Abstract
We examined the effect of introducing type I or IV staphylococcal cassette chromosome mec (SCCmec) elements on the growth yield of Staphylococcus aureus in glucose-limited continuous culture. Type I showed increased glucose consumption and ATP demand per gram of cells synthesized and decreased cell yield compared to those of the parent strain. In contrast, type IV SCCmec elements had no adverse energetic effect.
New strains of methicillin-resistant Staphylococcus aureus (MRSA), have emerged in the community, causing infections in young, otherwise healthy people, such as children and military personnel (15, 16), and these strains can achieve a higher infection burden than nosocomial strains (10). In New Zealand, a community-acquired MRSA (CA-MRSA) strain designated Western Samoan phage pattern led to a 15-fold increase in MRSA cases between 1993 and 2000 (14).
The origins of CA-MRSA strains remain elusive. Compared to hospital-acquired MRSA, CA-MRSA strains appear to have higher growth rates, but the basis for this is unknown (11). These strains compete in an environment that is largely devoid of antibiotic selective pressure, and therefore, the selection for factors that contribute to ecological fitness may outweigh the need for multiple resistance determinants. Moreover, genetic factors seem to determine the permissiveness of S. aureus strain lineages to accommodate mecA and maintain methicillin resistance (8).
In this study, we sought to determine the fitness cost of the staphylococcal cassette chromosome mec (SCCmec) elements by measuring various physiological parameters between three isogenic S. aureus strains carrying either SCCmec type I or SCCmec type IV elements or no SCCmec element, by way of glucose-limited continuous culture.
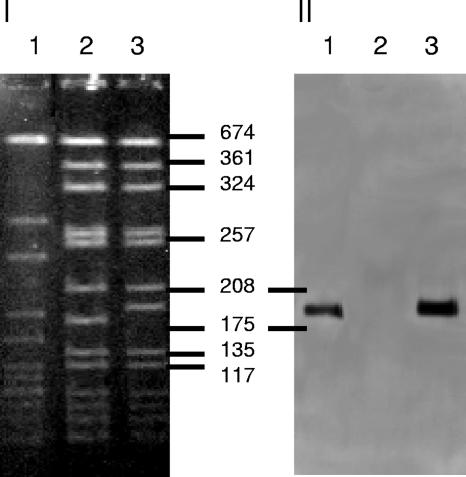
The susceptible parent strain BB255 is of NCTC8325 background. Strain RA120 is BB255 transformed with an SCCmec type I element (4). Strain RA2 was constructed in this study by transformation of strain BB255 with chromosomal DNA obtained from ST92/398, a CA-MRSA isolate harboring a SCCmec element type IV (1), as described earlier (4). In vitro transformation frequencies were extremely low (≤4 × 10−1/μg of chromosomal DNA). One representative transformant from eight analyzed, designated RA2, was chosen for further study. Figure 1 illustrates that the SmaI-G fragment of strain BB255 harboring orfX, the integration site for SCCmec, shifted by about 25 kb in the transformant RA2 due to the integration of the SCCmec type IV element, which was confirmed by Southern blotting with a mecA probe and by SCCmec typing (5, 12, 13).
FIG. 1.
(I) Pulsed-field gel electrophoresis patterns of Western Samoan phage pattern strain ST92/398 (lane 1), recipient strain BB255 (lane 2), and transformant RA2 (lane 3) after SmaI digestion. (II) Southern hybridization of the pulsed-field gel in panel I with a mecA probe. The sizes of the fragments (in kilobases) are shown.
Growth experiments were performed in a medium containing the following: Na2HPO4·2H2O, 6.0 g/liter; KH2PO4, 3.0 g/liter; NaCl, 0.5 g/liter; NH4Cl, 5.0 g/liter; CaCl2, 15 mg/liter; MgSO4, 247 mg/liter; MnSO4, 1.0 mg/liter; citric acid, 0.06 mg/liter; tryptone, 1.0 g/liter; and glucose, 0.9 g/liter. The cell yield for all strains was proportional to the glucose concentration, indicating that this medium was glucose limited. The doubling times for each strain at 37°C were as follows: 39 ± 3 min for BB255, 41 ± 3 min for RA2, and 54 ± 5 min for RA120. On the basis of these results, the SCCmec type I element appeared to have a negative effect on the growth rate of BB255 compared to SCCmec type IV in batch culture.
To study the roles of the different SCCmec elements on bacterial fitness at precisely controlled growth rates, all strains were grown in glucose-limited continuous culture, with a working volume of approximately 350 ml at a dilution rate of 0.1 h−1 (doubling time of 6.9 h). The growth medium was inoculated with exponential-phase cells and permitted to undergo batch growth to densities of approximately 0.3 to 0.8 (optical density at 600 nm) with constant agitation at 200 rpm. Concomitant with the initiation of continuous growth, agitation was then increased to 400 rpm to provide adequate aeration during steady-state conditions. The desired dilution rate was maintained for at least four residence times to allow the culture to reach steady state prior to sampling. At each steady state, 90-ml samples were removed and analyzed for residual glucose (UV method; R-Biopharm), end products (3), total viable CFU on LB agar plates, and cell weight (dry weight). After each sampling, the culture was again maintained for at least four residence times to reach steady state prior to replicate sampling. The values reported are the means of two to four independent experiments with triplicate determinations performed per sampling for each parameter measured. The standard experimental error of the mean associated with these determinations is shown.
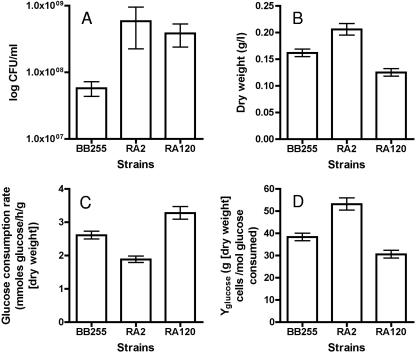
At a dilution rate of 0.1 h−1, cells reached a steady-state optical density at 600 nm of 0.39 to 0.56 (data not shown) and the CFU ranged from 5.75 × 107 to 5.86 × 108 (Fig. 2A). To determine the cell yield on glucose (Yglucose), the dry weight and glucose consumption rate of all strains were measured (Fig. 2B and C). Strain RA120 had the lowest dry weight but exhibited the greatest rate of glucose consumption, resulting in the lowest Yglucose for the three strains studied (Fig. 2D). In contrast, strain RA2 had the highest dry weight and the lowest rate of glucose consumption, resulting in a relatively high Yglucose. Strain BB255 was intermediate for both parameters. End product analysis from glucose consumption for each strain indicated that glucose was oxidized to CO2 at each dilution rate, i.e., no incomplete oxidation to acetate or lactate was detected.
FIG. 2.
Glucose-limited continuous culture of S. aureus strains at a dilution rate of 0.1 h−1. Strain BB255 is the susceptible parent, RA2 is the BB255 strain transformed with SCCmec type IV element, and RA120 is the BB255 strain transformed with SCCmec type I element. (A) Log CFU/ml, (B) cell weight (dry weight), (C) glucose consumption rate, and (D) Yglucose. The values reported are the means of two to four independent experiments with triplicate determinations performed per sampling for each parameter measured. The standard experimental error of the mean (error bars) associated with these determinations is shown.
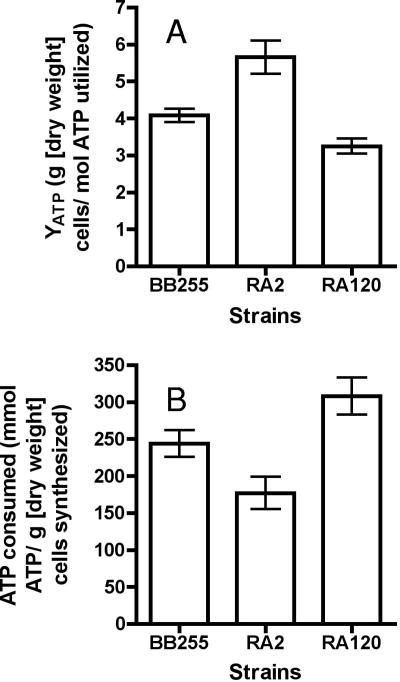
When S. aureus is grown on glucose as the sole carbon and energy source under aerobic growth conditions, the theoretical ATP yield is 9.4 mol of ATP produced per mol of glucose consumed (6). On the basis of this value, we calculated the weight in grams (dry weight) of cells produced per mole of ATP utilized (YATP) for each strain (Fig. 3A). For example, at a dilution rate of 0.1 h−1, the average Yglucose value for strain BB255 was 38.4 g (dry weight) of cells/mol glucose utilized (Fig. 2D) or 38.4 g (dry weight) of cells/9.4 mol of ATP utilized, which is equivalent to a YATP value of 4.10 ± 0.3 g (dry weight) cells/mol ATP utilized. The calculated YATP value for strain RA2 was 5.64 ± 0.45 g (dry weight) cells/mol ATP utilized, and for RA120, it was 3.24 ± 0.2 g (dry weight) cells/mol ATP utilized (Fig. 3A). These values are equivalent to an ATP demand per gram of newly synthesized biomass for each strain as follows: BB255, 244 ± 18 mmol ATP/g cells synthesized; RA2, 177 ± 22 mmol ATP/g cells synthesized; and for RA120, 308 ± 25 mmol ATP/g cells synthesized (Fig. 3B).
FIG. 3.
(A) YATP and (B) ATP consumed per gram of newly synthesized biomass for S. aureus strains BB255, RA2, and RA120 grown in glucose-limited continuous culture at a dilution rate of 0.1 h−1. Calculations are based on data presented in Fig. 2D and a theoretical ATP yield of 9.4 mol of ATP produced per mol of glucose consumed (6).
A number of studies have suggested that the size of the SCCmec element plays a major role in the fitness of S. aureus strains (2, 7, 16). Our data suggest that the SCCmec type IV element from CA-MRSA did not impose an energetic cost to its naïve host in terms of the maximum growth rate, cell yield, and the amount of cells that can be produced per mole of ATP consumed. However, the SCCmec type I element reduced the fitness of its host in terms of growth rate and cell yield. It is unlikely that the difference in size of the SCCmec elements is responsible for decreased fitness. Genes expressed from this DNA, such as pls coding for the plasmin-sensitive cell wall protein Pls, located on type I but not on type IV SCCmec elements, may affect the performance of MRSA. Notwithstanding this, the most likely factor may be the ease at which PBP2a was integrated into the existing cell wall synthesis complex, and the possible secondary compensatory events accompanying this event. Naïve cells, such as strain BB255 used here as a recipient for SCCmec, represent a host barrier for PBP2a, and were postulated to have to adapt in order to accept PBP2a (9). The altered ribosome binding site of mecA in the type IV SCCmec element (unpublished results) predicts lower PBP2a production compared to type I mecA, the latter of which is known to produce large and constitutive amounts of PBP2. It is possible that differences in the levels of PBP2a may have triggered different compensatory events and that these have caused the negative effect of SCCmec type I elements on growth yield.
Acknowledgments
Sui Mae Lee was supported by the University of Otago Dr Sulaiman Daud Jubilee 125 Postgraduate Scholarship. The financial assistance of the Otago Medical Research Foundation and the Deans Fund of the Otago School of Medical Sciences is gratefully acknowledged. M. Ender was supported by a grant of the Swiss National Science Foundation NF31-105390/1 to B. Berger-Bächi.
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Adhikari, R. P., G. M. Cook, I. Lamont, S. Lang, H. Heffernan, and J. M. Smith. 2002. Phenotypic and molecular characterization of community occurring, Western Samoan phage pattern methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 50:825-831. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, D. I., and B. R. Levin. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 3.Cook, G. M. 2000. The intracellular pH of the thermophilic bacterium Thermoanaerobacter wiegelii during growth and production of fermentation acids. Extremophiles 4:279-284. [DOI] [PubMed] [Google Scholar]
- 4.Ender, M., N. McCallum, R. Adhikari, and B. Berger-Bächi. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:2295-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geha, D. J., J. R. Uhl, C. A. Gustaferro, and D. H. Persing. 1994. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 32:1768-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinemann, M., A. Kummel, R. Ruinatscha, and S. Panke. 2005. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol. Bioeng. 92:850-864. [DOI] [PubMed] [Google Scholar]
- 7.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama, Y., D. A. Robinson, M. C. Enright, and H. F. Chambers. 2005. Genetic background affects stability of mecA in Staphylococcus aureus. J. Clin. Microbiol. 43:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katayama, Y., H. Z. Zhang, D. Hong, and H. F. Chambers. 2003. Jumping the barrier to β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 185:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laurent, F., H. Lelievre, M. Cornu, F. Vandenesch, G. Carret, J. Etienne, and J. P. Flandrois. 2001. Fitness and competitive growth advantage of new gentamicin-susceptible MRSA clones spreading in French hospitals. J. Antimicrob. Chemother. 47:277-283. [DOI] [PubMed] [Google Scholar]
- 11.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi, W., M. Ender, F. O'Brien, A. Imhof, C. Ruef, N. McCallum, and B. Berger-Bächi. 2005. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Zürich, Switzerland (2003): prevalence of type IV SCCmec and a new SCCmec element associated with isolates from intravenous drug users. J. Clin. Microbiol. 43:5164-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 14.Smith, J. M., and G. M. Cook. 2005. A decade of community MRSA in New Zealand. Epidemiol. Infect. 133:899-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber, J. T. 2005. Community-associated methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 41(Suppl. 4):S269-S272. [DOI] [PubMed] [Google Scholar]
- 16.Zetola, N., J. S. Francis, E. L. Nuermberger, and W. R. Bishai. 2005. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5:275-286. [DOI] [PubMed] [Google Scholar]