Abstract
Previous microarray data (E. Mongodin, J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer, J. Bacteriol. 185:4638-4643, 2003) noted an association in two vancomycin-intermediate Staphylococcus aureus (VISA) strains between high-level, passage-induced vancomycin resistance, a marked increase in the transcription of purine biosynthetic genes, and mutation of the putative purine regulator purR. Initial studies to report on the possible association between vancomycin resistance and alterations in purine metabolism in one of these strains (VP-32) confirmed, by Western analysis, an increase in the translation of PurH and PurM, two purine pathway enzymes. In addition, PurR was identified, by knockout and complementation in a vancomycin-susceptible strain, as a repressor of the purine biosynthetic operon in S. aureus, and the PurR missense mutation was shown to inactivate the repressor. However, despite the apparent relationship between increased purine biosynthesis and increased vancomycin resistance in VP-32, neither the addition of exogenous purines to a defined growth medium nor the truncation or inactivation of purR improved the growth of vancomycin-susceptible S. aureus in the presence of vancomycin. Furthermore, the passage of additional vancomycin-susceptible and VISA strains to high-level vancomycin resistance occurred without changes in cellular purine metabolism or mutation of purR despite the development of thickened cell walls in passaged strains. Thus, we could confirm neither a role for altered purine metabolism in the development of vancomycin resistance nor its requirement for the maintenance of a thickened cell wall. The failure of biochemical and physiological studies to support the association between transcription and phenotype initially found in careful microarray studies emphasizes the importance of follow-up investigations to confirm microarray observations.
Methicillin-resistant Staphylococcus aureus (MRSA) is a causative agent of serious infections in both the hospital and the community. Increased resistance to β-lactam antibiotics such as methicillin has increased the use of the glycopeptide antibiotic vancomycin to treat S. aureus infections. In the past decade, two different mechanisms of vancomycin resistance have emerged. The first, noted initially in Japan and subsequently in the United States, is not well defined but involves common phenotypic changes such as a thickened peptidoglycan layer, decreased peptidoglycan cross-linking, and slower growth rates (6, 8, 18, 19, 29, 30). All strains with this resistance mechanism are of intermediate resistance according to Clinical Laboratory Standards Institute classifications and are known as vancomycin-intermediate Staphylococcus aureus (VISA) strains. The second mechanism, noted in 2002, involves the van genes and mimics the vancomycin resistance mechanism in Enterococcus strains with the production of an altered peptidoglycan stem peptide conferring high-level resistance to glycopeptides (7).
Currently, intermediate resistance mediated by a thickened cell wall is more widespread (11, 18). Resistance likely develops via a multistep process involving many alterations in cell wall metabolism to allow for increased peptidoglycan production (17, 29, 34). Difficulties in the detection of early genotypic or phenotypic changes in S. aureus, indicative of later vancomycin resistance, result in the clinical failure of vancomycin treatment (10, 36).
Using two clinical VISA isolates, 5827 and Mu50, Mongodin et al. (25) compared isogenic pairs of either back-passaged vancomycin-susceptible or parent strain and a vancomycin passage-derived highly resistant strain. These strain pairs, P100/VP-32 and Mu50/Mu50-32, were then compared by DNA microarray. Those investigators found that the transcription of all genes in the pur operon was up-regulated 5- to 30-fold in the vancomycin-resistant strains compared to their more sensitive counterpart. The up-regulated transcription of purM was confirmed by quantitative reverse transcription-PCR. Those authors sequenced the putative purine repressor purR and found a single base pair change in each of the resistant strains at nucleotide 140 (T→A) producing a single amino acid change of isoleucine to lysine at amino acid 47. This mutant gene has been designated purR(I47K).
Mongodin et al. proposed that increased cell wall thickness likely requires additional cellular energy and that this additional energy is supplied by ATP from an up-regulation of enzymes in the purine pathway. Little is known about purine metabolism in Staphylococcus species. Most information about purine metabolism in S. aureus is assumed from comparisons with other bacterial purine metabolism systems that have been well characterized, such as those of Escherichia coli and Bacillus subtilis.
In S. aureus, the putative purine biosynthesis operon purEKCSQLFMNHD, purine biosynthesis-associated genes purA and purB, and the purine operon repressor purR most closely resemble the purine biosynthesis genes and gene organization found in Bacillus (12, 13). PurR has a 54% identity and a 78% similarity with PurR in B. subtilis, and the gene organization is very similar in both bacteria. However, in Lactococcus lactis, PurR, despite being 80% similar and 51% identical to PurR in B. subtilis, acts as a transcriptional activator and not a repressor as described previously for B. subtilis (21). In addition, purB is encoded within the pur operon in B. subtilis, while it is separate from the operon in S. aureus.
Sinha et al. (35) crystallized PurR from Bacillus, defined its functional domains, and compared its sequence to the putative purR sequences of 20 other gram-positive bacteria. This allowed those authors to define conserved and homologous regions within the protein structure. The structure revealed a two-domain protein dimer with a winged-helix domain at the N terminus and a 5-phosphoribosyl-1-pyrophosphate binding site in the C-terminal domain. The N-terminal domain consists of an N-terminal flag, a helix-turn-helix motif, and a wing. Amino acid 47 falls within alpha-helix 3 in the helix-turn-helix motif. Once folded, this residue is closely associated with seven other nonpolar amino acids forming the N-terminal dimer interface including alpha-helix 1 and alpha-helix 3. The mutant purR of VP-32 and Mu50-32, purR(I47K), contains a charged polar amino acid in this normally conserved hydrophobic region.
The current study, described below, is the follow-up to the initial microarray observations described previously by Mongodin et al. and attempts to identify any effect that changes in purine metabolism might have in the development of vancomycin resistance in Staphylococcus aureus. Specifically, the role of PurR mutation and inactivation is examined.
MATERIALS AND METHODS
Strains, plasmids, and primers.
Lists of the strains, plasmids, and primers used appear in Tables 1 and 2.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant characteristic(s) | Description (source or reference) |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | recA1 lacZΔM15 | Host for lacZ-containing cloning vectors (Invitrogen) |
| BL21(DE3) | Host for T7 promoter-based expression systems (Novagen) | |
| S. aureus | ||
| RN4220 | Mcs Vms | Restriction deficient mutagenized RN450 |
| RN450 | Mcs Vms | 8325-4 (27) |
| 450M | Mcr (he) Vms | RN450 transformed with COL mec region DNA |
| 450MΔpurR | Mcr (he) Vms Gmr Ems | purR interrupted by Gmr cassette (this study) |
| N315 | Mcr (he) Vms | Clinical isolate, Japan, 1982 (23) |
| COL | Mcr (ho) Vms | England, 1965 (2) |
| 3130 | Mcr (ho) Vms | Clinical isolate, SCOPE surveillance study (40) |
| 3134 | Mcr (he) Vms | Clinical isolate, SCOPE surveillance study (40) |
| 5836b | Mcr (he) Vmi | Clinical isolate, New Jersey, 1997 (377) |
| 5827c | Mcr (ho) Vmi | Clinical isolate, Michigan, 1997 (37) |
| 5827VR | Mcr (ho) Vmr | 5827 passaged in BHI broth containing increasing concentrations of vancomycin to a vancomycin MIC of 32 μg/ml (this study) |
| P100 | Mcr (ho) Vms | 5827 passaged 100 times on antibiotic-free medium (25) |
| V20 | Mcr (ho) Vmr | 5827 passaged on increasing concentrations of vancomycin to a vancomycin MIC of 20 μg/ml (this study) |
| V25 | Mcr (ho) Vmr | 5827 passaged on increasing concentrations of vancomycin to a vancomycin MIC of 25 μg/ml (this study) |
| VP-32 | Mcr (ho) Vmr | 5827 passaged on increasing concentrations of vancomycin to a vancomycin MIC of 32 μg/ml (25) |
| Mu50 | Mcr Vmi | Clinical isolate, Japan, 1996 (37) |
| Mu50-32 | Mcr Vmr | Mu50 passaged on increasing concentrations of vancomycin to a vancomycin MIC of 32 μg/ml (25) |
| 5836 | Mcr (he) Vmi | Clinical isolate, New Jersey, 1997 (37) |
| NRS12 | Mcs Vmi | Isolated on 6 μg/ml vancomycin from clinical human VISA strain, France, 1998 (5) |
| NRS52 | Mcs Vmi | Clinical isolate, California, 2000 (15) |
| Plasmids | ||
| E. coli | ||
| pCR2.1 | Apr Kmr | Cloning vector (Invitrogen) |
| pET24d(+) | Kmr | Expression vector (Novagen) |
| pMC01 | Kmr | purM in MCS of pET24d(+) (this study) |
| pMC02 | Kmr | purH in MCS of pET24d(+) (this study) |
| pMC04 | Apr Kmr | purR in MCS of pCR2.1 (this study) |
| pMC08 | Apr, Kmr, Gmr | Gmr in BstBI site of pMC04 (this study) |
| S. aureus | ||
| pCN51 | Emr Pcad | E. coli-S. aureus shuttle vector with a cadmium-inducible promoter (9) |
| pE194(Ts) | Emr | E. coli-S. aureus shuttle vector with temperature-sensitive replicon (20) |
| pGO1 | Gmr | Conjugative staphylococcal plasmid (1) |
| pMC10 | Emr Gmr | pMC08 ligated into the XbaI site of pE194(Ts) (this study) |
| pR | Emr Pcad | purR in MCS of pCN51 (this study) |
| pRm | Emr Pcad | purR(I47K) in MCS of pCN51 (this study) |
Mc, methicillin; Vm, vancomycin; Gm, gentamicin; Em, erythromycin; Ap, ampicillin; Km, kanamycin; r, resistant; s, sensitive; i, intermediate; ho, homotypic; he, heterotypic; Pcad, cadmium promoter; MCS, multiple cloning site.
Also known as S. aureus 992.
Also known as S. aureus 966.
TABLE 2.
Primers used
| Primer | Sequence (5′-3′) | Gene target |
|---|---|---|
| PJM01 | GAT CCC ATG GCT AAA GCA TAT GAA CAA TCT GG | purM |
| PJM02 | ACG TGG ATC CTA CCC CCA ACA ATT CAA TTG C | purM |
| PJM03 | GAT CCC ATG GTG AAG AAA GCT ATT TTG AGC | purH |
| PJM04 | ACG TGG ATC CGT GTT TAA AAT GTC GAG TGC | purH |
| PJM11 | ACG TGT CGA CGA ATA TGG TCC AAG TGC TTC CGG | purR |
| PJM14 | GAT CGG ATC CGA TGG TGC GTT AAT GAG TGG | purR or purR(I47K) |
| PJM15 | GAT CGG ATC CTG CTG GCG CAA GTG GTG G | purR |
| PJM24 | ACG TTC GAA ACA CAG GAG TCT GGA CTT GAC TCA C | Gentamicin cassette |
| PJM27 | ACG TGG TAC CGA ATA TGG TCC AAG TGC TTC CGG | purR or purR(I47K) |
| PJM30 | GAT CTT CGA ACA TCA ATT TTG ATA AGT AGA AAT GG | Gentamicin cassette |
Cloning, transformation, electroporation, and transduction.
Restriction endonuclease digestions and ligations were carried out according to manufacturers' (Promega [Madison, WI] or New England Biolabs [Ipswich, MA]) specifications. Plasmid DNA was transformed into chemically competent E. coli cells according to manufacturers' (Invitrogen [Carlsbad, CA] or Novagen [Madison, WI]) specifications. Cells were plated onto selective agar for incorporation of the plasmid. Vectors were moved from E. coli to restriction-deficient S. aureus strain RN4220 by electroporation using 2-mm cuvettes in a Gene Pulser apparatus (Bio-Rad, Richmond, CA) at settings of 100 Ω, 25 μF, and 2.5 kV as previously described (32). Plasmids were introduced into S. aureus strains other than RN4220 using the general transduction phage 80α adapted from the method described previously by Thompson and Pattee (38).
Passage technique.
Strains were grown with shaking at 220 rpm at 37°C in brain heart infusion (BHI) (Becton Dickinson, Cockeysville, MD) broth with increasing levels of vancomycin (Sigma-Aldrich, St. Louis, MO) until a thick culture was grown (up to 48 h). Strains were plated onto BHI agar with the same concentration of vancomycin as that used for their most recent growth in broth to check stability and resistance. A single colony was then selected for continued passage. The MIC of all strains was determined by Etest (AB Biodisk, Piscataway, NJ) for vancomycin passages.
Etest.
A single colony was inoculated into 5 ml Mueller-Hinton broth (Becton Dickinson) and shaken at 37°C at 220 rpm overnight. The culture was then diluted to a McFarland standard of 0.5 (unless otherwise indicated) and swabbed onto an Mueller-Hinton agar (MHA) plate, and an Etest strip was placed according to the manufacturer's instructions. The plate was incubated at 37°C, and MICs were determined at 24 and 48 h.
PCR.
Primers for PCR were made by the Nucleic Acids Research Facility at Virginia Commonwealth University. Amplifications using Taq Master Mix (QIAGEN, Valencia, CA) were carried out using a MiniCycler (MJ Research, Watertown, MA) according to standard protocols (3).
Sequencing.
Sequencing was performed on PCR-amplified products by the Nucleic Acids Research Facility at Virginia Commonwealth University.
Population analysis profiling.
Phenotypic expression of vancomycin resistance was determined by population analysis profiling adapted from the procedures described previously by Hackbarth et al. and Wootton et al. (14, 41). A single colony of the strain of interest was selected and grown in 5 ml of BHI or Mueller-Hinton broth overnight at 37°C with shaking at 220 rpm. A 1-ml aliquot of the culture grown overnight was serially diluted in the same medium used for growth. Ten microliters of each dilution was dropped into one of 36 squares marked on a square agar plate. Each dilution, from 10−2 to 10−10, of each strain was plated in duplicate at each antibiotic concentration. The following concentrations of vancomycin in BHI agar or MHA were used: 0, 0.5, 1, 2, 4, 8, 16, and 32 μg/ml. Drops were allowed to dry before inverting plates at 37°C. Colonies were counted after 24 and/or 48 h of growth.
PFGE.
Methods for the preparation of genomic DNA and separation by pulsed-field gel electrophoresis (PFGE) were adapted from methods described previously by Bannerman et al. (4). Briefly, agarose plugs containing intact bacterial cells were digested with SmaI (Promega) overnight at 25°C according to the manufacturer's instructions. Digested plugs were separated on a 1% agarose gel run in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1.3 mM EDTA) under the following parameters: 6 V/cm, an initial switching time of 1 s, a final switching time of 30 s, a run time of 22 h, and a run temperature of 14°C. Bands were visualized with UV light after ethidium bromide staining. PFGE was used to confirm strain identity after passage.
Construction of PurH and PurM overexpression vectors.
purH and purM were amplified from N315 using primers PJM03 and PJM04 for purH and PJM01 and PJM02 for purM. PCR products were run on a 1% agarose gel in 0.5× TBE, excised, and purified using the QIAquick PCR purification kit (QIAGEN). Purified PCR products were ligated at 14°C overnight into the pET24d(+) vector (Novagen), which contains an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter and a C-terminal His tag sequence for protein purification. Both constructs pMC02 (containing purH) and pMC01 (containing purM) were transformed into chemically competent TOP10 E. coli cells (Invitrogen). Positive clones were selected on BHI agar containing 15 μg/ml kanamycin (Sigma-Aldrich). Plasmids were prepared from positive clones using the QIAprep Spin Miniprep kit (QIAGEN). pMC02 and pMC01 were then transformed into chemically competent BL21(DE3) E. coli cells (Novagen) for overexpression and protein purification.
Protein purification.
E. coli BL21(DE3) cells containing either pMC02 or pMC01 were grown to the mid-logarithmic phase of growth (optical density at 600 nm [OD600] of ≈0.6) in BHI broth with 15 μg/ml kanamycin and 1% glucose with shaking at 37°C. Cultures were then induced with 1 mM IPTG and grown for an additional 3 h. Cells were pelleted and proteins were purified on a nickel column using the QIAexpressionist denaturing protocol (QIAGEN). The identity of purified proteins was confirmed by N-terminal sequencing (Iowa State Protein Facility, Ames, IA). Purified proteins were dialyzed against 1× phosphate-buffered saline (150 mM NaCl, 2.7 mM KCl, 12 mM NaHPO4, 2 mM KH2PO4 [pH 7.4]) at 4°C overnight.
Antibodies.
Antibodies to PurM and PurH recombinant proteins were made in chickens by Alpha Diagnostic International (San Antonio, TX) using their standard 63-day protocol.
Western analysis.
Western analysis procedures were adapted from methods described previously by McKinney et al. (24). Briefly, strains were grown to the mid-logarithmic phase of growth in BHI broth, pelleted, and washed in 1/2× phosphate-buffered saline. Cells were lysed in triple detergent saline (50 mM Tris-HCl [pH 8.5], 150 mM NaCl, 0.02% sodium azide, 0.1% sodium dodecyl sulfate [SDS], 1% IGEPAL CA-630, 0.5% Na-deoxycholate) by bead beating for 30 s at 6.0 in a FastPrep apparatus (Thermo Savant, Holbrook, NY). Five micrograms of protein per sample was separated on a NuPAGE 12% Bis-Tris gel (Invitrogen) in NuPAGE MOPS SDS running buffer (Invitrogen). The gel was transferred onto a Hybond-HCl nitrocellulose membrane (Amersham Biosciences, Piscataway, NJ) using a Bio-Rad semidry transfer in transfer buffer (25 mM Tris, 150 mM glycine, 10% methanol). Blots were blocked with 5% skim milk in TTBS (10 mM Tris-HCl [pH 7.2 to 7.4], 150 mM NaCl, 2.7 mM KCl, 0.05% Tween 20). Rabbit anti-PurH (or anti-PurM) polyclonal antiserum was used at a 1:100 dilution. The secondary antibody (goat anti-chicken) was used at a 1:10,000 dilution. Bound antibodies were detected using the ECL Plus Western blotting detection system (Amersham Biosciences) followed by film exposure.
Disruption of purR using allelic replacement.
purR was amplified from 450 M by PCR using primers PJM15 and PJM11 and ligated into pCR2.1 (Invitrogen). The vector was cloned into chemically competent TOP10 E. coli cells and plated onto BHI plates containing 100 μg/ml ampicillin (Fisher Scientific) to select clones containing the vector of interest, pMC04. A gentamicin resistance cassette was amplified from pGO1 by PCR using PJM30 and PJM24, both containing BstBI sites. The gentamicin cassette was subcloned into the unique BstBI site of purR in pMC04 to create pMC08. The temperature-sensitive staphylococcal vector pE194(Ts), which carries an erythromycin resistance cassette, was added to the unique XbaI site of pMC08 to create pMC10. The resulting vector was electroporated into RN4220 and transduced into strain 450 M using 80α as described above. Clones containing pMC10, resistant to erythromycin and gentamicin, were grown overnight in MHA containing 5 μg/ml gentamicin (Fisher Scientific) at 30°C. Cultures were diluted 1:1,000 in fresh MHA with gentamicin and placed at 42°C. Cultures were grown and diluted back for 5 days to select for allelic replacement of purR with purR containing the gentamicin cassette and loss of the plasmid (confirmed by PCR). The strain of interest, 450 MΔpurR, was gentamicin resistant and erythromycin sensitive.
Inducible purR and purR(I47K) vectors.
purR and purR(I47K) were amplified from N315 and VP-32, respectively, using PJM14 and PJM27. The resulting PCR products were ligated into the multiple cloning site of the cadmium-inducible vector pCN51, creating pR and pRm (9). The vectors were transformed into chemically competent TOP10 E. coli cells, electroporated into RN4220, and then transduced into 450 M and 450 MΔpurR using 80α. pR and pRm were induced by growth in 10 μM cadmium (Fisher Scientific).
CDM growth curves.
N315 was grown overnight at 37°C in chemically defined medium (CDM) with or without supplementation as described previously (39). Cultures grown overnight were diluted to an OD600 of <0.1, and 150-μl aliquots were placed into each well of a 96-well plate with or without vancomycin at the given concentration. Plates were incubated at 37°C with shaking for 18 h in a Multiskan Ascent apparatus (Thermo Lab Systems, Franklin, MA). The OD595 was taken every 15 min. Doubling times were determined by averaging the time required for OD595 doubling at at least two time intervals within log-phase growth. A Student's t test was used to compare differences in doubling times between groups. A P value of <0.05 was considered to be statistically significant.
Transmission electron microscopy.
A single colony was inoculated into BHI broth and grown overnight at 37°C with shaking at 220 rpm. Cultures were diluted back to an OD600 of ∼0.075 in fresh BHI broth and grown at 37°C with shaking to the mid-logarithmic phase of growth. Three milliliters of cells was pelleted and resuspended in fix buffer (2% glutaraldehyde in 0.1 M sodium cacodylate). Cells were stored at 4°C until use. Section preparation and transmission electron microscopy image acquisition were performed by the Virginia Commonwealth University Department of Neurobiology and Anatomy Microscopy Facility.
RESULTS
Purine enzyme translation in S. aureus made highly resistant to vancomycin by passage.
The increased transcription seen in microarray and reverse transcription-PCR studies reported previously by Mongodin et al. (25) results in the increased translation of the enzymes in the purine pathway as shown in Fig. 1. Vancomycin-susceptible S. aureus (VSSA) strain P100 and VISA strain 5827 do not show increased translation of PurH and PurM by Western analysis. In contrast, vancomycin-resistant S. aureus strains V20, V25, and V32, created by passage of strain 5827 in the presence of increasing concentrations of vancomycin, have increased translation of both enzymes.
FIG. 1.
Western blot analysis of purine enzyme levels. Soluble proteins prepared from cells grown in antibiotic-free medium to the mid-logarithmic phase of growth were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred onto nitrocellulose membranes and probed with antibodies to PurH or antibodies to PurM as indicated. P100, strain 5827 passaged 100 times in the absence of vancomycin; strain 5827, first clinical VISA isolate in the United States; strains V20, V25, and VP-32, strain 5827 passaged on vancomycin to MICs of 20, 25, and 32 μg/ml, respectively.
Exogenously increased purine levels.
To determine whether increasing purine concentrations alone increase vancomycin resistance, N315, an MRSA/VSSA strain, was grown in chemically defined medium with and without purine or pyrimidine supplementation in increasing levels of vancomycin (26, 39). Generally, doubling times, determined during logarithmic growth, increased in all media with increasing vancomycin concentrations (Fig. 2). The addition of adenine, guanine, or xanthine to CDM significantly (P < 0.05) slowed the growth of N315 in the presence of 2 μg/ml vancomycin, indicating that exogenous purine supplementation does not promote N315 growth in vancomycin.
FIG. 2.
Growth curve analysis of N315 grown in supplemented or unsupplemented CDM in the presence of increasing vancomycin concentrations. N315 was grown in CDM with or without supplementation in increasing concentrations of vancomycin for 18 h. The OD595 was taken every 15 min. Doubling times were calculated between multiple points within the log phase of growth. Each bar is the average of two or more replicates. The CDM bar represents N315 grown in unsupplemented medium. All other bars are N315 grown in CDM with the specified supplement(s). A, adenine (0.037 mM); G, guanine (0.033 mM); C, cytosine (0.045 mM); T, thymine (0.160 mM); U, uracil (0.045 mM); A2, doubled concentration of A; X, xanthine (0.033 mM); Vit2, doubled concentration of vitamins (0.006 mM thiamine, 0.02 mM nicotinic acid, 0.002 mM calcium pantothenate, 0.00004 mM biotin).
Endogenously increased purine biosynthetic enzyme levels.
A panel of N315 isolates was gathered from laboratory stocks frozen over the past 20 years. The levels of PurH and PurM translation were determined by Western analysis, and purR was sequenced and compared to the published sequence (GenBank accession no. NP373706). One strain, designated N315mutant, has a spontaneous nonsense mutation that truncates the protein by approximately 14%. The remaining four strains, designated N315A to N315D, were determined to have wild-type purR. The Western analysis results shown in Fig. 3 demonstrated that PurM and PurH levels can be elevated in an isolate with a wild-type purR sequence (N315A). N315mutant also displays elevated translation of PurM and PurH. Regardless of purine pathway enzyme levels or the purR sequence, all isolates showed the same survival profile at multiple vancomycin concentrations compared by population analysis profiling (data not shown).
FIG. 3.
Western analysis of the N315 panel. Equal amounts of soluble protein were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. Membranes were probed with antibodies to (A) PurH or (B) PurM. The negative and positive control lanes represent strains known to have normal and up-regulated pur operon translation, respectively. The mutant lane is an N315 strain with a spontaneous nonsense mutation at amino acid 236 of 274. Lanes A, B, C, and D are N315 strains from a laboratory collection from various sources with wild-type purR sequences. All five N315 strains are identical by PFGE.
Inactivation of purR and overexpression of PurR(I47K).
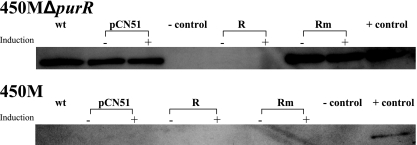
Allelic replacement was used to exchange the wild-type chromosomal copy of purR in strain 450 M, an MRSA/VSSA strain, with purR interrupted by a gentamicin resistance cassette producing an inactive PurR. The inactivation of purR resulted in increased levels of PurM (Fig. 4) by Western analysis. Translation of PurM in 450 MΔpurR mimics the level seen in VP-32. However, 450 MΔpurR did not show increased resistance to vancomycin compared to that of 450 M by vancomycin Etest or population analysis profiling on increasing levels of vancomycin (data not shown).
FIG. 4.
Western analysis of PurM translation changes due to alterations in PurR. Soluble proteins were prepared from strains grown to the mid-logarithmic phase growth in the presence (+) or absence (−) of 10 μM cadmium. Proteins were separated by SDS-PAGE and transferred onto a nitrocellulose membrane, which was probed with antibodies to PurM. Strains contained either no plasmid (wt), empty vector (pCN51), pR (R), or pRm (Rm). Plasmids were induced by growth in cadmium in both backgrounds, 450 MΔpurR and 450 M.
The mutant purR sequence found in VP-32 and Mu50-32 (25), purR(I47K), and the wild-type sequence were introduced into and overexpressed in 450 MΔpurR using the cadmium-inducible vector pCN51, creating pRm and pR (9). Purine enzyme levels were reduced to undetectable levels by pR, whereas they remained elevated following the introduction of pRm, confirming the inactivation of the repressor by the mutation at nucleotide 140 (T→A) (Fig. 4). The specific effect on vancomycin resistance of PurR(I47K) versus that of an inactive PurR, such as that expressed by 450 MΔpurR, was examined by population analysis profiling, which demonstrated little to no difference between the two profiles or compared to wild-type 450 M (data not shown).
Additionally, the overexpression of PurR(I47K) from pCN51 in 450 M carrying wild-type purR did not increase PurM or PurH levels upon Western analysis (Fig. 4). This finding suggests that wild-type PurR is dominant compared to PurR(I47K). We were unable to introduce wild-type PurR into VP-32 because the thickened cell wall precluded the introduction of plasmids by either electroporation or transduction.
Passage to high-level vancomycin resistance in diverse parental backgrounds.
To investigate whether purine biosynthesis alterations were common in highly vancomycin-resistant strains (in the absence of the van genes), three clinical MRSA/VSSA isolates (3130, 3134, and COL), two clinical MRSA/VISA isolates (5836 and 5827), and two clinical methicillin-sensitive S. aureus/VISA isolates (NRS12 and NRS52) were passaged in BHI broth with increasing concentrations of vancomycin to a vancomycin MIC of >16 μg/ml (determined by Etest). Strains are detailed in Table 1. Sequence analysis of purR and Western analysis of purine enzyme levels showed neither a mutated purR nor an increase in purine pathway enzymes in any of the strains examined following passage to high-level vancomycin resistance (data not shown).
5827VR, created by the passage of 5827 in increasing concentrations of vancomycin in BHI broth, was examined by transmission electron microscopy to confirm the development of a thickened cell wall in the absence of a change in purine enzyme levels (data not shown). Despite no increase in purine enzyme quantity, 5827VR demonstrated a thickened cell wall similar to that of VP-32 and those described for other VISA and vancomycin-resistant passage isolates (16, 22, 25, 33). Taken together, these data indicate that in S. aureus, a mutation in purR is required for neither the development of vancomycin resistance by in vitro passage nor the construction and maintenance of a thickened cell wall.
DISCUSSION
Previous work demonstrated that a mutation of purR occurred during the passage of two VISA strains, 5827 and Mu50, from low-level to high-level vancomycin resistance (25, 28). Purine metabolism was normal in the two parent VISA strains, with vancomycin MICs of 6 to 8 μg/ml, but the transcription of genes in the pur operon was markedly increased when the MICs increased to 32 μg/ml following prolonged passage in vancomycin. Two hypotheses were formed as a result of these data. The first hypothesis was that there might be some direct association between purine metabolism and vancomycin resistance. The second was that the increase in cellular purine concentrations provided ATP for energy, a requirement to meet the demands of increased cell wall biosynthesis.
The current study examined these hypotheses in multiple ways and found no link between purine metabolism and the development of vancomycin resistance. First, exogenous purine supplementation in CDM failed to improve the growth of N315 in the presence of vancomycin. Second, a spontaneous increase in PurM and PurH in N315, in both the presence and absence of purR mutations, had no effect on vancomycin susceptibility. Third, the inactivation of PurR and the expression of PurR(I47K) in a vancomycin-susceptible strain failed to decrease the strain's vancomycin susceptibility even though the amounts of PurM and PurH were similar to those seen in VP-32.
Finally, a panel of strains including VSSA and VISA strains was passaged in BHI broth with increasing concentrations of vancomycin to achieve an MIC of >16 μg/ml. Purine enzyme levels were not elevated in any of the vancomycin-passaged strains, nor was the purR sequence altered. Additionally, the same strain used by Mongodin et al. (25), strain 5827, was passaged to high-level resistance and achieved a thickened cell wall, as detected by transmission electron microscopy, similar to that seen in VP-32. This passage occurred without any change in purine biosynthesis. It is important that while we could not replicate the findings of Mongodin et al. (25), we used a different passage technique from that used by those authors. Mongodin and coworkers used agar passage on vancomycin gradient plates with occasional passage in broth to create VP-32 and Mu50-32. We, however, used passage in broth containing vancomycin with occasional plating onto vancomycin-containing BHI agar to check resistance levels. The difference between growth primarily on agar (the technique described by Mongodin et al.) versus broth (our technique) containing vancomycin could have imposed different physiological demands on the organism, one of which led to alterations in purine metabolism. However, what the current study makes clear is that these metabolic alterations were not related to the development of vancomycin resistance. The alteration of purine metabolism and the development of vancomycin resistance were independent processes.
However, a by-product of this investigation was the first molecular investigation into purine metabolism in S. aureus. Western analysis of purine enzyme levels in VP-32, allelic replacement and complementation studies, as well as Western analysis of N315 strains with various purR sequences and protein expression levels allow several inferences to be made about purine regulation in S. aureus. First, purR encodes the repressor of the purine pathway. Second, PurR function is sensitive to a single nucleotide alteration in a hydrophobic region that has been conserved in gram-positive bacteria (35) (Fig. 1) as well as a C-terminal truncation (Fig. 3). Third, there are likely other regulators of purine biosynthesis in S. aureus. This can be inferred from Fig. 3, where, despite a wild-type purR sequence, N315A displays elevated translation of PurM and PurH. Last, wild-type PurR appears to be dominant compared to a mutant PurR, PurR(I47K) (Fig. 4).
These findings create a platform for further analysis of purine biosynthesis in S. aureus. In B. subtilis, transcription of the pur operon, purA, purR, and a number of other genes is repressed by the binding of PurR to two PurBox sequences. The consensus PurBoxes are pairs of 14-nucleotide stretches of inverted sequence separated by 16 or 17 nucleotides. There is a conserved set of four bases in the middle of each box, —CGAA— (31). The affinity of PurR for the PurBox is influenced by the intracellular concentration of 5-phosphoribosyl-1-pyrophosphate, the starting material for purine biosynthesis. A search for PurBox sequences in S. aureus found that sequences within 250 nucleotides of the start codons for purA and purB have proper spacing and only two or three deviations from the consensus, while those upstream of purE and purR have six and nine differences, respectively, greater than any differences among PurBoxes in B. subtilis. Further studies are needed to investigate the PurR binding sequences in S. aureus.
This study illustrates the importance of performing detailed follow-up investigations to confirm findings from microarray analyses that associate transcriptional alterations with phenotype. In our initial study, we were careful to control variables that may have led to false results. We investigated two different strains, we imposed stringent cutoff parameters for severalfold increases in transcription, we performed the analysis multiple times and then “flipped” the labeling dyes between comparator samples, and we used appropriate statistical analysis to confirm the significance of results. We were confident that the results were “true” for the given strains. However, further biochemical and physiological analyses confirmed that while the results were true, the phenotype and metabolic alterations were unrelated.
Footnotes
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Archer, G. L., J. P. Coughter, and J. L. Johnston. 1986. Plasmid-encoded trimethoprim resistance in staphylococci. Antimicrob. Agents Chemother. 29:733-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L., D. M. Niemeyer, J. A. Thanassi, and M. J. Pucci. 1994. Dissemination among staphylococci of DNA sequences associated with methicillin resistance. Antimicrob. Agents Chemother. 38:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2002. The polymerase chain reaction, p. 15-1 to 15-40. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Short protocols in molecular biology, 5th ed., vol. 2. John Wiley & Sons, Inc., Hoboken, NJ. [Google Scholar]
- 4.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobin-Dubreux, S., M. E. Reverdy, C. Nervi, M. Rougier, A. Bolmstrom, F. Vandenesch, and J. Etienne. 2001. Clinical isolate of vancomycin-heterointermediate Staphylococcus aureus susceptible to methicillin and in vitro selection of a vancomycin-resistant derivative. Antimicrob. Agents Chemother. 45:349-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., S. K. Berke, J. C. Lee, and R. S. Daum. 2000. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 44:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 1997. Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morb. Mortal. Wkly. Rep. 46:765-766. [PubMed] [Google Scholar]
- 9.Charpentier, E., A. I. Anton, P. Barry, B. Alfonso, Y. Fang, and R. P. Novick. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70:6076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven, D. E., and D. S. Shapiro. 2006. Staphylococcus aureus: times, they are a-changin'. Clin. Infect. Dis. 42:179-180. [DOI] [PubMed] [Google Scholar]
- 11.de Lassence, A., N. Hidri, J. F. Timsit, M. L. Joly-Guillou, G. Thiery, A. Boyer, P. Lable, A. Blivet, H. Kalinowski, Y. Martin, J. P. Lajonchere, and D. Dreyfuss. 2006. Control and outcome of a large outbreak of colonization and infection with glycopeptide-intermediate Staphylococcus aureus in an intensive care unit. Clin. Infect. Dis. 42:170-178. [DOI] [PubMed] [Google Scholar]
- 12.Ebbole, D. J., and H. Zalkin. 1989. Bacillus subtilis pur operon expression and regulation. J. Bacteriol. 171:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebbole, D. J., and H. Zalkin. 1987. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 262:8274-8287. [PubMed] [Google Scholar]
- 14.Hackbarth, C. J., C. Miick, and H. F. Chambers. 1994. Altered production of penicillin-binding protein 2a can affect phenotypic expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2568-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hageman, J. C., D. A. Pegues, C. Jepson, R. L. Bell, M. Guinan, K. W. Ward, M. D. Cohen, J. A. Hindler, F. C. Tenover, S. K. McAllister, M. E. Kellum, and S. K. Fridkin. 2001. Vancomycin-intermediate Staphylococcus aureus in a home health-care patient. Emerg. Infect. Dis. 7:1023-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 17.Hiramatsu, K. 1998. Vancomycin resistance in staphylococci. Drug Resist. Updat. 1:135-150. [DOI] [PubMed] [Google Scholar]
- 18.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 19.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 20.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J. Bacteriol. 150:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kilstrup, M., and J. Martinussen. 1998. A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. J. Bacteriol. 180:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koehl, J. L., A. Muthaiyan, R. K. Jayaswal, K. Ehlert, H. Labischinski, and B. J. Wilkinson. 2004. Cell wall composition and decreased autolytic activity and lysostaphin susceptibility of glycopeptide-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 48:3749-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 24.McKinney, T. K., V. K. Sharma, W. A. Craig, and G. L. Archer. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and β-lactamase regulators. J. Bacteriol. 183:6862-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niemeyer, D. M., M. J. Pucci, J. A. Thanassi, V. K. Sharma, and G. L. Archer. 1996. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 178:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick, R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 28.Nygaard, P. 1993. Purine and pyrimidine salvage pathways, p. 359-378. In A. Sonenshein, J. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 29.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotun, S. S., V. McMath, D. J. Schoonmaker, P. S. Maupin, F. C. Tenover, B. C. Hill, and D. M. Ackman. 1999. Staphylococcus aureus with reduced susceptibility to vancomycin isolated from a patient with fatal bacteremia. Emerg. Infect. Dis. 5:147-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxild, H. H., K. Brunstedt, K. I. Nielsen, H. Jarmer, and P. Nygaard. 2001. Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO. J. Bacteriol. 183:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma, V. K., C. J. Hackbarth, T. M. Dickinson, and G. L. Archer. 1998. Interaction of native and mutant MecI repressors with sequences that regulate mecA, the gene encoding penicillin binding protein 2a in methicillin-resistant staphylococci. J. Bacteriol. 180:2160-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sieradzki, K., and A. Tomasz. 2003. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 185:7103-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha, S. C., J. Krahn, B. S. Shin, D. R. Tomchick, H. Zalkin, and J. L. Smith. 2003. The purine repressor of Bacillus subtilis: a novel combination of domains adapted for transcription regulation. J. Bacteriol. 185:4087-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover, F. C. 1999. Implications of vancomycin-resistant Staphylococcus aureus. J. Hosp. Infect. 43:S3-S7. [DOI] [PubMed] [Google Scholar]
- 37.Tenover, F. C., M. V. Lancaster, B. C. Hill, C. D. Steward, S. A. Stocker, G. A. Hancock, C. M. O'Hara, S. K. McAllister, N. C. Clark, and K. Hiramatsu. 1998. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J. Clin. Microbiol. 36:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, N. E., and P. A. Pattee. 1981. Genetic transformation in Staphylococcus aureus: demonstration of a competence-conferring factor of bacteriophage origin in bacteriophage 80α lysates. J. Bacteriol. 148:294-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson, B. J. 1997. Biology, p. 1-38. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, NY.
- 40.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]
- 41.Wootton, M., R. A. Howe, R. Hillman, T. R. Walsh, P. M. Bennett, and A. P. MacGowan. 2001. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47:399-403. [DOI] [PubMed] [Google Scholar]