofloxacin. Error bars, standard deviations for three determinations.
Analysis of vqsR-gfp expression using flow cytometry.
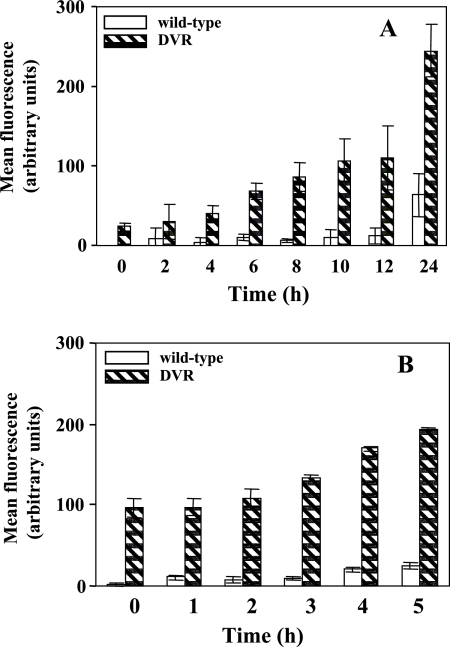
For these studies, we transformed both the wild type (PAO1) and the rpoN mutant (DVR) with the PvqsR-gfp reporter contained on an MMB67-based shuttle vector (p67GFP) and measured GFP expression over time by flow cytometry. As shown in Fig. 7A, in the rpoN mutant, the expression of vqsR-gfp increased about 10-fold at the 24-h time point. Despite the lower expression, an increase in vqsR-gfp expression at the stationary phase was also observed for the wild-type strain. When we measured the expression of vqsR-gfp in the stationary-phase cultures in the presence of 8 μg/ml of ofloxacin, the rpoN mutant, compared to the parental strain PAO1, exhibited a notable increase in vqsR-gfp expression (Fig. 7B). At time zero, the rpoN mutant displayed an approximately 45-fold increase in the expression of vqsR-gfp in comparison to the wild-type strain. Five hours after the addition of ofloxacin, the expression of vqsR-gfp in the rpoN mutant increased about eightfold relative to that in the wild-type strain. The results presented suggest that in P. aeruginosa a regulatory link between rpoN, pvdS, and vqsR exists.
FIG. 7.
Expression of the vqsR-gfp transcriptional fusion. Samples from the wild type (PAO1) and the rpoN mutant (DVR) were transformed with the indicated reporter construct, p67GFP, and analyzed by flow cytometry. vqsR-gfp expression was analyzed throughout growth (A) and in the stationary phase in the presence of 8 μg/ml of ofloxacin (B). Mean fluorescence intensities for vqsR-gfp were plotted against time on the graphs. Each bar represents the average of three experiments.
DISCUSSION
In this study, we have shown that insertional inactivation of the rpoN gene of P. aeruginosa affects the activities of quinolones and carbapenems, leading to decreased susceptibility to these antimicrobial agents. The results presented suggest that the rpoN mutant, depending on the growth phase and the nutritional balance in its environment, allows different triggers for survival under stress conditions such as antibiotic addition. Studies done with Pseudomonas putida reveal that while RpoN does not play a key role in survival under various nutritional and environmental stresses, the rpoN mutant maintained a significantly higher viability than the wild-type strain when exposed to stress conditions, such as oxidative damage and a hyperosmotic environment (1). In support of the involvement of RpoN in the antibiotic stress response in P. aeruginosa, we offer the following arguments.
The transition to stationary phase and prolonged stationary phase led to increased pyoverdine production in the rpoN mutant. Using qRT-PCR, we followed the accumulation pattern of pyoverdine by quantifying pvdS transcripts in the absence or presence of an antimicrobial agent. During growth in the antibiotic-free medium, pvdS transcript levels in the rpoN mutant were 5 to 140 times higher than those in the wild type. The addition of ofloxacin significantly increased pyoverdine synthesis in the rpoN mutant. At first glance, our findings were surprising in light of the role of RpoN in pyoverdine synthesis during the stationary phase. In P. aeruginosa, pyoverdine synthesis is regulated by PvdS, an alternative sigma factor. The role of pyoverdine extends beyond its role in chelating and transporting iron in the cells during iron limitation; it also serves as a signaling molecule representing a mechanism by which P. aeruginosa cells may respond to increased cell density (7). Moreover, pyoverdine expression positively regulates the expression of genes for synthesis of exotoxin A, PrpL protease, and pyoverdine itself (7). Several pieces of evidence are consistent with the notion that increased cell density might control pyoverdine synthesis (7, 21). The involvement of RpoN in the regulation of quorum sensing (2, 22) and our observations of pyoverdine production solely in the stationary phase led us to propose that a high cell density and an altered nutrient supply may induce a certain metabolic imbalance in the rpoN mutant, followed by upregulation of the stationary-phase survival genes. On account of its role as a quorum-sensing regulator and its impact on iron homeostasis in P. aeruginosa (5), it was possible that VqsR actually regulated pvdS expression and played a role in antibiotic stress response in the rpoN mutant. While the real-time PCR and flow cytometry studies presented here clearly demonstrated VqsR upregulation, we do not yet know if the siderophore trigger seen in the rpoN mutant is regulated by VqsR, quorum sensing, or some other, unknown pathway. It is unlikely that the increased pyoverdine production presented in this study is due to upregulation of VqsR alone, because the levels of vqsR transcripts seem to be unaltered in the rpoN mutant with FeCl3 addition (data not shown), suggesting that additional levels of regulation are probably implicated. In addition, the transcription pattern of pvdS during growth in the antibiotic-free medium parallels that seen for vqsR and provides for possible pvdS upregulation by VqsR. In contrast, antibiotic addition demonstrated that pvdS upregulation is probably mediated by some other antibiotic stress-regulated gene and is not only VqsR dependent. At the level of regulation, an rpoN vqsR double mutant would probably shed further light on the requirement and role of VqsR in pyoverdine synthesis and survival during antibiotic stress. The upregulation of quorum-sensing-related genes such as vqsR in the rpoN mutant suggests that the rpoN gene downregulates quorum-sensing gene expression in the wild-type strain, either directly or indirectly.
Another interest in this work was to assess the importance of pyoverdine in the antimicrobial stress response of the rpoN mutant by constructing a pyoverdine-deficient mutant of P. aeruginosa PAO1. Killing curve assays suggested that the pvdS gene is involved in the tolerance of stationary-phase cells to biapenem. At this point, it is worth considering that the mexAB-oprM operon has been implicated in the uptake of pyoverdine and also in the transport of certain β-lactam antibiotics (14), suggesting that a possible selectivity for uptake between pyoverdine and carbapenems might exist, thus explaining the partially tolerant phenotype of the rpoN mutant with imipenem and FeCl3. While the wild-type susceptibility of the pvdS mutant to ofloxacin complicates the explanation of increased survival of the rpoN mutant by overproduction of pyoverdine, it also suggests that pyoverdine probably requires another regulator for direct interaction with antimicrobials, such as quinolones. Nutrient limitation (absence of glutamine) in the medium allowed log-phase cells to overcome the action of biapenem. We hypothesized that the physiological adaptation of the logarithmic-phase cells of the rpoN mutant to environmental conditions could be attributed to ppGpp. In support of our observations, Powell and Court (15) proposed that ppGpp controls glutamine metabolism in cells lacking adequate nitrogen metabolism. Recent studies by Merrell et al. (9) demonstrated for Helicobacter pylori a strong level of coregulation of genes involved in nitrogen metabolism and iron starvation. The loss of σ54 activity leads to diminished glutamine metabolism; therefore, we presume that this selectivity in nitrogen usage may in some instances affect the uptake of iron in the cell or that some other pathways for iron uptake may not be available. The data presented here suggest that the link between pyoverdine production and vqsR gene expression is a participant, but not the only one, in rendering the rpoN mutant tolerant to antimicrobials. While the mechanism behind the observed pyoverdine and its role in the antibiotic stress response is obviously complex, it seems that iron alters the outcome of the P. aeruginosa response to antimicrobial agents. Further studies will be aimed at answering the questions that lie in the background of the tolerance to antimicrobial agents seen in the rpoN mutant.
Acknowledgments
We thank Herbert P. Schweizer for providing plasmids pACΩGm and pMOB3 and M. Tsuda for providing plasmid pMMB67EH.
This work was supported by a grant-in-aid for scientific research (no. 17591914) to T.O. from the Japan Society for Promotion of Science.
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Cases, I., and V. de Lorenzo. 2001. The limits to genomic predictions: role of σN in environmental stress survival of Pseudomonas putida. FEMS Microbiol. Ecol. 35:217-221. [DOI] [PubMed] [Google Scholar]
- 2.Heurlier, K., V. Denervaud, G. Pessi, C. Reimmann, and D. Haas. 2003. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:2227-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hohnadel, D., D. Haas, and J.-M. Meyer. 1986. Mapping of mutations affecting pyoverdine production in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 36:195-199. [Google Scholar]
- 4.Jovanovic, M., M. Lilic, R. Janjusevic, G. Jovanovic, and D. J. Savic. 1999. tRNA synthetase mutants of Escherichia coli K-12 are resistant to the gyrase inhibitor novobiocin. J. Bacteriol. 181:2979-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juhas. M., L. Wiehlmann, B. Huber, D. Jordan, J. Lauber, P. Salunkhe, A. S. Limpert, F. von Gotz, I. Steinmetz, L. Eberl, and B. Tümmler. 2004. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology 150:831-841. [DOI] [PubMed] [Google Scholar]
- 6.Juhas, M., L. Wiehlmann, P. Salunkhe, J. Lauber, J. Buer, and B. Tümmler. 2005. GeneChip expression analysis of the VqsR regulon of Pseudomonas aeruginosa TB. FEMS Microbiol. Lett. 242:287-295. [DOI] [PubMed] [Google Scholar]
- 7.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and σ54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 9.Merrell, D. S., L. J. Thompson, C. C. Kim, H. Mitchell, L. S. Tompkins, A. Lee, and S. Falkow. 2003. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect. Immun. 71:6510-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyake, Y., S. Fujiwara, T. Usui, and H. Suginaka. 1992. Simple method for measuring the antibiotic concentration required to kill adherent bacteria. Chemotherapy 38:286-290. [DOI] [PubMed] [Google Scholar]
- 11.Murakami, K., T. Ono, D. Viducic, S. Kayama, M. Mori, K. Hirota, K. Nemoto, and Y. Miyake. 2005. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol. Lett. 242:161-167. [DOI] [PubMed] [Google Scholar]
- 12.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 13.Ohman, E. D., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 15.Powell, B. S., and C. L. Court. 1998. Control of ftsZ expression, cell division, and glutamine metabolism in Luria-Bertani medium by the alarmone ppGpp in Escherichia coli. J. Bacteriol. 180:1053-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Schweizer, H. P. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 18.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 19.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 20.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stintzi, A., K. Evans, J.-M. Meyer, and K. Poole. 1998. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol. Lett. 166:341-345. [DOI] [PubMed] [Google Scholar]
- 22.Thompson, L. S., J. S. Webb, S. A. Rice, and S. Kjelleberg. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220:187-195. [DOI] [PubMed] [Google Scholar]
- 23.Tomasz, A., A. Albino, and E. Zanati. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138-140. [DOI] [PubMed] [Google Scholar]
- 24.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viducic, D., T. Ono, K. Murakami, H. Susilowati, S. Kayama, K. Hirota, and Y. Miyake. 2006. Functional analysis of spoT, relA and dksA genes on quinolone tolerance in Pseudomonas aeruginosa under nongrowing condition. Microbiol. Immunol. 50:349-357. [DOI] [PubMed] [Google Scholar]
- 26.Woods, D. E., D. C. Straus, W. G. Johanson, Jr., W. K. Berry, and J. A. Bass. 1980. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect. Immun. 29:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]