Abstract
The genetic structures surrounding the plasmid-carried blaOXA-23 oxacillinase gene, encoding resistance to carbapenems, were studied in Acinetobacter baumannii. ISAba1 and the novel element ISAba4 were detected upstream of the blaOXA-23 gene, providing promoter sequences for its expression. These insertion elements were likely involved in transposition processes at the origin of acquisition of this β-lactamase gene.
Acinetobacter baumannii is a typical opportunistic pathogen, often involved in nosocomial outbreaks, for which resistance to carbapenems is increasingly reported (23) and may be linked to the production of Ambler class B metallo-β-lactamases (30) but also to the production of carbapenem-hydrolyzing class D β-lactamases (CHDLs) (18, 31). Although integrons are associated with many metallo-β-lactamase or oxacillinase genes, they are likely not the genetic vehicles for acquisition of CHDL genes since these genes are not in the form of gene cassettes (10, 22, 23). Three main acquired CHDL gene clusters have been described for A. baumannii, namely, the blaOXA-23-, blaOXA-24-, and blaOXA-58-like genes, whereas the blaOXA-51 gene cluster is naturally occurring and chromosomally located in A. baumannii (5, 6, 11, 16, 23, 29). The blaOXA-23 gene has been identified worldwide in A. baumannii (4, 8, 9, 13, 15, 27) and in Proteus mirabilis in France (3). Whereas the blaOXA-24-like genes have been identified as chromosomally encoded, the blaOXA-23 and blaOXA-58 genes are mostly found on plasmids (23). The acquisition of blaOXA-58 in an A. baumannii isolate from France has been associated with a homologous recombination process (21, 22).
ISAba1 has been found upstream of blaOXA-58, blaOXA-51-like, and blaampC genes in A. baumannii and is involved in their expression (7, 12, 23, 26, 28). ISAba1 belongs to the IS4 family of insertion sequences, possesses two 16-bp imperfect inverted repeats (IRs), and generates a 9-bp target site duplication upon transposition. Its transposase is made of two open reading frames, encoding 189 and 178 amino acids, leading to a functional protein when a frameshift occurs during the translation process (12).
The aim of this study was to analyze the genetics of acquisition and expression of the blaOXA-23 gene in unrelated A. baumannii isolates recovered from different countries (Table 1). Genes coding for CHDLs were searched for by PCR, using primers specific for the blaOXA-23-like, blaOXA-24-like, and blaOXA-58 genes (15). Two blaOXA-23-positive A. baumannii isolates (Ab13 and Ab14) resistant to all β-lactams, including carbapenems, were studied in detail (Table 1). Genetic structures surrounding the blaOXA-23 gene in A. baumannii Ab13 and Ab14 were cloned by restricting total DNA by BamHI or SacI, ligating it into BamHI- or SacI-restricted plasmid pBK-CMV, and transforming the recombinant plasmids into Escherichia coli DH10B, as described previously (11). Recombinant plasmids were selected on Trypticase soy agar plates containing amoxicillin (50 μg/ml) and kanamycin (30 μg/ml). The cloned DNA fragments of several recombinant plasmids (pAB13B, pAB13S, and pAB14B) were sequenced and analyzed as described previously (21).
TABLE 1.
Characteristics of blaOXA-23-positive Acinetobacter baumannii clinical isolates
| Strain | Date of isolation (mo/yr) | Location | MIC of imipenem (μg/ml)a | Transposonb | Reference |
|---|---|---|---|---|---|
| Ab13 | 06/2004 | Bicêtre, France | 32 | Tn2006 | This study |
| TN585 | 08/2004 | Bicêtre, France | 32 | Tn2006 | This study |
| DOS | 05/2004 | Paris, France | 32 | Tn2007 | This study |
| TN273 | 12/2003 | Paris, France | 16 | Tn2006 | This study |
| TN508 | 03/2004 | Papeete, Tahiti | 64 | Tn2006 | 17 |
| TN712 | 03/2005 | Papeete, Tahiti | 64 | Tn2006 | 17 |
| TN479 | 04/2004 | Tarbes, France | 32 | Tn2006 | This study |
| Ab14 | 12/2004 | Algeria | 16 | Tn2007 | This study |
| TN802 | 02/2004 | Noumea, New Caledonia | >64 | Tn2006 | This study |
| TN850 | 06/2006 | Ankara, Turkey | 32 | Tn2006 | This study |
| TN761 | 05/2005 | Vietnam | 32 | Tn2006 | This study |
| TN614 | 11/2004 | Libya | 64 | Tn2006 | This study |
| RF10 | 11/2003 | Romania | 32 | Tn2006 | 15 |
MICs of meropenem were very similar to those of imipenem.
As presented in Fig. 1.
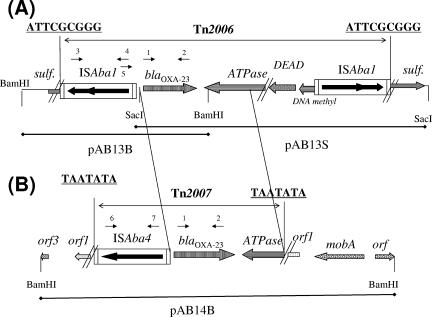
Recombinant plasmids pAB13B and pAB13S, obtained from A. baumannii Ab13, revealed that blaOXA-23 was bracketed by two copies of an identical ISAba1 element that were in opposite orientations (Fig. 1). Detailed DNA sequence analysis revealed a 9-bp target site duplication at the inverted repeat right (IRR) extremities of the two ISAba1 elements, suggesting that both copies formed a putative composite transposon, Tn2006, likely at the origin of blaOXA-23 acquisition (Fig. 1). The Tn2006 insertion occurred inside a gene encoding a putative sulfonamide resistance protein sharing 89% amino acid identity with a protein identified in A. baumannii AYE (accession no. CAJ31116). Sequencing of the internal sequence of that transposon revealed a 2,445-bp sequence containing, in addition to the blaOXA-23 gene, two other genes, encoding part of a putative AAA ATPase (83% amino acid identity with that of Acinetobacter baylyi ADP-1 [2] [accession no. YP_046025] but lacking the first 70 N-terminal amino acids) and part of a putative DEAD helicase sharing 69% identity with that of Ralstonia solanacearum but lacking its 493-amino-acid C-terminal extremity (accession no. ZP_00943964) (Fig. 1). These two proteins were truncated at the exact same position, suggesting that a recombination event had occurred. Even though ISAba1 has been shown to be very prevalent in A. baumannii and might be “customized” for that species (25), this is the first description of an ISAba1-based putative composite transposon.
FIG. 1.
Schematic maps of inserts of recombinant plasmids harboring the blaOXA-23 gene. (A) Two recombinant plasmids obtained from isolate Ab13. (B) Recombinant plasmid obtained from isolate Ab14. BamHI and SacI restriction sites are indicated, as well as the corresponding recombinant plasmids obtained (pAB13B, pAB13S, and pAB14B). The boundaries of Tn2006 and Tn2007 are indicated, together with the target site duplications likely generated by the transposition events (bold and underlined). The sequences that are identical between Tn2006 and Tn2007 are bracketed by two diagonal, parallel lines. Primer positions are indicated, and their sequences are available in Table 2. orf, orf1, and orf3, genes of unknown function; sulf, sulfonamide resistance gene; ATPase, gene encoding the putative AAA ATPase; DEAD, gene encoding the putative DEAD helicase; and mob, gene encoding the putative MobB mobilization protein.
Sequencing of the recombinant plasmid pAB14B obtained from isolate Ab14 identified a novel ISAba4 element upstream of the blaOXA-23 gene. ISAba4 belongs to the IS982 family, is 975 bp long, possesses two 18-bp IRs, and encodes a 292-amino-acid putative transposase. No target site duplication was observed on either end of ISAba4. PCR mapping with different sets of primers did not detect any extra copy of ISAba4 downstream of the blaOXA-23 gene. Detailed analysis of sequences located downstream of the blaOXA-23 gene identified a 1,497-bp sequence that was identical to that identified in Tn2006, with the same gene encoding a putative AAA ATPase. This gene was truncated at its 5′ extremity, leading to a protein lacking the first 108 N-terminal amino acids. Detailed analysis of the site of truncation identified a 7-bp sequence with an A+T-rich content (TAATATA) that was also identified at the extremity of the IRR of ISAba4 (Fig. 1). This feature was very likely the signature of a transposition process mediated by ISAba4 that occurred at the origin of acquisition of the blaOXA-23 gene. This potential transposon, termed Tn2007, was 2,471 bp long and included ISAba4 and blaOXA-23. The ISAba4-mediated mobilization process likely corresponded to a one-ended transposition mechanism, in contrast to what has been observed with ISEcp1, which during its mobilization process uses a wide range of DNA sequences as IRRs, which are, however, not absolutely random (19, 20). In this case, the sequence identified at the right end of Tn2007 did not exhibit homology with the IRs of ISAba4. Alternatively, it could be hypothesized that a second copy of ISAba4 in a likely ISAba4-made composite transposon might have been lost by excision. At the left extremity of Tn2007, an open reading frame (ORF) (Fig. 1B, orf1) encoding 62 N-terminal amino acids sharing 68% amino acid identity with a plasmid maintenance killer protein of Photorabdus luminescens (accession no. NP_928205) was identified. At the right extremity of Tn2007, an ORF was identified that encoded 309 amino acids that shared 51% amino acid identity with a MobA/MobL mobilization protein found on the pP plasmid of Salmonella enterica serovar Enteritidis (accession no. NP_604396).
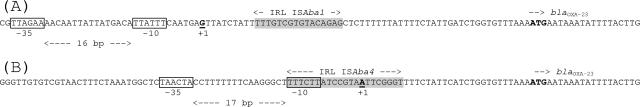
Using 5′ rapid amplification of cDNA ends-PCR (22), the sites of initiation of transcription of the blaOXA-23 gene were mapped in both A. baumannii isolates Ab13 and Ab14, possessing ISAba1 and ISAba4, respectively, upstream of blaOXA-23. In Ab13, the ISAba1 element was located 25 bp upstream of blaOXA-23, and the +1 transcription start was identified 60 bp upstream of the start codon of the blaOXA-23 gene, located inside the ISAba1 element. The corresponding promoter, made of a −35 sequence (TTAGAA) and separated by 16 bp from the −10 sequence (TTATTT), was identical to that identified previously in ISAba1, located at the origin of overexpression of the naturally occurring blaampC and blaOXA-51-like genes in A. baumannii (7, 12) (Fig. 2). The ISAba4 element was located 25 bp upstream of blaOXA-23 in Ab14, and the +1 transcription start was located 31 bp upstream of the start codon of the blaOXA-23 gene, just inside the left IRs of ISAba4. The −35 sequence (TAACTA) and a −10 sequence (TTTCTT) separated by 17 bp acted as promoter sequences (Fig. 2).
FIG. 2.
Promoter structures for expression of the blaOXA-23 genes of A. baumannii Ab13 (A) and Ab14 (B). The +1 initiation sites of transcription are shown in bold and underlined, and the promoter sequences are boxed. The ATG start codon of the blaOXA-23 gene is shown in bold. The left IRs (IRL) of the ISAba1 and ISAba4 elements are shaded.
Subsequent PCR mapping was performed to evaluate the presence of these transposons in 11 blaOXA-23-positive A. baumannii isolates, using the primers detailed in Table 2. Pulsed-field gel electrophoresis analysis revealed that the 12 isolates studied were unrelated (data not shown). In these isolates, the blaOXA-23 gene was plasmid borne, according to the results of the Kieser technique (14) and of hybridization experiments (data not shown). Tn2006-like elements were identified in 10 of 12 isolates, whereas the 2 others harbored the Tn2007 structure.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Nucleotide sequence (5′→3′) | Location | No. in Fig. 1 |
|---|---|---|---|
| OXA-23A | GGAATTCCATGAATAAATATTTTACTTGC | blaOXA-23 | 2 |
| OXA-23B | CGGGATCCCGTTAAATAATATTCAGGTC | blaOXA-23, reverse primer | 1 |
| ISAba1A | GTGCTTTGCGCTCATCATGC | tnpA of ISAba1 | 3 |
| ISAba1B | CATGTAAACCAATGCTCACC | tnpA of ISAba1, reverse primer | 4 |
| ISAba1-ext | AAGCACTTGATGGGCAAGGC | tnpA of ISAba1, external primer | 5 |
| ISAba4A | ATTTGAACCCATCTATTGGC | tnpA of ISAba4 | 7 |
| ISAba4B | ACTCTCATATTTTTTCTTGG | tnpA of ISAba4, reverse primer | 6 |
ISAba1, which enhances the expression of several unrelated resistance genes, seems to be quite an important factor of genetic plasticity in A. baumannii (25). The large distribution and high copy number of ISAba1 may facilitate the creation of composite transposons (24). On the other hand, a single copy of ISAba4 might mobilize sequences located at its right-end extremity in what is considered a one-ended transposition process (1). Further in vitro experiments are now required to establish under which conditions and at what frequency these transposition mechanisms occur. Interestingly, we have identified sequences located just downstream of the blaOXA-23 gene possessing significant homology with those of A. baylyi. Together with the fact that ISAba1 could be considered widespread in the Acinetobacter genus, this could suggest that blaOXA-23 originated from an Acinetobacter-like species. It could also be hypothesized that the Tn2006 putative composite transposon might have been formed in A. baumannii after a previous acquisition process not related to ISAba1.
Nucleotide sequence accession numbers.
The nucleotide sequences of the insertion elements reported in this paper have been submitted to the IS Finder website (http://www-is.biotoul.fr). The entire sequences identified and described in this study have been assigned accession no. EF127491, for Tn2006, and EF059914, for Tn2007.
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France, and mostly by a grant from the European community (LSHM-CT-2005-018705). L.P. is a researcher from INSERM, France.
We thank A. Carattoli for precious advice on plasmid feature analysis. We also thank I. Podglajen and D. Colak for their gifts of isolates DOS and TN850, respectively.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Avila, P., J. Grinsted, and F. de la Cruz. 1988. Analysis of the variable endpoints generated by one-ended transposition of Tn21. J. Bacteriol. 170:1350-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 28:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, R., H. Marchandin, C. Chanal, D. Sirot, R. Labia, C. De Champs, E. Jumas-Bilak, and J. Sirot. 2002. Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boo, T. W., F. Walsh, and B. Crowley. 2006. First report of OXA-23 carbapenemase in clinical isolates of Acinetobacter species in the Irish Republic. J. Antimicrob. Chemother. 58:1101-1102. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S., and S. G. Amyes. 2005. The sequences of seven class D β-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin. Microbiol. Infect. 11:326-329. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S., H. K. Young, and S. G. Amyes. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11:15-23. [DOI] [PubMed] [Google Scholar]
- 7.Corvec, S., N. Caroff, E. Espaze, C. Giraudeau, H. Drugeon, and A. Reynaud. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52:629-635. [DOI] [PubMed] [Google Scholar]
- 8.Dalla-Costa, L. M., J. M. Coelho, H. A. Souza, M. E. Castro, C. J. Stier, K. L. Bragagnolo, A. Rea-Neto, S. R. Penteado-Filho, D. M. Livermore, and N. Woodford. 2003. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J. Clin. Microbiol. 41:3403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donald, H. M., W. Scaife, S. G. Amyes, and H. K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Héritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Héritier, C., L. Poirel, and P. Nordmann. 2006. Cephalosporinase overexpression as a result of insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123-130. [DOI] [PubMed] [Google Scholar]
- 13.Jeon, B. C., S. H. Jeong, I. K. Bae, S. B. Kwon, K. Lee, D. Young, J. H. Lee, J. S. Song, and S. H. Lee. 2005. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 β-lactamase in Korea. J. Clin. Microbiol. 43:2241-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieser, T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 15.Marqué, S., L. Poirel, C. Héritier, S. Brisse, M. D. Blasco, R. Filip, G. Coman, T. Naas, and P. Nordmann. 2005. Regional occurrence of plasmid-mediated carbapenem-hydrolyzing oxacillinase OXA-58 in Acinetobacter spp. in Europe. J. Clin. Microbiol. 43:4885-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merkier, A. K., and D. Centron. 2006. blaOXA-51-type β-lactamase genes are ubiquitous and vary within a strain in Acinetobacter baumannii. Int. J. Antimicrob. Agents 28:110-113. [DOI] [PubMed] [Google Scholar]
- 17.Naas, T., M. Levy, C. Hirschauer, H. Marchandin, and P. Nordmann. 2005. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J. Clin. Microbiol. 43:4826-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naas, T., and P. Nordmann. 1999. OXA-type β-lactamases. Curr. Pharm. Des. 5:865-879. [PubMed] [Google Scholar]
- 19.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel, L., M. F. Lartigue, J. W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., S. Marqué, C. Héritier, C. Segonds, G. Chabanon, and P. Nordmann. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 24.Prentki, P., B. Teter, M. Chandler, and D. J. Galas. 1986. Functional promoters created by the insertion of transposable element IS1. J. Mol. Biol. 191:383-393. [DOI] [PubMed] [Google Scholar]
- 25.Segal, H., S. Garny, and B. G. Elisha. 2005. Is ISAba1 customized for Acinetobacter? FEMS Microbiol. Lett. 243:425-429. [DOI] [PubMed] [Google Scholar]
- 26.Segal, H., E. C. Nelson, and B. G. Elisha. 2004. Genetic environment of ampC in Acinetobacter baumannii clinical isolate. Antimicrob. Agents Chemother. 48:612-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turton, J. F., E. M. K. Kaufmann, J. Glover, J. M. Coelho, M. Warner, R. Pike, and T. L. Pitt. 2005. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 43:3074-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 29.Turton, J. F., N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walther-Rasmussen, J., and N. Høiby. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373-383. [DOI] [PubMed] [Google Scholar]