Abstract
In a previous study, we detected unexpectedly high levels of acquired antibiotic resistance in commensal Escherichia coli isolates from a remote Guaraní Indian (Bolivia) community with very low levels of antibiotic exposure and limited exchanges with the exterior. Here we analyzed the structure of the resistant E. coli population from that community and the resistance mechanisms. The E. coli population (113 isolates from 72 inhabitants) showed a high degree of genetic heterogeneity, as evidenced by phylogenetic grouping (77% group A, 10% group B1, 8% group D, 5% group B2) and genotyping by randomly amplified polymorphic DNA (RAPD) analysis (44 different RAPD types). The acquired resistance genes were always of the same types as those found in antibiotic-exposed settings [blaTEM, blaPSE-1, catI, cmlA6, tet(A), tet(B), dfrA1, dfrA7, dfrA8, dfrA17, sul1, sul2, aphA1, aadA1, aadA2, aadA5, aadB, and sat-1]. Class 1 and class 2 integrons were found in 12% and 4% of the isolates, respectively, and harbored arrays of gene cassettes similar to those already described. The cotransferability of multiple-resistance traits was observed from selected isolates and was found to be associated with resistance conjugative plasmids of the F, P, and N types. Overall, these data suggest that the resistance observed in this remote community is likely the consequence of the dissemination of resistant bacteria and resistance genes from antibiotic-exposed settings (rather than of an independent in situ selection) which involved both the clonal expansion of resistant strains and the horizontal transfer/recombination of mobile genetic elements harboring resistance genes.
The notion that the global dissemination of microbial drug resistance observed in the antibiotic era is related to the selective pressure generated by the use of antibiotics in clinical and veterinary practices, animal husbandry, and agriculture is supported by studies that have clearly correlated the emergence and dissemination of resistance with the use of antibiotics (1, 10, 19) and by the absence of acquired resistance in clinical isolates from the preantibiotic era (13, 14).
Surprisingly, however, antibiotic-resistant bacteria have also recently been detected in humans and wild animals living in remote areas where antibiotic exposure has been absent or minimal (2, 9, 11, 22, 30, 32), raising a question about the mechanisms of resistance spread in those settings. To the best of our knowledge, the most isolated human context thus far investigated is represented by a very remote community of Guaraní Indians in the Bolivian Chaco, where we detected high levels of acquired antibiotic resistance in commensal Escherichia coli isolates (2). In that community, exchanges with inhabitants of other areas were very limited; antibiotic exposure at the time of the study had been minimal; and locally collected rainwater was the only water source, ruling out the possibility of sustained contamination of drinking water from the exterior (2).
In this work we have analyzed the population structure of resistant bacteria collected from that remote setting and the nature of the acquired resistance genes and of the cognate genetic elements to gather insights into the mechanisms involved in the spread of acquired antibiotic resistance in similar settings.
MATERIALS AND METHODS
Bacterial strains.
In a previous study, we detected antibiotic-resistant E. coli isolates in 72 of 108 members of the Guaraní Indian community of Alto Los Zarzos (Bolivia) (2). Detailed information on ethical permission, the sampling strategy, and the demographic characteristics of the study population are reported elsewhere (2). A total of 113 E. coli isolates were investigated in this work. These included all the isolates with acquired resistance traits collected from the 72 individuals who exhibited fecal carriage of antibiotic-resistant E. coli (from each individual, all the isolates showing different resistance phenotypes were included in the study) (2). Among the resistant isolates, traits of acquired resistance to the following antibiotics were represented: ampicillin, chloramphenicol, tetracycline, trimethoprim, sulfonamides, kanamycin, and tobramycin. The vast majority of the isolates (76%) were resistant to more than one drug, and the most common multidrug resistance pattern (found in 38 [34%] of the isolates) included ampicillin, chloramphenicol, tetracycline, trimethoprim, and sulfonamides (2).
Molecular analysis techniques.
Basic procedures for DNA extraction, analysis, and manipulation were performed as described by Sambrook and Russell (28). The nucleotide sequences of both strands of the PCR amplification products were determined as described previously (24). Analysis and comparisons of the nucleotide sequences were carried out with the help of programs available at the NCBI web interface (http://www.ncbi.nlm.nih.gov).
Population genetics analysis.
Phylogenetic grouping of the E. coli isolates was performed by the multiplex PCR-based method developed by Clermont et al. (5). Genotyping was performed by randomly amplified polymorphic DNA (RAPD) analysis by using, separately, two decamer primers, primer 1290 (5′-GTGGATGCGA) and primer 1254 (5′-CCGCAGCCAA), as described previously (23). The RAPD patterns were considered to be different when the profiles differed by at least one band. Analysis of the RAPD patterns was performed with Diversity Database fingerprinting software (version 2; Bio-Rad Laboratories, Hercules, CA). The similarity between the RAPD patterns, based on band position, was determined by using the Dice similarity coefficient, and a dendrogram was constructed by using the unweighted pair-group method with arithmetic averages.
Characterization of acquired resistance genes and integrons.
The PCR methodology was used, as described previously, for detection of the following resistance genes: blaTEM-like and blaSHV-like genes for β-lactam resistance (20); tet(A), tet(B), tet(C), and tet(D) for tetracycline resistance (12); catI and cmlA for phenicol resistance (16, 20); dfrA1, dfrA5, dfrA7, dfrA8, dfrA12, dfrA13, dfrA14, and dfrA17 for trimethoprim resistance (18); sul1, sul2, and sul3 for sulfonamide resistance (20, 25); and aphA1 and aadB for aminoglycoside resistance (20). Colony blot hybridization with intI1- and intI2-specific probes, generated as described previously (31), was used to investigate the isolates for the presence of class 1 and class 2 integrons, respectively. The variable regions of the integrons were amplified by PCR, as described previously (27, 33).
Plasmid analysis.
Replicon typing was carried out by a recently developed PCR-based method (4), and positive results were confirmed by colony blot hybridization and/or sequence analysis of the replicons. Conjugative transfer of the resistance determinants was assayed in Mueller-Hinton broth by using E. coli J53 (pro met Rifr Nalr) as a recipient and an initial donor/recipient ratio of 0.1. The mating tubes were incubated at 30°C for 14 h. Transconjugants were selected on Mueller-Hinton agar containing rifampin (400 μg/ml) and nalidixic acid (32 μg/ml) plus one of the following antibiotics: ampicillin (200 μg/ml), chloramphenicol (30 μg/ml), tetracycline (5 μg/ml), trimethoprim (40 μg/ml), or sulfamethoxazole (200 μg/ml). Under the conditions described above, the detection sensitivity of the mating assay was ≥1 × 10−8 transconjugants/recipient. The antimicrobial susceptibilities of the transconjugants were determined by the disk diffusion method (6), and the results were interpreted according to the approved standards of CLSI (7). Antibiotic disks were from Oxoid (Milan, Italy). E. coli ATCC 25922 was used for quality control purposes.
Colicin production.
Colicin production was assayed by the overlay method described by Pugsley (26).
RESULTS
Population structure and demographic distribution of resistant E. coli isolates.
The 113 E. coli isolates were tested for their phylogenetic group of origin (group A, B1, B2, or D) and for their genomic relatedness by RAPD analysis.
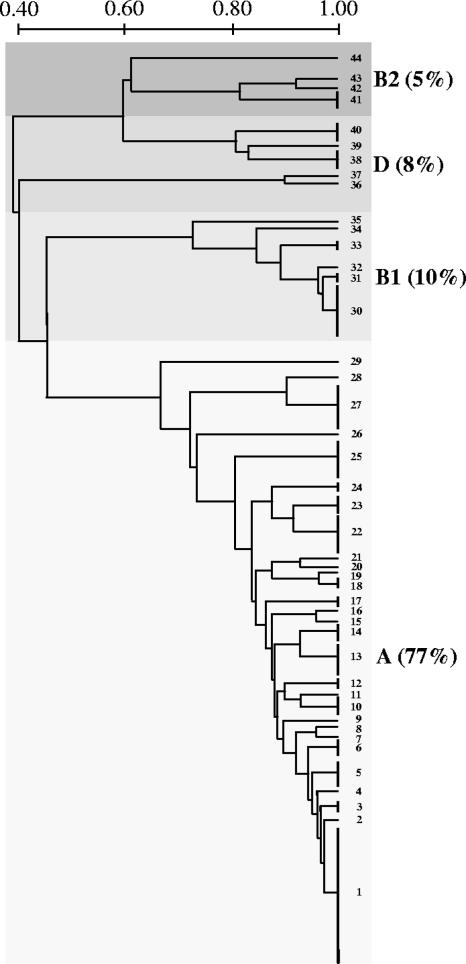
Phylogenetic grouping revealed the presence of isolates from each of the four groups. Isolates of group A were the most prevalent (77%), followed by those of groups B1 (10%), D (8%), and B2 (5%) (Fig. 1).
FIG. 1.
Genetic structure of 113 antibiotic-resistant commensal E. coli isolates from 72 members of a remote community with very low levels of antibiotic exposure. Capital letters refer to phylogenetic groups, and numbers identify RAPD types. Scale bar, 0.2 Dice similarity.
RAPD analysis identified 44 different types, of which 1 (RAPD type 1) included 22 isolates, 20 included 2 to 7 isolates, and the remaining 23 types were singletons (Fig. 1 and Table 1). RAPD type diversity was observed within each phylogenetic group, and the same RAPD type was never observed for isolates of different phylogenetic groups (Fig. 1 and Table 1). Isolates of the same RAPD type usually showed the same resistance phenotype, although differences in individual resistance traits were noticed in a few cases (Table 1). On the other hand, identical resistance phenotypes were represented in different RAPD types and phylogenetic groups (Table 1).
TABLE 1.
RAPD types, phylogenetic groups, colicin production, antibiotic resistance phenotypes, patterns of resistance genes, and integrons of the 113 antibiotic-resistant E. coli isolates
| RAPD type | Phylogenetic group | No. of isolates | Colicin production | Resistance phenotypea | Resistance gene(s) | Class 1 integron (VRb) | Class 2 integron (VR) |
|---|---|---|---|---|---|---|---|
| 44 | B2 | 1 | − | AMP/CHL/TET/TMP/SUL | catI, tet(B), sul1, sul2 | + (blaPSE-1, aadA2) | − |
| 43 | B2 | 1 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 42 | B2 | 1 | − | AMP/CHL/TMP/SUL/KAN/NN | blaTEM, sul1 | + (dfrA17, aadA5), (aadB, aadA1, cmlA6) | − |
| 41 | B2 | 3 | + | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(B), dfrA8, sul2 | − | − |
| 40 | D | 1 | − | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(B), sul1, sul2 | + (dfrA17, aadA5) | − |
| 1 | − | AMP/CHL/TET/SUL | blaTEM, catI, tet(B), sul2 | − | + | ||
| 1 | − | AMP/TMP/SUL | blaTEM, sul2 | − | − | ||
| 39 | D | 1 | − | AMP/TMP/SUL | blaTEM, dfrA8, sul2 | − | − |
| 38 | D | 3 | + | TET | tet(A) | − | − |
| 37 | D | 1 | + | AMP/CHL/TET/TMP/SUL/KAN | blaTEM, catI, tet(A), dfrA8, sul2, aphA1 | − | − |
| 36 | D | 1 | + | AMP/TET/TMP/SUL/KAN | blaTEM, tet(B), sul2, aphA1 | − | + (dfrA1, sat-1, aadA1) |
| 35 | B1 | 1 | − | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(B), sul2 | − | + (dfrA1, sat-1, aadA1) |
| 34 | B1 | 1 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), sul1, sul2 | + (dfrA1, aadA1) | − |
| 33 | B1 | 2 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 32 | B1 | 1 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 31 | B1 | 2 | + | TET/SUL | tet(A), sul2 | − | − |
| 30 | B1 | 3 | − | AMP/TET/TMP/SUL | blaTEM, tet(A), sul1, sul2 | + (dfrA7) | − |
| 2 | − | AMP/TET/SUL | blaTEM, tet(A), sul2 | − | − | ||
| 29 | A | 1 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8 | − | − |
| 28 | A | 1 | − | AMP/TET | blaTEM, tet(B) | − | − |
| 27 | A | 7 | + | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(A), sul1, sul2 | + (dfrA1, aadA1) | − |
| 26 | A | 1 | − | TET | tet(B) | − | − |
| 25 | A | 6 | + | TET | tet(B) | − | − |
| 24 | A | 1 | − | AMP/TET | blaTEM, tet(B) | − | − |
| 1 | − | AMP/TET/SUL | blaTEM, tet(B), sul2 | − | − | ||
| 23 | A | 3 | − | AMP | blaTEM | − | − |
| 22 | A | 2 | − | AMP/CHL/TET/TMP/SUL/KAN | blaTEM, catI, tet(B), sul2, aphA1 | − | + (dfrA1, sat-1, aadA1) |
| 3 | + | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − | ||
| 1 | − | TMP/SUL | sul2 | − | + (dfrA1, sat-1, aadA1) | ||
| 21 | A | 1 | − | AMP/TMP/SUL | blaTEM, dfrA8, sul2 | − | − |
| 20 | A | 1 | + | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 19 | A | 1 | − | CHL/TET/TMP/SUL | tet(A), dfrA8, sul2 | − | − |
| 18 | A | 2 | − | TET | tet(B) | + | − |
| 17 | A | 1 | + | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(B), dfrA8, sul2 | − | − |
| 1 | + | AMP/TMP/SUL | blaTEM, dfrA8, sul2 | − | − | ||
| 16 | A | 1 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 15 | A | 1 | − | AMP/TET/TMP/SUL | blaTEM, tet(A), dfrA8, sul2 | − | − |
| 14 | A | 3 | − | TET | tet(A) | − | − |
| 13 | A | 5 | − | TET | tet(B) | − | − |
| 12 | A | 2 | − | AMP/TET/SUL | blaTEM, tet(B), sul2 | − | − |
| 11 | A | 1 | − | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(B), dfrA8, sul2 | − | − |
| 10 | A | 2 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 1 | + | AMP/SUL | blaTEM, sul2 | − | − | ||
| 9 | A | 1 | + | TET | tet(B) | − | − |
| 8 | A | 1 | − | TET | tet(B) | − | − |
| 7 | A | 1 | − | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(B), dfrA8, sul2 | − | − |
| 6 | A | 3 | − | AMP/TET/TMP/SUL | blaTEM, tet(A), dfrA8, sul2 | − | − |
| 5 | A | 4 | − | CHL/TET/TMP | catI, tet(A), dfrA8 | − | − |
| 4 | A | 1 | − | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 3 | A | 2 | − | TET | tet(B) | − | − |
| 2 | A | 1 | + | AMP/TET/TMP/SUL | blaTEM, tet(B), dfrA8, sul2 | − | − |
| 1 | A | 22 | + | AMP/CHL/TET/TMP/SUL | blaTEM, catI, tet(A), dfrA8, sul2 | − | − |
AMP, ampicillin; CHL, chloramphenicol; TET, tetracycline; TMP, trimethoprim; SUL, sulfamethoxazole; KAN, kanamycin; NN, tobramycin. Acquired resistance genes not identified are underlined. Phenotypic assays for investigation of resistance to streptomycin and spectinomycin (conferred by aadA-type genes) and to streptothricin (conferred by sat-1 genes) were not performed.
VR, integron variable region. The cassette arrays of class 1 integrons from RAPD type 30, RAPD types 27 and 34, RAPD types 40 and 42, and RAPD type 44 were identical to the sequences with GenBank accession nos. AF139109, AJ884723, AF169041, and AB207867, respectively. The second integron from RAPD type 42 showed 99% identity with the sequence with GenBank accession no. DQ836058, from which it differed by a single mutation (C485T) in the aadA1 gene, resulting in the replacement of proline with leucine (as in the sequence with GenBank accession no. DQ517526). All class 2 integrons showed a gene cassette array identical to that of the integron in Tn7 (GenBank accession no. AB188272).
Identical RAPD types were observed in members of different families (e.g., the dominant RAPD type 1 was detected in members of 8 of the 22 families), revealing the interindividual circulation of antibiotic-resistant clones within the community (data not shown).
Characterization of acquired resistance genes.
The 113 E. coli isolates were investigated for the presence of several acquired resistance genes that could account for the observed resistance phenotypes. Of the 81 ampicillin-resistant isolates, 80 harbored a blaTEM β-lactamase gene, while the remaining ampicillin-resistant isolate did not contain either blaTEM or blaSHV. Of the 48 chloramphenicol-resistant isolates, 46 harbored a catI gene and 1 harbored a cmlA gene. Of the 103 tetracycline-resistant isolates, 52 harbored a tet(A) gene and 51 harbored a tet(B) gene, while tet(C) and tet(D) were never detected. Of the 75 trimethoprim-resistant isolates, 73 harbored one of the dfrA genes for which investigations were conducted, including dfrA8 (55 isolates), dfrA1 (13 isolates), dfrA7 (3 isolates), and dfrA17 (2 isolates). Of the 80 sulfonamide-resistant isolates, 78 harbored a sul2 gene, and 13 of them also harbored a sul1 gene. sul1 alone was detected in one of the remaining isolates, while the other was negative for all three sul genes. The four isolates resistant to kanamycin carried an aphA1 gene, while the isolate resistant to kanamycin and tobramycin carried an aadB gene (Table 1).
The isolates were also investigated for the presence of class 1 and class 2 integrons, which usually carry mobile cassettes with resistance genes, by DNA hybridization. Class 1 integrons were found in 14 isolates (12%), while class 2 integrons were found in 5 isolates (4%) (Table 1). Characterization of the variable regions of the integrons revealed, in all cases, the presence of inserted cassettes. Class 1 integrons carried 5 different cassette arrays, with a total of 10 different resistance cassettes (and three minor allelic variants of the aadA1-carrying gene cassette) (Table 1). Class 2 integrons carried a single cassette array with the three resistance cassettes commonly found in this class of integrons (Table 1). Some of the integron-associated resistance genes (cmlA, dfrA1, dfrA7, dfrA17, aadB) corresponded to those already detected in the initial screening (see above). The sequence of the cmlA-like determinant revealed a cmlA6 gene. Additional integron-associated resistance cassettes carried a blaPSE-1 β-lactamase gene (accounting for the ampicillin resistance in the blaTEM-negative isolate); three aminoglycoside nucleotidyltransferase genes (aadA1, aadA2, and aadA5), which confer resistance to streptomycin and spectinomycin; and a streptothricin acetyltransferase gene (sat-1), which confers resistance to streptothricin (Table 1). All the gene cassettes arrays found in this peculiar bacterial population were identical to those already described in isolates from other settings, even though a new aadA1 gene cassette was detected (Table 1).
Isolates of the same RAPD type and with the same resistance phenotype always carried an identical pattern of resistance genes, supporting the occurrence of clonal expansion (Table 1). However, the presence of isolates of the same RAPD type showing a different resistance phenotype due to the acquisition/loss of individual resistance genes revealed the occurrence of recombination of resistance genes within some clones (Table 1).
Transferability of resistance genes and plasmid analysis.
The conjugative transfer of resistance genes was investigated in the 38 isolates exhibiting the most prevalent resistance phenotype (resistance to ampicillin, chloramphenicol, tetracycline, trimethoprim, and sulfonamides). These isolates belonged to each of the four phylogenetic groups and comprised nine different RAPD types (including the two most prevalent ones) (Table 2 and Table 1). Conjugative transfer of resistance traits was observed for five of the nine RAPD types tested. With RAPD type 1 (22 isolates), conjugative transfer could not be assessed due to killing of the recipient by the colicin activity produced by the donor strain (Table 2 and Table 1). In most cases, the cotransfer of several resistance traits was observed (Table 2), suggesting the linkage of the resistance genes in the same transferable plasmid.
TABLE 2.
Transferability of resistance plasmids from 38 multidrug-resistant E. coli isolates and genetic features of resistance conjugative plasmids
| RAPD type | No. of isolates | Results for E. coli J-53 transconjugants
|
||
|---|---|---|---|---|
| Resistance gene(s)a | Colicin production | Replicon type(s)b | ||
| 1 | 22 | —c | — | — |
| 7 | 1 | blaTEM, catI, tet(B), dfrA8, sul2 | No | FIA, FIB, FII, N |
| 11 | 1 | blaTEM, tet(B), dfrA8, sul2 | No | FII |
| 17 | 1 | — | — | — |
| 27 | 7 | blaTEM, catI, tet(A), Int 1 (dfrA1, aadA1, sul1) | Yes | FIB, FII, P |
| 35 | 1 | — | — | — |
| 40 | 1 | blaTEM, catI, tet(B), Int 1 (dfrA17, aadA5, sul1) | No | FIB, FII |
| 41 | 3 | tet(B) | No | FIB, FII |
| 44 | 1 | — | — | — |
Int 1 and Int 2, class 1 and class 2 integrons, respectively.
When more than one replicon was identified in the transconjugants, plasmid extraction and DNA hybridization experiments suggested the presence of a single multireplicon plasmid (data not shown). DNA sequence of the FII replicons further evidenced the heterogeneity among FII replicons (data not shown).
—, transfer of resistance plasmids was not observed.
Plasmids contain genes essential for their replication and, often, accessory genes (e.g., antibiotic resistance genes and virulence genes), and the plasmid replication system is used for plasmid classification and identification. We investigated the replicon types in the resistance plasmids from all the transconjugants. Several plasmid replicon types were detected in this subpopulation, including the F, P, and N types (Table 2). Such heterogeneity was also reflected in the different plasmid sizes and restriction fragment length polymorphism profiles (data not shown).
Colicin production.
Colicin production was observed in 42% of the isolates (Table 1). Noteworthy was the fact that colicinogenic activity was often detected in isolates showing phenomena of clonal expansion (Table 1).
DISCUSSION
Two models could be considered to explain the unexpected finding of a high prevalence of antibiotic-resistant bacteria in remote settings where antibiotic exposure has been minimal and there are no obvious sources of a sustained contamination from the exterior: (i) a primitive selection of resistance in the remote setting, due to peculiar environmental conditions (e.g., a consistent exposure to natural products with antibiotic activity in food or water), and (ii) the introduction of resistant strains (via occasional travelers and/or animals) from antibiotic-exposed settings, followed by the local dissemination and maintenance of resistance in the absence of antibiotic exposure (by unknown mechanisms). In the first model one would expect a limited number of resistance traits (restricted to natural compounds) and, possibly, the evolution of some original resistance genes that differ from those selected in antibiotic-exposed settings. In the second model, one would rather expect the dissemination of a limited number of resistant clones (or mobile genetic elements) carrying resistance genes typical of antibiotic-exposed settings.
The molecular characterization of resistant isolates from a remote human community, carried out in this study, provided some insights into this phenomenon. Resistant bacteria isolated from the remote community carried a remarkable variety of acquired resistance genes (even for resistance to synthetic agents, such as sulfonamides and trimethoprim) which were entirely like those encountered in isolates from antibiotic-exposed settings (3, 18, 20). Specialized elements carrying the resistance genes, such as integrons and backbones of conjugative plasmids, were also like those described elsewhere (4, 21). Even the arrays of gene cassettes carried on integrons and the linkage of different resistance genes on conjugative plasmids mostly reflected those described already (3, 15, 29, 30, 33). Overall, this scenario appears to be consistent with the model ascribing antibiotic resistance, observed in remote areas not exposed to antibiotic use, to the dissemination of resistant bacteria and resistance genes from antibiotic-exposed settings rather than to an independent in situ selection. This view is further supported by the finding that similar resistance patterns and resistance genes are highly prevalent in commensal E. coli isolates from urban areas of the same region of Bolivia (3).
Despite the evidence for the expansion of some resistant clones, a remarkable heterogeneity was observed in the population structure of resistant bacteria from the remote setting and in the repertoire of specialized elements carrying resistance genes. Similar findings do not support a simple model in which a few resistant strains are occasionally introduced in the remote setting and expand to replace the existing susceptible population but, rather, suggest either a sustained flow of diverse resistant strains from the exterior (despite the very limited exchanges) or the occurrence of substantial horizontal gene transfer and recombination phenomena involving resistance genes following the occasional introduction of resistance strains from the exterior into the remote setting.
Whichever the mechanism responsible for this genetic diversity is, the reasons for maintaining the high prevalence of resistance in the absence of antibiotic use remain unexplained. Substantial exposure to natural antibiotics seems unlikely, since it would imply the presence of natural products that select for the same patterns of resistance traits that are common in commensal E. coli isolates from antibiotic-exposed areas of that region of Latin America (3). A possible explanation could be the selective advantage conferred by genetic determinants linked to resistance genes on the same plasmids (e.g., genes for colicins, iron uptake systems, and/or intestinal colonization factors, which could increase the fitness of the organisms for the intestinal ecosystem, or determinants for heavy metal resistance and/or additional metabolic pathways, which could facilitate survival in the environment of the remote setting). The clonal expansion phenomena observed with some resistant strains and the finding of colicin determinants, sometimes associated with resistance genes within conjugative plasmids, in antibiotic-resistant E. coli isolates that showed clonal expansion would support this hypothesis.
The maintenance of different types of acquired resistance genes in such an “antibiotic-free” setting is consistent with recent reports suggesting that expression of acquired antibiotic resistance in bacteria might not always involve a fitness cost (otherwise, resistance genes would rapidly be lost) (8, 17). This has implications relevant for the control of microbial drug resistance in the community and suggests that although strategies based on antibiotic restriction policies are important for decreasing the emergence and dissemination of resistance, strategies based only on antibiotic restriction policies are unlikely to fully succeed for these types of resistant strains and resistance genes.
Acknowledgments
We are indebted to the members of the community of Alto Los Zarzos for their cooperation and willingness to take part in the study. We thank the local health authorities and the Asamblea del Pueblo Guaraní for their encouragement and cooperation.
This work was supported by Ente Cassa di Risparmio (Florence, Italy) and by a grant from the Italian Ministry for University and Research (grant PRIN 2005).
The authors declare no competing financial interests.
Footnotes
Published ahead of print on 12 January 2006.
REFERENCES
- 1.Barbosa, T. M., and S. B. Levy. 2000. The impact of antibiotic use on resistance development and persistence. Drug Resist. Update 3:303-311. [DOI] [PubMed] [Google Scholar]
- 2.Bartoloni, A., F. Bartalesi, A. Mantella, E. Dell'Amico, M. Roselli, M. Strohmeyer, H. Gamboa Barahona, V. P. Barron, F. Paradisi, and G. M. Rossolini. 2004. High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. J. Infect. Dis. 189:1291-1294. [DOI] [PubMed] [Google Scholar]
- 3.Bartoloni, A., L. Pallecchi, M. Benedetti, C. Fernandez, Y. Vallejos, E. Guzman, A. L. Villagran, A. Mantella, C. Lucchetti, F. Bartalesi, M. Strohmeyer, A. Bechini, H. Gamboa, H. Rodriguez, T. Falkenberg, G. Kronvall, E. Gotuzzo, F. Paradisi, and G. M. Rossolini. 2006. Multidrug-resistant commensal Escherichia coli in children, Peru and Bolivia. Emerg. Infect. Dis. 12:907-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 5.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; 6th ed. Approved standard M07-A6. Clinical Laboratory Standards Institute, Wayne, PA.
- 7.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. Clinical Laboratory Standards Institute, Wayne, PA.
- 8.Enne, V. I., A. A. Delson, G. R. Davis, S. L. Haywars, J. M. Roe, and P. M. Benett. 2005. Assessment of the fitness impacts on Escherichia coli of acquisition of antibiotic resistance genes encoded by different types of genetic element. J. Antimicrob. Chemother. 56:544-551. [DOI] [PubMed] [Google Scholar]
- 9.Gilliver, M., M. Bennett, M. Begon, S. Hazel, and C. Hart. 1999. Antibiotic resistance found in wild rodents. Nature 401:233-234. [DOI] [PubMed] [Google Scholar]
- 10.Goossens, H., M. Ferech, R. Vander Stichele, M. Elseviers, and the ESAC Project Group. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579-587. [DOI] [PubMed] [Google Scholar]
- 11.Grenet, K., D. Guillemot, V. Jarlier, B. Moreau, S. Dubourdieu, R. Ruimy, L. Armand-Lefevre, P. Bau, and A. Andremont. 2004. Antibacterial resistance, Wayampis Amerindians, French Guyana. Emerg. Infect. Dis. 10:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman, A. B., I. I. Essiet, D. W. Isenbarger, and L. E. Lindler. 2003. Epidemiology of tetracycline resistance determinants in Shigella spp. and enteroinvasive Escherichia coli: characterization and dissemination of tet(A)-1. J. Clin. Microbiol. 41:1023-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houndt, T., and H. Ochman. 2000. Long-term shifts in patterns of antibiotic resistance in enteric bacteria. Appl. Environ. Microbiol. 66:5406-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, V. M., and N. Datta. 1983. Conjugative plasmids in bacteria of the ‘pre-antibiotic’ era. Nature 302:725-726. [DOI] [PubMed] [Google Scholar]
- 15.Kang, H. Y., Y. S. Jeong, J. Y. Oh, S. H. Tae, C. H. Choi, D. C. Moon, W. K. Lee, Y. C. Lee, S. Y. Seol, D. T. Cho, and J. C. Lee. 2005. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J. Antimicrob. Chemother. 55:639-644. [DOI] [PubMed] [Google Scholar]
- 16.Keyes, K., C. Hudson, J. J. Maurer, S. Thayer, D. G. White, and M. D. Lee. 2000. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob. Agents Chemother. 44:421-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2006. Antimicrobial drug resistance genes do not convey a secondary fitness advantage to calf-adapted Escherichia coli. Appl. Environ. Microbiol. 72:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, J. C., J. Y. Oh, J. W. Cho, J. C. Park, J. M. Kim, S. Y. Seol, and D. T. Cho. 2001. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 47:599-604. [DOI] [PubMed] [Google Scholar]
- 19.Levy, S. B. 2002. Factors impacting on the problem of antibiotic resistance. J. Antimicrob. Chemother. 49:25-30. [DOI] [PubMed] [Google Scholar]
- 20.Maynard, C., S. Bekal, F. Sanschagrin, R. C. Levesque, R. Brousseau, L. Masson, S. Lariviere, and J. Harel. 2004. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol. 42:5444-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazel, D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608-620. [DOI] [PubMed] [Google Scholar]
- 22.Osterblad, M., K. Norrdahl, E. Korpimaki, and P. Huovinen. 2001. How wild are wild animals? Nature 409:37-38. [DOI] [PubMed] [Google Scholar]
- 23.Pacheco, A. B., B. E. Guth, K. C. Soares, L. Nishimura, D. F. de Almeida, and L. C. Ferreira. 1997. Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J. Clin. Microbiol. 35:1521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perilli, M., E. Dell'Amico, B. Segatore, M. R. De Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum beta-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perreten, V., and P. Boerlin. 2003. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47:1169-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pugsley, A. P. 1985. Escherichia coli K12 for use in the identification and characterization of colicins. J. Gen. Microbiol. 131:369-376. [DOI] [PubMed] [Google Scholar]
- 27.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Solberg, O. D., R. M. Ajiboye, and L. W. Riley. 2006. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J. Clin. Microbiol. 44:1347-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural population of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunde, M. 2005. Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. J. Antimicrob. Chemother. 56:1019-1024. [DOI] [PubMed] [Google Scholar]
- 32.Walson, J. L., B. Marshall, B. M. Pokhrel, K. K. Kafle, and S. B. Levy. 2001. Carriage of antibiotic-resistant fecal bacteria in Nepal reflects proximity to Kathmandu. J. Infect. Dis. 184:1163-1169. [DOI] [PubMed] [Google Scholar]
- 33.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]