Abstract
PD 0305970 and PD 0326448 are new bacterial gyrase and topoisomerase inhibitors (quinazoline-2,4-diones) that possess outstanding in vitro and in vivo activities against a wide spectrum of bacterial species including quinolone- and multidrug-resistant gram-positive and fastidious organism groups. The respective MICs (μg/ml) for PD 0305970 capable of inhibiting ≥90% of bacterial strains tested ranged from 0.125 to 0.5 versus staphylococci, 0.03 to 0.06 versus streptococci, 0.25 to 2 versus enterococci, and 0.25 to 0.5 versus Moraxella catarrhalis, Haemophilus influenzae, Listeria monocytogenes, Legionella pneumophila, and Neisseria spp. PD 0326448 MIC90s were generally twofold higher versus these same organism groups. Comparative quinolone MIC90 values were 4- to 512-fold higher than those of PD 0305970. In testing for frequency of resistance, PD 0305970 and levofloxacin showed low levels of development of spontaneous resistant mutants versus both Staphylococcus aureus and Streptococcus pneumoniae. Unlike quinolones, which target primarily gyrA and parC, analysis of resistant mutants in S. pneumoniae indicates that the likely targets of PD 0305970 are gyrB and parE. PD 0305970 demonstrated rapid bactericidal activity by in vitro time-kill testing versus streptococci. This bactericidal activity carried over to in vivo testing, where PD 0305970 and PD 0326448 displayed outstanding Streptococcus pyogenes 50% protective doses (PD50s) (oral dosing) of 0.7 and 3.6 mg/kg, respectively (ciprofloxacin and levofloxacin PD50s were >100 and 17.7 mg/kg, respectively). PD 0305970 was also potent in a pneumococcal pneumonia mouse infection model (PD50 = 3.2 mg/kg) and was 22-fold more potent than levofloxacin.
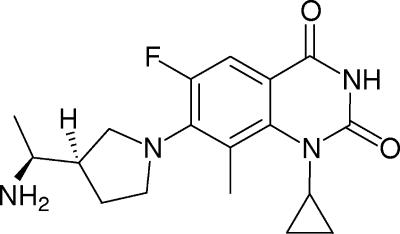
The continuing emergence and development of bacterial resistance to existing antibacterial agents (fluoroquinolones, macrolides, and vancomycin) in gram-positive organisms have created the need for new compounds that retain activity against these resistant strains (1, 4, 9, 16). PD 0305970 and PD 0326448 are new bacterial gyrase and topoisomerase inhibitors developed as part of a program to introduce an orally active quinazolinedione (QD), displaying highly potent in vitro and in vivo activities versus susceptible and resistant gram-positive and fastidious organism groups. The structure of PD 0305970, 3-amino-7-{(R)-3-[(S)-1-amino-ethyl]-pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methyl-1H-quinazoline-2,4-dione, is displayed in Fig. 1. PD 0326448 is the des-3-amino version of PD 0305970 (Fig. 2). The data presented here describe the in vitro susceptibilities, in vivo efficacies, and frequencies of resistance for these compounds compared with quinolones in medically significant bacterial species.
FIG. 1.
PD 0305970.
FIG. 2.
PD 0326448.
MATERIALS AND METHODS
Antimicrobial agents.
The antibacterial compounds used in these studies were obtained from the following sources: PD 0305970, PD 0326448, and gatifloxacin were obtained from the Chemistry Department at Pfizer Global Research and Development, Ann Arbor, MI; garenoxacin was obtained from Bristol Myers, Princeton, NJ; levofloxacin was obtained from R. W. Johnson Pharmaceutical Research Institute, Springhouse, PA; and ciprofloxacin was obtained from Bayer (Miles Pharmaceuticals), West Haven, CT. Details of PD 0305970 synthesis were described previously by Ellsworth et al. (6).
Bacterial strains.
Most of the 1,036 bacterial strains employed in testing were obtained from the Pfizer culture collection and consisted primarily of recent (received within the last 3 years) clinical isolates (>95%). Representative strains ranged from newly acquired clinical isolates to established cultures with special characteristics (e.g., mouse virulent or drug resistant). Several reference strains were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). Bacterial culture identifications were confirmed using a MicroScan Walkaway 40 SI instrument (Dade Behring, Deerfield, IL) and by standard microbiological methods (11).
Antimicrobial susceptibility testing.
Determination of MICs was done according to guidelines of the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) (13-15) or according to the procedures described below. Broth microdilution susceptibility testing was performed using a BioMek FX robotic workstation (Beckman-Coulter, Fullerton, CA). Nonfastidious organisms were tested in cation-adjusted Mueller-Hinton broth (CAMHB; Becton Dickinson, Sparks, MD); streptococci, Staphylococcus haemolyticus, Alloiococcus otitidis, Listeria monocytogenes, and Corynebacterium spp. were tested in CAMHB containing 3% lysed horse blood (Remel, Lenexa, KS); Haemophilus strains were tested in Haemophilus test medium (PML Microbiologicals, Wilsonville, OR); Neisseria strains were tested in gonococcal broth (18); anaerobes were tested in supplemented Brucella broth (13); and Legionella pneumophila was tested in buffered yeast extract α-ketoglutarate broth (5). All incubations were performed at 35°C. Legionella pneumophila, anaerobes, and A. otitidis MICs were read after 48 h of incubation. Neisseria strains were incubated in 5% CO2. Anaerobic MIC testing was performed using a Bactron IV anaerobe chamber (Sheldon manufacturing, Cornelius, OR) containing a gas mixture of 5% CO2, 5% H2, and 90% N2.
Determinations of frequency of resistance.
Streptococcus pneumoniae 7785 (SP-2870) and Staphylococcus aureus UC-76 (SA-1) were grown for 18 to 22 h in appropriate media and concentrated 100 times by centrifugation. Agar plates were prepared on the day of testing by adding PD 0305970, ciprofloxacin, gatifloxacin, or levofloxacin to molten Mueller-Hinton agar at 55°C (5% lysed horse blood was added for S. pneumoniae) and poured immediately. Compound MICs were confirmed by agar dilution (14). The single-step frequency of resistance was determined (150-mm plates) at 0.5×, 1×, 2×, 4×, or 8× MIC concentrations. Specifically, 200 μl of organisms (containing 107 to 1010 CFU) was spread onto agar plates and grown at 35°C for 48 to 72 h. Streptococcus pneumoniae cells were incubated in 5% CO2. Resistant clones were replated onto fresh drug plates to confirm the resistant phenotype. The frequency of resistance was reported as the number of confirmed phenotypically resistant mutants per total organisms plated.
Using S. aureus UC-76 and broth macrodilution methodologies (14), a series of quinolone-resistant mutants was generated by multistep passage in the presence of ciprofloxacin. The MICs of these mutants were confirmed and genotypes of resistant mutants were characterized by sequence analysis (gyrA, gyrB, parC, and parE genes). PD 0305970 was then evaluated by MIC for cross-resistance to these mutants.
Analysis of spontaneous mutants.
The frequency of resistance was determined as described above. The genotype for resistant mutants was characterized by sequence analysis of the gyrA, gyrB, parC, and parE genes. Genomic DNA was isolated from cells grown to log phase using the DNeasy tissue kit (QIAGEN Inc., Valencia, CA). The quality of the DNA was confirmed by visualization on an agarose gel. PCR primers were designed (Table 1) to bind approximately 100 bp upstream from the start site for each gene based on the S. pneumoniae R6 genome (GenBank accession number AE007317). The primers for gyrA, gyrB, parC, and parE were synthesized by MWG Biotech Inc. (Highpoint, NC). PCRs were carried out using a Peltier Thermal Cycler 225 (MJ Research, Waltham, MA.). The 100-μl reaction mix consisted of PCR SuperMix High Fidelity (Invitrogen, Carlsbad, CA), PCR primers (100 pmol), and DNA template (20 to 200 ng).
TABLE 1.
Streptococcus pneumoniae quinolone resistance-determining-region primers
| Primer and description | Sequence |
|---|---|
| gyrA PCR primer, forward (bp 32-54) | 5′-CTGACAAAGGAGATGAAGGCAAGT-3′ |
| gyrA PCR/sequencing primer, reverse (bp 488-511) | 5′-CAGTTGCTCCATTAACCAAAAGGT-3′ |
| gyrA sequencing primer, forward (bp 148-161) | 5′-ATGAATGAATTGGGTGTGACCCCA-3′ |
| gyrB PCR/sequencing primer, forward (bp 1113-1130) | 5′-TTCTCCGATTTCCTCATG-3′ |
| gyrB PCR/sequencing primer, reverse (bp 1518-1536) | 5′-AGAAGGGTACGAATGTGG-3′ |
| parC PCR/sequencing primer, forward (bp 105-122) | 5′-TGGGTTGAAGCCGGTTCA-3′ |
| parC PCR/sequencing primer, reverse (bp 454-471) | 5′-TTGAAGGTTCAGACTATCCG-3′ |
| parE PCR primer, forward (bp 943-961) | 5′-TTGAAGGTTCAGACTATCG-3′ |
| parE PCR/sequencing primer, reverse (bp 1686-1702) | 5′-CGGAGTTCTTCTAGCTC-3′ |
| parE sequencing primer, forward (bp 1167-1183) | 5′-TGAAGCAGCACGTAAGG-3′ |
In vitro time-kill studies.
PD 0305970 and ciprofloxacin were considered to be bactericidal at the lowest concentration that reduced original inoculum levels by ≥3 log10 in 24 h. Time-kill testing was monitored using 10-ml volumes (without agitation) of CAMHB supplemented with 3% lysed horse blood (Remel). PD 0305970 was tested at 0.25×, 1×, 2×, 4×, and 8× MIC (0.5×, 2×, and 8× for ciprofloxacin), and samples were plated for bacterial colony counts at 0, 2, 4, 6, and 24 h (2). Aliquots of 100 μl were cultured on 25 ml tryptic soy agar plates (Becton Dickinson) containing 5% sheep blood (limit of detection of 10 CFU/ml). Drug carryover was reduced by ≥250-fold in the sample dilution. Viable counts were recorded after 24 h of incubation at 35°C.
Murine infection models.
Animal infection model procedures were carried out in compliance with NIH Guidelines for the Care and Use of Laboratory Animals under a protocol approved by the Pfizer Global Research and Development Animal Use Committee.
PD50 determinations.
Six- to eight-week-old (18- to 22-g) CD1 female mice (Charles River, Portage, MI) were used for animal studies. Acute lethal infection was induced by intraperitoneal (i.p.) injection (0.5 ml) with ≥100-fold median lethal challenge doses (3, 17). Todd-Hewitt broth (Difco Laboratories, BD, Sparks, MD) was used as the vehicle for i.p. challenge versus streptococci, and 20% hog gastric mucin (Pfaltz and Bauer, Waterbury, CT) was employed as an adjuvant with the remaining organisms. Treatment was oral at the time of challenge by gavage with 0.5 ml drug solution suspended in 0.5% methylcellulose (Sigma, St. Louis, MO). In untreated controls (10 mice per group), 100% lethality was generally observed within 24 h. The drug dose protecting 50% of challenged mice from lethal bacterial infection (PD50) was expressed in mg/kg and calculated by probit analysis (10).
Pneumococcal pneumonia model.
Streptococcus pneumoniae strains SVI and SP-4333 were grown on plates of tryptic soy agar containing 5% sheep blood for 6 to 8 h at 35°C in 5% CO2. Colonies were inoculated into tryptic soy broth (BD) containing 5% goat serum (Rockland, Gilbertville, PA) and grown to 108 to 109 CFU/ml. The optical density (0.70) at 600 nm was measured using a Beckman DU 530 spectrophotometer (Beckman-Coulter, Fullerton, CA). Lung infection was induced by intranasal instillation of 50 μl undiluted bacterial suspension administered via pipette to mice (n = 8) held upright and under anesthesia by i.p. injection with 0.2 ml of 12.5 mg/kg ketamine HCl (Ketaset; Fort Dodge Animal Health, Fort Dodge, IA). This inoculum ensures 100% mortality in untreated mice. Final inoculum concentrations were confirmed by standard plate counts. Therapy (oral dosing) was initiated 24 h postchallenge and consisted of either a single day of therapy (QD dose) or 3 days of treatment (twice-daily [BID] dosing). PD50s were determined as described previously (10).
RESULTS
In vitro antibacterial activity.
The in vitro antibacterial activities of PD 0305970 and PD 0326448 versus 1,036 clinically significant bacterial strains are shown in Table 2 and compared with those of relevant quinolones. These data are displayed as MICrange, MIC50, and MIC90. PD 0305970 and PD 0326448 demonstrate exceptional antibacterial potency against gram-positive (including multidrug-resistant strains) and fastidious organism groups. PD 0305970 MIC90 values ranged from 0.008 to 0.5 μg/ml versus staphylococci, streptococci, Corynebacterium spp., and fastidious organism groups including Moraxella catarrhalis, Haemophilus influenzae, Listeria monocytogenes, Legionella pneumophila, A. otitidis, and Neisseria spp.; corresponding PD 0326448 MIC90 values were generally two- to fourfold higher against these organism groups. Compared to garenoxacin, gatifloxacin, and levofloxacin, the outstanding antibacterial potency of PD 0305970 is exemplified by an 8- to 512-fold MIC advantage against multidrug-resistant gram-positive strains including: oxacillin-, levofloxacin-, and/or vancomycin-resistant staphylococci; penicillin-, macrolide-, and/or levofloxacin-resistant Streptococcus pneumoniae; and vancomycin- and/or levofloxacin-resistant Enterococcus faecalis and Enterococcus faecium.
TABLE 2.
Antibacterial activities of PD 0305970 and PD 0326448
| Organism (no. of strains), description | Antimicrobial agent | MIC (μg/ml)a
|
||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| Staphylococcus aureus (20), oxacillin susceptible | PD 0305970 | 0.06-0.125 | 0.125 | 0.125 |
| PD 0326448 | 0.125-0.5 | 0.25 | 0.25 | |
| Garenoxacin | 0.015-2 | 0.06 | 0.06 | |
| Gatifloxacin | 0.03-2 | 0.125 | 0.125 | |
| Levofloxacin | 0.125-4 | 0.25 | 0.25 | |
| Staphylococcus aureus (47), oxacillin resistant | PD 0305970 | 0.06-0.5 | 0.125 | 0.25 |
| PD 0326448 | 0.125-1 | 0.25 | 0.5 | |
| Garenoxacin | 0.03-4 | 2 | 2 | |
| Gatifloxacin | 0.06-8 | 4 | 8 | |
| Levofloxacin | 0.125-8 | 8 | 8 | |
| Staphylococcus aureus (35), oxacillin resistant, levofloxacin resistant | PD 0305970 | 0.25-1 | 0.25 | 0.5 |
| PD 0326448 | 0.25-4 | 1 | 4 | |
| Garenoxacin | 0.25-32 | 2 | 4 | |
| Gatifloxacin | 1-64 | 8 | 16 | |
| Levofloxacin | 16->64 | 32 | 64 | |
| Staphylococcus aureus (3), vancomycin intermediate | PD 0305970 | 0.125-0.25 | ||
| PD 0326448 | 0.25-1 | |||
| Garenoxacin | 1-4 | |||
| Gatifloxacin | 8-16 | |||
| Levofloxacin | 16-64 | |||
| Vancomycin | 4-8 | |||
| Staphylococcus epidermidis (5), oxacillin susceptible | PD 0305970 | 0.06-0.125 | ||
| PD 0326448 | 0.125-0.25 | |||
| Garenoxacin | 0.06-4 | |||
| Gatifloxacin | 0.125-2 | |||
| Levofloxacin | 0.25-16 | |||
| Staphylococcus epidermidis (23), oxacillin resistant | PD 0305970 | 0.06-0.125 | 0.125 | 0.125 |
| PD 0326448 | 0.125-0.5 | 0.25 | 0.25 | |
| Garenoxacin | 0.06-32 | 2 | 32 | |
| Gatifloxacin | 0.125-64 | 2 | 32 | |
| Levofloxacin | 0.25->64 | 8 | 64 | |
| Staphylococcus haemolyticus (10) | PD 0305970 | 0.06-0.5 | 0.125 | 0.5 |
| PD 0326448 | 0.125-1 | 0.25 | 1 | |
| Garenoxacin | 0.03-8 | 0.06 | 4 | |
| Gatifloxacin | 0.06-8 | 0.125 | 4 | |
| Levofloxacin | 0.06-64 | 0.25 | 32 | |
| Staphylococcus saprophyticus (10) | PD 0305970 | 0.03-0.125 | 0.06 | 0.125 |
| PD 0326448 | 0.125-0.5 | 0.125 | 0.5 | |
| Garenoxacin | 0.03-0.06 | 0.03 | 0.06 | |
| Gatifloxacin | 0.25-0.5 | 0.25 | 0.25 | |
| Levofloxacin | 0.125-1 | 0.25 | 0.5 | |
| Staphylococcus warneri (3) | PD 0305970 | 0.06 | ||
| PD 0326448 | 0.125 | |||
| Garenoxacin | 0.03-0.125 | |||
| Gatifloxacin | 0.125 | |||
| Levofloxacin | 0.125-0.25 | |||
| Alloiococcus otitidis (15) | PD 0305970 | 0.004-0.008 | 0.004 | 0.008 |
| PD 0326448 | 0.004-0.008 | 0.008 | 0.008 | |
| Garenoxacin | ≤0.001 | ≤0.001 | ≤0.001 | |
| Gatifloxacin | 0.03-0.125 | 0.06 | 0.06 | |
| Levofloxacin | 0.125-0.25 | 0.25 | 0.25 | |
| Streptococcus pyogenes (28) | PD 0305970 | 0.015-0.06 | 0.03 | 0.03 |
| PD 0326448 | 0.015-0.06 | 0.03 | 0.03 | |
| Garenoxacin | 0.03-0.25 | 0.125 | 0.25 | |
| Gatifloxacin | 0.125-1 | 0.25 | 0.5 | |
| Levofloxacin | 0.25-2 | 0.5 | 2 | |
| Range | 50% | 90% | ||
| Streptococcus agalactiae (21) | PD 0305970 | 0.03-0.06 | 0.03 | 0.03 |
| PD 0326448 | 0.015-0.125 | 0.015 | 0.03 | |
| Garenoxacin | 0.03-0.125 | 0.125 | 0.125 | |
| Gatifloxacin | 0.25 | 0.25 | 0.25 | |
| Levofloxacin | 0.5-1 | 1 | 1 | |
| Streptococcus bovis (14) | PD 0305970 | 0.015-0.06 | 0.03 | 0.03 |
| PD 0326448 | 0.03-0.06 | 0.06 | 0.06 | |
| Garenoxacin | 0.125-0.5 | 0.25 | 0.25 | |
| Gatifloxacin | 0.5-1 | 1 | 1 | |
| Levofloxacin | 1-4 | 2 | 2 | |
| Streptococcus spp. (22), viridans groupb | PD 0305970 | 0.008-0.06 | 0.03 | 0.03 |
| PD 0326448 | 0.015-0.06 | 0.03 | 0.06 | |
| Garenoxacin | 0.06-0.25 | 0.125 | 0.25 | |
| Gatifloxacin | 0.125-1 | 0.5 | 0.5 | |
| Levofloxacin | 0.25-1 | 1 | 1 | |
| Streptococcus pneumoniae (23), penicillin susceptible | PD 0305970 | 0.015-0.06 | 0.03 | 0.03 |
| PD 0326448 | 0.015-0.125 | 0.03 | 0.06 | |
| Garenoxacin | 0.03-1 | 0.125 | 0.125 | |
| Gatifloxacin | 0.125-4 | 0.25 | 0.5 | |
| Levofloxacin | 0.5-16 | 0.5 | 1 | |
| Streptococcus pneumoniae (27), penicillin intermediate | PD 0305970 | 0.03-0.06 | 0.03 | 0.06 |
| PD 0326448 | 0.03-0.125 | 0.06 | 0.125 | |
| Garenoxacin | 0.06-1 | 0.25 | 0.25 | |
| Gatifloxacin | 0.125-4 | 0.25 | 0.5 | |
| Levofloxacin | 0.25-16 | 1 | 1 | |
| Streptococcus pneumoniae (40), penicillin resistant | PD 0305970 | 0.008-0.125 | 0.03 | 0.03 |
| PD 0326448 | 0.015-0.25 | 0.03 | 0.06 | |
| Garenoxacin | 0.004-0.5 | 0.03 | 0.125 | |
| Gatifloxacin | 0.125-2 | 0.25 | 1 | |
| Levofloxacin | 0.25-4 | 0.5 | 1 | |
| Streptococcus pneumoniae (26), levofloxacin resistant | PD 0305970 | 0.03-0.25 | 0.03 | 0.06 |
| PD 0326448 | 0.03-0.5 | 0.06 | 0.06 | |
| Garenoxacin | 0.25-2 | 1 | 2 | |
| Gatifloxacin | 2-8 | 4 | 4 | |
| Levofloxacin | 8-32 | 16 | 16 | |
| Enterococcus faecalis (19) | PD 0305970 | 0.125-0.25 | 0.125 | 0.25 |
| PD 0326448 | 0.125-0.5 | 0.25 | 0.5 | |
| Garenoxacin | 0.06-4 | 2 | 4 | |
| Gatifloxacin | 0.25-16 | 8 | 16 | |
| Levofloxacin | 1-32 | 16 | 32 | |
| Enterococcus faecalis (6), VanA | PD 0305970 | 0.125-0.25 | ||
| PD 0326448 | 0.25 | |||
| Garenoxacin | 2-8 | |||
| Gatifloxacin | 8-16 | |||
| Levofloxacin | 16-32 | |||
| Enterococcus faecalis (22), VanB | PD 0305970 | 0.06-0.5 | 0.25 | 0.25 |
| PD 0326448 | 0.125-0.5 | 0.25 | 0.5 | |
| Garenoxacin | 0.06-8 | 2 | 8 | |
| Gatifloxacin | 0.25-16 | 8 | 16 | |
| Levofloxacin | 0.25-32 | 16 | 32 | |
| Enterococcus faecalis (7), β-lactamase positive | PD 0305970 | 0.06-0.125 | ||
| PD 0326448 | 0.06-0.25 | |||
| Garenoxacin | 0.125-0.25 | |||
| Gatifloxacin | 0.25-0.5 | |||
| Levofloxacin | 0.5-1 | |||
| Enterococcus faecium (13) | PD 0305970 | 0.015-2 | 1 | 2 |
| Range | 50% | 90% | ||
| PD 0326448 | 0.03-4 | 1 | 4 | |
| Garenoxacin | 0.03-64 | 8 | 32 | |
| Gatifloxacin | 0.06->64 | 8 | 64 | |
| Levofloxacin | 0.125->64 | 32 | 64 | |
| Enterococcus faecium (46), VanA | PD 0305970 | 0.25-8 | 1 | 2 |
| PD 0326448 | 0.25-8 | 1 | 2 | |
| Garenoxacin | 2-32 | 8 | 16 | |
| Gatifloxacin | 0.5->64 | 32 | 64 | |
| Levofloxacin | 1->64 | 64 | 64 | |
| Enterococcus faecium (3), VanB | PD 0305970 | 0.5-2 | ||
| PD 0326448 | 1-2 | |||
| Garenoxacin | 4-32 | |||
| Gatifloxacin | 2-64 | |||
| Levofloxacin | 4-64 | |||
| Enterococcus casseliflavus (10) | PD 0305970 | 0.125-0.5 | 0.25 | 0.25 |
| PD 0326448 | 0.125-0.5 | 0.25 | 0.25 | |
| Garenoxacin | 0.125-0.5 | 0.25 | 0.5 | |
| Gatifloxacin | 0.25-1 | 0.5 | 1 | |
| Levofloxacin | 0.25-2 | 2 | 2 | |
| Enterococcus gallinarum (11) | PD 0305970 | 0.125-0.25 | 0.25 | 0.25 |
| PD 0326448 | 0.125-0.5 | 0.25 | 0.5 | |
| Garenoxacin | 0.125-32 | 0.25 | 2 | |
| Gatifloxacin | 0.25-32 | 0.5 | 1 | |
| Levofloxacin | 0.25-32 | 1 | 2 | |
| Listeria monocytogenes (10) | PD 0305970 | 0.125-0.25 | 0.125 | 0.25 |
| PD 0326448 | 0.125-0.25 | 0.25 | 0.25 | |
| Garenoxacin | 0.125-1 | 0.25 | 0.5 | |
| Gatifloxacin | 0.5-1 | 0.5 | 1 | |
| Levofloxacin | 0.5-1 | 1 | 1 | |
| Corynebacterium jeikeium (10) | PD 0305970 | 0.125-0.5 | 0.25 | 0.25 |
| PD 0326448 | 0.25-2 | 0.5 | 1 | |
| Garenoxacin | 0.125-0.5 | 0.25 | 0.25 | |
| Gatifloxacin | 0.125-0.5 | 0.125 | 0.5 | |
| Levofloxacin | 0.25-0.5 | 0.25 | 0.5 | |
| Corynebacterium spp. (20) | PD 0305970 | 0.03-0.5 | 0.25 | 0.25 |
| PD 0326448 | 0.06-1 | 0.5 | 1 | |
| Garenoxacin | 0.015->64 | 0.25 | 64 | |
| Gatifloxacin | 0.03-32 | 0.25 | 32 | |
| Levofloxacin | 0.125-64 | 0.5 | 32 | |
| Haemophilus influenzae (31), β-lactamase negative | PD 0305970 | 0.06-0.25 | 0.125 | 0.25 |
| PD 0326448 | 0.125-0.25 | 0.25 | 0.5 | |
| Garenoxacin | 0.004-0.06 | 0.008 | 0.03 | |
| Gatifloxacin | 0.004-0.03 | 0.008 | 0.015 | |
| Levofloxacin | 0.004-0.06 | 0.015 | 0.03 | |
| Haemophilus influenzae (36), β-lactamase positive | PD 0305970 | 0.125-0.5 | 0.125 | 0.25 |
| PD 0326448 | 0.25-1 | 0.5 | 1 | |
| Garenoxacin | 0.004-0.06 | 0.015 | 0.03 | |
| Gatifloxacin | 0.008-0.25 | 0.015 | 0.015 | |
| Levofloxacin | 0.004-0.5 | 0.008 | 0.015 | |
| Legionella pneumophila (10) | PD 0305970 | 0.25-0.5 | 0.25 | 0.25 |
| PD 0326448 | 0.25-1 | 0.5 | 0.5 | |
| Garenoxacin | ≤0.001-0.015 | 0.002 | 0.008 | |
| Gatifloxacin | ≤0.001-0.008 | 0.008 | 0.008 | |
| Levofloxacin | 0.002-0.015 | 0.008 | 0.015 | |
| Moraxella catarrhalis (15), β-lactamase negative | PD 0305970 | 0.125-0.5 | 0.25 | 0.5 |
| PD 0326448 | 0.25-1 | 0.5 | 1 | |
| Range | 50% | 90% | ||
| Garenoxacin | 0.015-0.03 | 0.03 | 0.03 | |
| Gatifloxacin | 0.03-0.06 | 0.06 | 0.06 | |
| Levofloxacin | 0.03-0.06 | 0.03 | 0.06 | |
| Moraxella catarrhalis (15), β-lactamase positive | PD 0305970 | 0.25-0.5 | 0.25 | 0.5 |
| PD 0326448 | 0.5-1 | 0.5 | 0.5 | |
| Garenoxacin | 0.03-0.125 | 0.06 | 0.06 | |
| Gatifloxacin | 0.03-0.06 | 0.03 | 0.06 | |
| Levofloxacin | 0.03-0.06 | 0.03 | 0.06 | |
| Neisseria spp.c (16) | PD 0305970 | 0.125-0.5 | 0.25 | 0.5 |
| PD 0326448 | 0.125-1 | 0.5 | 1 | |
| Garenoxacin | ≤0.001-0.008 | 0.004 | 0.004 | |
| Gatifloxacin | ≤0.001-0.125 | 0.004 | 0.008 | |
| Levofloxacin | 0.004-0.125 | 0.008 | 0.008 | |
| Citrobacter diversus (10) | PD 0305970 | 0.5-8 | 1 | 4 |
| PD 0326448 | 1-8 | 2 | 8 | |
| Garenoxacin | 0.03-0.5 | 0.125 | 0.25 | |
| Gatifloxacin | 0.015-0.125 | 0.03 | 0.06 | |
| Levofloxacin | 0.015-0.125 | 0.03 | 0.06 | |
| Citrobacter freundii (20) | PD 0305970 | 0.5-64 | 4 | 16 |
| PD 0326448 | 1->64 | 8 | >64 | |
| Garenoxacin | 0.06->64 | 1 | >64 | |
| Gatifloxacin | 0.03-64 | 0.25 | 16 | |
| Levofloxacin | 0.015->64 | 0.125 | 16 | |
| Enterobacter aerogenes (17) | PD 0305970 | 0.5-8 | 4 | 8 |
| PD 0326448 | 1-64 | 8 | 64 | |
| Garenoxacin | 0.125->64 | 2 | >64 | |
| Gatifloxacin | 0.03-32 | 1 | 32 | |
| Levofloxacin | 0.06-32 | 1 | 32 | |
| Enterobacter cloacae (21) | PD 0305970 | 0.5-16 | 1 | 4 |
| PD 0326448 | 1-32 | 2 | 16 | |
| Garenoxacin | 0.06->64 | 0.125 | 2 | |
| Gatifloxacin | 0.015-32 | 0.06 | 1 | |
| Levofloxacin | 0.008-64 | 0.06 | 1 | |
| Escherichia coli (31) | PD 0305970 | 0.5-16 | 1 | 2 |
| PD 0326448 | 1-64 | 2 | 8 | |
| Garenoxacin | 0.015->64 | 0.06 | 1 | |
| Gatifloxacin | 0.015-32 | 0.03 | 1 | |
| Levofloxacin | 0.008-64 | 0.03 | 1 | |
| Klebsiella oxytoca (13) | PD 0305970 | 0.5-4 | 1 | 2 |
| PD 0326448 | 1-16 | 2 | 16 | |
| Garenoxacin | 0.125-2 | 0.125 | 2 | |
| Gatifloxacin | 0.03-1 | 0.03 | 1 | |
| Levofloxacin | 0.03-2 | 0.06 | 2 | |
| Klebsiella pneumoniae (30) | PD 0305970 | 0.5-16 | 2 | 4 |
| PD 0326448 | 2-64 | 4 | 16 | |
| Garenoxacin | 0.06-64 | 0.25 | 1 | |
| Gatifloxacin | 0.03-16 | 0.06 | 0.5 | |
| Levofloxacin | 0.03-16 | 0.06 | 0.5 | |
| Klebsiella pneumoniae (14), extended-spectrum β-lactamase positive | PD 0305970 | 0.5-16 | 2 | 8 |
| PD 0326448 | 4-64 | 8 | 64 | |
| Garenoxacin | 0.06-64 | 1 | 64 | |
| Gatifloxacin | 0.06-16 | 1 | 16 | |
| Levofloxacin | 0.03-32 | 0.5 | 32 | |
| Morganella morganii (13) | PD 0305970 | 0.5-8 | 2 | 8 |
| PD 0326448 | 1-32 | 2 | 32 | |
| Garenoxacin | 0.25-64 | 2 | 32 | |
| Range | 50% | 90% | ||
| Gatifloxacin | 0.06-32 | 0.25 | 32 | |
| Levofloxacin | 0.03-64 | 0.125 | 16 | |
| Proteus mirabilis (11) | PD 0305970 | 1-2 | 1 | 2 |
| PD 0326448 | 2-8 | 2 | 8 | |
| Garenoxacin | 0.125-16 | 0.5 | 16 | |
| Gatifloxacin | 0.06-4 | 0.25 | 4 | |
| Levofloxacin | 0.03-4 | 0.125 | 2 | |
| Proteus vulgaris (6) | PD 0305970 | 1-2 | ||
| PD 0326448 | 2 | |||
| Garenoxacin | 0.25-0.5 | |||
| Gatifloxacin | 0.06-0.125 | |||
| Levofloxacin | 0.03-0.06 | |||
| Salmonella/Shigella spp.d (12) | PD 0305970 | 0.25-1 | 0.5 | 1 |
| PD 0326448 | 0.5-4 | 2 | 4 | |
| Garenoxacin | 0.03-0.125 | 0.03 | 0.125 | |
| Gatifloxacin | 0.015-0.06 | 0.03 | 0.06 | |
| Levofloxacin | 0.015-0.06 | 0.03 | 0.06 | |
| Serratia marcescens (15) | PD 0305970 | 1-16 | 2 | 16 |
| PD 0326448 | 2-64 | 4 | 32 | |
| Garenoxacin | 0.25-32 | 1 | 8 | |
| Gatifloxacin | 0.06-16 | 0.25 | 4 | |
| Levofloxacin | 0.06-16 | 0.125 | 4 | |
| Acinetobacter spp.e (19) | PD 0305970 | 0.5-8 | 1 | 8 |
| PD 0326448 | 0.5-64 | 4 | 64 | |
| Garenoxacin | 0.015-32 | 0.125 | 16 | |
| Gatifloxacin | 0.015-16 | 0.03 | 16 | |
| Levofloxacin | 0.03-16 | 0.125 | 16 | |
| Burkholderia cepacia (24) | PD 0305970 | 0.125->64 | 32 | 64 |
| PD 0326448 | 0.25->64 | 32 | >64 | |
| Garenoxacin | 0.03->64 | 32 | >64 | |
| Gatifloxacin | 0.125-64 | 4 | 16 | |
| Levofloxacin | 0.25-32 | 4 | 16 | |
| Pseudomonas aeruginosa (20) | PD 0305970 | 2-64 | 4 | 32 |
| PD 0326448 | 2->64 | 8 | >64 | |
| Garenoxacin | 0.5->64 | 2 | >64 | |
| Gatifloxacin | 0.5-64 | 1 | 64 | |
| Levofloxacin | 0.5->64 | 1 | 64 | |
| Stenotrophomonas maltophilia (17) | PD 0305970 | 4->64 | 8 | 64 |
| PD 0326448 | 8->64 | 32 | 64 | |
| Garenoxacin | 2->64 | 4 | 8 | |
| Gatifloxacin | 0.5-16 | 1 | 8 | |
| Levofloxacin | 0.5-16 | 1 | 4 | |
| Actinomyces spp. (4) | PD 0305970 | 0.5-1 | ||
| PD 0326448 | 1-2 | |||
| Garenoxacin | 0.125-2 | |||
| Gatifloxacin | 0.125-1 | |||
| Levofloxacin | 0.25-1 | |||
| Bacteroides fragilis (13) | PD 0305970 | 0.5-4 | 2 | 4 |
| PD 0326448 | 2-16 | 8 | 16 | |
| Garenoxacin | 0.25-4 | 0.5 | 2 | |
| Gatifloxacin | 0.5-32 | 1 | 8 | |
| Levofloxacin | 1-64 | 4 | 32 | |
| Bacteroides fragilis groupf (13) | PD 0305970 | 1-16 | 4 | 16 |
| PD 0326448 | 4-64 | 16 | 32 | |
| Garenoxacin | 0.25-4 | 2 | 4 | |
| Gatifloxacin | 1-16 | 4 | 16 | |
| Levofloxacin | 2->64 | 8 | 32 | |
| Range | 50% | 90% | ||
| Clostridium spp.g (11) | PD 0305970 | 0.25-2 | 0.5 | 2 |
| PD 0326448 | 0.5-8 | 2 | 8 | |
| Garenoxacin | 0.125-2 | 0.5 | 1 | |
| Gatifloxacin | 0.125-1 | 0.5 | 0.5 | |
| Levofloxacin | 0.25-4 | 0.5 | 0.5 | |
| Fusobacterium spp.h (8) | PD 0305970 | 0.06-0.125 | ||
| PD 0326448 | 0.125-0.25 | |||
| Garenoxacin | 0.125-1 | |||
| Gatifloxacin | 0.015-0.5 | |||
| Levofloxacin | 0.03-1 | |||
| Peptostreptococcus spp.i (8) | PD 0305970 | 0.03-1 | ||
| PD 0326448 | 0.06-2 | |||
| Garenoxacin | 0.015-32 | |||
| Gatifloxacin | 0.06-16 | |||
| Levofloxacin | 0.125-64 | |||
| Prevotella spp.j (11) | PD 0305970 | 0.03-1 | 0.25 | 1 |
| PD 0326448 | 0.03-4 | 0.5 | 2 | |
| Garenoxacin | 0.008-1 | 0.25 | 1 | |
| Gatifloxacin | 0.06-2 | 0.25 | 0.5 | |
| Levofloxacin | 0.125-4 | 1 | 2 | |
| Propionibacterium acnes (13) | PD 0305970 | 0.5-4 | 2 | 4 |
| PD 0326448 | 2-16 | 4 | 8 | |
| Garenoxacin | 0.5-8 | 1 | 2 | |
| Gatifloxacin | 0.25-16 | 1 | 8 | |
| Levofloxacin | 0.125->64 | 2 | 64 | |
50% and 90%, MICs for 50 and 90% of isolates tested, respectively.
Includes 1 S. sanguis, 1 S. salivarius, 1 S. mitis, and 19 S. viridans strains.
Includes 4 N. gonorrhoeae and 12 N. meningitidis strains.
Includes one Salmonella enterica serovar Enteritidis, one S. enterica serovar Paratyphi, two S. enterica serovar Typhimurium, two S. enterica serovar Typhi, two Shigella dysenteriae, two Shigella flexneri, and two Shigella sonnei strains.
Includes 19 Acinetobacter strains.
Includes one Bacteroides distasonis, one B. ovatus, two B. thetaiotaomicron, and nine B. fragilis group strains.
Includes 1 C. difficile and 10 C. perfringens strains.
Includes six Fusobacterium nucleatum, one Fusobacterium necrophorum, and one Fusobacterium sp. strain.
Includes four Peptostreptococcus asaccharolyticus, two P. anaerobius, one P. magnus, and one P. prevotii strain.
Includes two Prevotella bivia, two P. buccae, one P. corporis, three P. melaninogenica, one P. oralis, and two Prevotella sp. strains
Activity versus gram-negative strains was more modest, with PD 0305970 MIC90s ranging from 1 to 2 μg/ml versus Escherichia coli, Klebsiella oxytoca, Proteus mirabilis, and Salmonella/Shigella spp. PD 0305970 was the most active compound tested (MIC90 = 8 μg/ml) versus Enterobacter aerogenes, K. pneumoniae (extend-spectrum β-lactamase positive), Morganella morganii, and Acinetobacter spp.
Spontaneous mutant selection.
Spontaneous mutation frequencies for PD 0305970, ciprofloxacin, gatifloxacin, and levofloxacin are reported in Table 3. At 2× the ciprofloxacin MIC, the frequency of resistance development for S. aureus UC-76 was high (too numerous to count [TNTC]). In contrast, the frequency of S. aureus resistance at 2× the MIC of PD 0305970 was 3 × 10−7. Ciprofloxacin resistance development (3 × 10−7 and <6 × 10−8) at 4× and 8× the MIC versus S. aureus UC-76 correlates with data reported in the literature (8). Ciprofloxacin resistance in S. pneumoniae 7785 showed low frequencies at 1× to 8× the MIC as reported previously (1 × 10−9 to 6 × 10−10) (7, 12). Spontaneous resistant mutants were not obtained for PD 0305970 (1× to 8× MIC, <6 × 10−10) and levofloxacin (4× MIC, <1.2 × 10−10) versus S. pneumoniae 7785. Likewise, against S. aureus UC-76 at 4× the MIC, PD 305970- and levofloxacin-resistant mutants were not obtained (<6 × 10−8 and 1.2 × 10−10, respectively).
TABLE 3.
Determinations of frequency of resistance to PD 0305970
| Organism and drug | MIC
|
|||
|---|---|---|---|---|
| 1× | 2× | 4× | 8× | |
| S. aureus UC-76 (SA-1) | ||||
| PD 0305970 (MIC = 0.03 μg/ml) | 5 × 10−7 | 3 × 10−7 | < 6 × 10−8 | <6 × 10−8 |
| Ciprofloxacin (MIC = 0.125 μg/ml) | TNTC | TNTC | 3 × 10−7 | <6 × 10−8 |
| Levofloxacin (MIC = 0.125 μg/ml) | Not tested | Not tested | 8.9 × 10−9 | Not tested |
| S. pneumoniae 7785 (SP-2870) | ||||
| PD 0305970 (MIC = 0.03 μg/ml) | <6 × 10−10 | <6 × 10−10 | <6 × 10−10 | <6 × 10−10 |
| Ciprofloxacin (MIC = 1 μg/ml) | 6 × 10−10 | 1 × 10−9 | <6 × 10−10 | <6 × 10−10 |
| Gatifloxacin (MIC = 0.25 μg/ml) | Not tested | 3 × 10−8 | <1 × 10−9 | <1 × 10−9 |
| Levofloxacin (MIC = 0.5 μg/ml) | Not tested | Not tested | 1.2 × 10−10 | Not tested |
Mechanism-of-action studies.
Resistance mutation analysis of S. pneumoniae 7785 (Table 4) suggests that PD 0305970 targets primarily bacterial gyrB and parE DNA topoisomerases. In contrast, quinolones target gyrA and parC. The proposed mechanism of action of PD 0305970 is corroborated by low MIC90 values against S. pneumoniae. PD 0305970 MIC90s were 4- to 32-fold lower than those of garenoxacin, gatifloxacin, and levofloxacin against penicillin-susceptible S. pneumoniae and 32- to 256-fold lower versus levofloxacin-resistant strains. This same trend was observed for Staphylococcus aureus. PD 0305970 MIC90s increased 4-fold, from 0.125 to 0.5 μg/ml (oxacillin-susceptible S. aureus compared to levofloxacin-resistant and oxacillin-resistant S. aureus), while garenoxacin, gatifloxacin, and levofloxacin MIC90s increased 64- to 256-fold. The fourfold MIC increase observed with PD 0305970 versus levofloxacin- and oxacillin-resistant S. aureus strains may be explained by the up-regulation of the norA efflux pump. Ciprofloxacin-resistant S. aureus mutants raised during multistep passage studies were tested for cross-resistance to PD 0305970. A fourfold MIC increase (0.06 to 0.25 μg/ml) was observed in strains containing the up-regulated norA efflux pump. No additional PD 0305970 MIC increases were observed in strains containing norA and mutations in grlA (Ser-80-Phe) and/or gyrA (Ser-84-Leu).
TABLE 4.
Mutations identified in the quinolone resistance-determining region (gyrA, gyrB, parC, and parE) of S. pneumoniae 7785 selected by PD 0305970
| Strain | MIC (μg/ml) of PD 0305970 | Mutation in QRDRa
|
|||
|---|---|---|---|---|---|
| gyrA | gyrB | parC | parE | ||
| 7785 | 0.03 | None | None | None | None |
| 1-970-1 | 0.06 | None | Asp435Glu | None | None |
| 1-970-2 | 0.125 | None | Asp435Glu Glu475Ala | None | Pro475Ser |
QRDR, quinolone resistance-determining region.
In vitro time-kill studies.
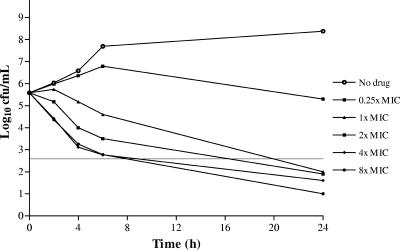
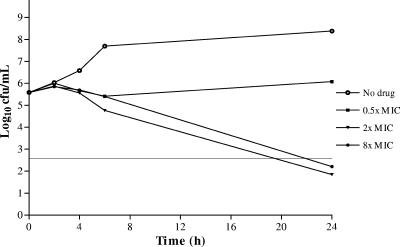
The bactericidal activities of PD 0305970 and ciprofloxacin were compared by in vitro time-kill versus S. pneumoniae SVI and S. pyogenes C203 (Fig. 3 to 6). PD 0305970 MICs were 128- and 256-fold higher than those of ciprofloxacin versus SVI and C203, respectively. Rapid and sustained bactericidal activity was demonstrated by PD 0305970 and ciprofloxacin versus both strains at concentrations greater than or equal to their respective MICs over the 24 h measured.
FIG. 3.
PD 0305970 (MIC = 0.008 μg/ml) versus S. pneumoniae SVI.
PD50 determinations in a sepsis model.
Median protective doses for PD 0305970, ciprofloxacin, and levofloxacin are presented in Table 5. Outstanding in vivo potency was observed for PD 0305970 versus Streptococcus pyogenes C203 and S. pneumoniae SVI (PD50s of 0.7 mg/kg and 2.7 mg/kg, respectively). Levofloxacin PD50s were 25 times higher, and ciprofloxacin treatment was ineffective (PD50 of >100 mg/kg). PD 0305970 PD50s versus S. aureus UC-76 (methicillin susceptible) were 2.4- and 4.8-fold greater than levofloxacin and ciprofloxacin. Against SA-1417 (methicillin-resistant Staphylococcus aureus [MRSA]), PD 0305970 was 7.2-fold more potent than ciprofloxacin. PD 0305970 retained in vivo activity (PD50 of 23 mg/kg) against a ciprofloxacin- and levofloxacin-resistant S. aureus strain (SA-2017) (MRSA).
TABLE 5.
Antibacterial efficacy in a murine acute lethal infection model
| Infecting organism | PD50a ± 95% confidence limit (mg/kg) (MIC [μg/ml])
|
||
|---|---|---|---|
| PD 0305970 | Ciprofloxacin | Levofloxacin | |
| S. aureus UC-76 (MSSAb) | 1.9 ± 0.7 (0.06) | 9.1 ± 2.2 (0.06) | 4.6 ± 1.2 (0.125) |
| S. aureus SA-1417 (MRSA) | 2.5 ± 1.8 (0.25) | 18.0 ± 6.9 (0.5) | Not tested (0.25) |
| S. aureus SA-2017 (CMRSAc) | 23 ± 5.2 (0.25) | >100 (>64) | Not tested (8) |
| Streptococcus pneumoniae SVI | 2.7 ± 0.8 (0.015) | >100 (2) | 69 ± 37 (0.5) |
| S. pyogenes C203d | 0.7 ± 0.1 (0.015) | >100 (0.5) | 17.7 ± 2.1 (0.5) |
| Enterococcus faecalis MGH-2 | 19 ± 6.1 (0.125) | Not tested (0.5) | Not tested (1) |
| Escherichia coli Vogel | 82 ± 31 (1) | Not tested (0.03) | Not tested (0.06) |
Oral drug dose protecting 50% of mice challenged with 100 50% lethal doses from a lethal bacterial infection.
MSSA, methicillin-susceptible Staphylococcus aureus.
CMRSA, ciprofloxacin- and methicillin-resistant Staphylococcus aureus.
PD 0326448 PD50 = 3.6 mg/kg (MIC = 0.015 μg/ml).
Efficacy in a pneumococcal pneumonia model.
PD 0305970 demonstrated excellent oral effectiveness against pneumococcal pneumonia (Table 6). PD 0305970 therapy versus S. pneumoniae SVI (levofloxacin susceptible, QD for 1 day) and SP-4333 (levofloxacin-resistant, BID for 3 days) resulted in PD50 values of 3.2 and 4.1 mg/kg, respectively. Levofloxacin PD50 values were considerably higher and ranged from 69 mg/kg (QD for 1 day versus SVI) to >200 mg/kg (BID for 3 days versus SP-4333).
TABLE 6.
Efficacy in a murine pneumococcal pneumonia model
| Infecting organism | Treatment | PD50a ± 95% confidence limit (mg/kg) (MIC [μg/ml])
|
|
|---|---|---|---|
| PD 0305970 | Levofloxacin | ||
| Streptococcus pneumoniae SVI | QD × 1 day | 3.2 ± 3.8 (0.008) | 69 ± 37 (0.5) |
| S. pneumoniae SP-4333 (quinolone resistant) | BID × 3 days | 4.1 ± 1.4 (0.03) | >200 (32) |
PD50, oral drug dose protecting 50% of challenged mice from lethal bacterial infection.
DISCUSSION
With the continuing emergence and development of antimicrobial resistance, the currently marketed quinolones now have significant gaps in their antibacterial spectra. PD 0305970 and PD 0326448 were developed to introduce an orally active quinazolinedione that fills this unmet medical need. Overall, antibacterial properties of the quinazolinediones (PD 0305970 and PD 0326448) versus gram-positive and fastidious organism groups were similar or superior to those of currently available quinolones (low MICs, bactericidal, low frequency of resistance development, and high in vivo potency). The advantages of PD 0305970 and PD 0326448 are clearly demonstrated by antibacterial activity reflected in their MICs versus gram-positive clinical isolates containing frequently encountered quinolone resistance mutations and the lack of cross-resistance to other drug classes. This high potency versus quinolone-resistant strains may be explained by the targeting of the gyrB and parE DNA topoisomerase subunits by PD 0305970 in S. pneumoniae. Additional studies of both S. aureus and S. pneumoniae will help to confirm the mechanism of action. This activity is noteworthy due to rising antibacterial resistance being observed in S. aureus (methicillin-resistant, ciprofloxacin- and methicillin-resistant, and vancomycin-intermediate isolates), enterococci (vancomycin), and S. pneumoniae (penicillin, macrolides, and quinolones). The low PD50s observed for PD 0305970 relative to ciprofloxacin and levofloxacin in both acute sepsis and pneumococcal pneumonia animal infection models suggest the potential clinical utility of this compound.
The activity versus quinolone-resistant strains, lack of cross-resistance to other drug classes, potent in vivo activity, and high degree of antibacterial activity associated with PD 0305970 and PD 0326448 against multidrug-resistant gram-positive and fastidious bacterial strains provide support for additional exploration into the quinazolinedione antibacterial drug class.
FIG. 4.
Ciprofloxacin (MIC = 1 μg/ml) versus S. pneumoniae SVI.
FIG. 5.
PD 0305970 (MIC = 0.008 μg/ml) versus S. pyogenes C203.
FIG. 6.
Ciprofloxacin (MIC = 2 μg/ml) versus S. pyogenes C203.
Footnotes
Published ahead of print on 29 January 2007.
REFERENCES
- 1.Bassetti, M., G. Melica, A. Di Biagio, E. Righi, R. Rosso, and D. Bassetti. 2004. New antibiotics for treatment of serious infections due to antibiotic-resistant gram-positive cocci. Rev. Med. Microbiol. 15:109-117. [Google Scholar]
- 2.Cohen, M. A., M. D. Huband, S. L. Yoder, J. W. Gage, and G. E. Roland. 1998. Bacterial eradication by clinafloxacin, CI-990, and ciprofloxacin employing MBC test, in-vitro time-kill and in vivo time-kill studies. J. Antimicrob. Chemother. 41:605-614. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, M. A., S. L. Yoder, M. D. Huband, G. E. Roland, and C. L. Courtney. 1995. In vitro and in vivo activities of clinafloxacin, CI-990 (PD 131112), and PD 138312 versus enterococci. Antimicrob. Agents Chemother. 39:2123-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman, K. 2004. Recent advances in the treatment of gram-positive infections. Drug Discov. Today 1:455-460. [Google Scholar]
- 5.Edelstein, P. H., M. A. C. Edelstein, J. Weidenfeld, and M. B. Dorr. 1990. In vitro activity of sparfloxacin (CI-978; AT-4140) for clinical Legionella isolates, pharmacokinetics in guinea pigs, and use to treat guinea pigs with L. pneumophila pneumonia. Antimicrob. Agents Chemother. 34:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellsworth, E. L., T. P. Tuan, H. D. Showalter, J. P. Sanchez, B. M. Watson, M. A. Stier, J. M. Domagala, S. J. Gracheck, E. T. Joannides, M. A. Shapiro, S. Dunham, D. L. Hanna, M. D. Huband, J. W. Gage, D. Q. Nguyen, and R. Singh. 2006. 3-Aminoquinazolinediones as a new class of antibacterial agents demonstrating excellent antibacterial activity against wild-type and multi-drug resistant organisms. J. Med. Chem. 49:6435-6438. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda, H., and K. Hiramatsu. 1999. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:410-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda, H., S. Hori, and K. Hiramatsu. 1998. Antibacterial activity of gatifloxacin (AM-1155, CG5501, BMS-206584), a newly developed fluoroquinolone, against sequentially acquired quinolone-resistant mutants and the norA transformant of Staphylococcus aureus. Antimicrob. Agents Chemother. 42:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges, and responses. Nat. Med. 10(Suppl. 12):S122-S129. [DOI] [PubMed] [Google Scholar]
- 10.Miller, L. C., and M. L. Tainter. 1944. Estimation of the ED50 and its error by means of logarithmic-probit graph paper. Proc. Soc. Exp. Biol. Med. 57:261-264. [Google Scholar]
- 11.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken. 2003. Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 12.Nagai, K., T. A. Davis, G. A. Pankuch, B. E. Dewasse, M. R. Jacobs, and P. C. Appelbaum. 2000. In vitro selection of resistance to clinafloxacin, ciprofloxacin, and trovafloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2740-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NCCLS. 2001. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard, 5th ed. NCCLS document M11-A5. NCCLS, Wayne, PA.
- 14.NCCLS. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 6th ed. NCCLS document M7-A6. NCCLS, Wayne, PA.
- 15.NCCLS. 2003. Performance standards for antimicrobial susceptibility testing, thirteenth informational supplement. NCCLS document M100-S13. NCCLS, Wayne, PA.
- 16.Norrby, R. S., C. E. Nord, and R. Finch. 2005. Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect. Dis. 5:15-119. [DOI] [PubMed] [Google Scholar]
- 17.Sesnie, J. C., P. W. Fritsch, T. J. Griffin, C. L. Heifetz, E. T. Leopold, T. E. Malta, M. A. Shapiro, and P. W. Vincent. 1989. Comparative chemotherapeutic activity of new fluorinated 4-quinolones and standard agents against a variety of bacteria in a mouse infection model. J. Antimicrob. Chemother. 23:729-736. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro, M. A., C. L. Heifetz, and J. C. Sesnie. 1984. Comparison of microdilution and agar dilution procedures for testing antibiotic susceptibility of Neisseria gonorrhoeae. J. Clin. Microbiol. 20:828-830. [DOI] [PMC free article] [PubMed] [Google Scholar]