Abstract
A study was designed to describe the molecular epidemiology of CTX-M-producing Escherichia coli over a 6-year period (2000 to 2005) in a large well-defined Canadian region with a centralized laboratory system. Molecular characterization was done by isoelectric focusing, PCR, and automated sequencing, while genetic relatedness was determined by pulsed-field gel electrophoresis with XbaI. Of the 552 viable extended-spectrum β-lactamase-producing E. coli isolates isolated, 354 (64%) were positive for blaCTX-M genes associated with ISEcp1; 211 produced CTX-M-14, 128 produced CTX-M-15, 5 produced CTX-M-2, 4 produced CTX-M-3, 4 produced CTX-M-24, and 2 produced CTX-M-27. CTX-M-positive isolates were significantly more resistant to the fluoroquinolones than CTX-M-negative isolates, while CTX-M-15 producers were more likely to be resistant to gentamicin and tobramycin. There was a predominance of CTX-M-14 during the first 4 years of the study period, with community outbreaks associated with cluster 14A during 2000, 2001, and 2003. A substantial increase in CTX-M-15 producers occurred during the last 18 months and was due to clusters 15A and 15AR (where AR indicates related to A) in the hospital and nursing home sectors. Our results demonstrate that the persistence and dissemination of CTX-M genes among E. coli populations in larger geographic health care regions is dynamic, with the continuous emergence of clonally related CTX-M-15. This study illustrates the importance of molecular surveillance in tracking CTX-M-producing E. coli strains in the community and investigating their influx into hospitals.
Members of the family Enterobacteriaceae, especially Klebsiella spp. producing extended-spectrum β-lactamases (ESBLs), such as the SHV and TEM types, have been recognized since the 1980s as major causes of hospital-acquired infections (20). These infections were mainly associated with epidemic clones in the intensive care setting. From the late 1990s to the present, Enterobacteriaceae (mostly Escherichia coli) producing different ESBLs, such as the CTX-M enzymes, have emerged within the hospital and community settings as important causes of urinary tract infections (UTIs) (1). The majority of studies that have investigated community-onset infections caused by CTX-M-producing E. coli isolates showed that these isolates are often not clonally related, although clusters have been described in the United Kingdom and Canada (25).
The CTX-M β-lactamases, of which there are now over 50 different types, have originated from Kluyvera spp. and can be divided into five groups on the basis of their amino acid identities (3): the CTX-M-1 group, the CTX-M-2 group, the CTX-M-8 group, the CTX-M-9 group, and the CTX-M-25 group.
CTX-M β-lactamases have become the most prevalent type of ESBLs described during the last 5 years, especially from certain European and South American countries (7). CTX-M-producing E. coli isolates isolated from hospital and community sites often exhibited coresistance to trimethoprim-sulfamethoxazole, tetracycline, gentamicin, tobramycin, and ciprofloxacin. The CTX-M enzymes have been associated with the presence of different qnr genes and the production of a novel AAC(6′)-Ib aminoglycoside-modifying enzyme that has the additional ability to modify certain fluoroquinolones (7).
Very limited data regarding the molecular epidemiology of ESBL-producing Enterobacteriaceae in large geographical areas are available (15). Previous studies in the Calgary Health Region of Canada have shown that E. coli is responsible for more than 95% of the ESBL prevalence in that region and that the majority of isolates are positive for blaCTX-M β-lactamases (23). We also noted that the majority of infections originated from the community and that a CTX-M-14-producing cluster was responsible for community outbreaks of UTIs during 2000 and 2001 (21, 23). We designed a study that investigated the molecular epidemiology of CTX-M-producing E. coli isolates isolated in the Calgary Health Region over a 6-year period (January 2000 to December 2005) and determined if these bacteria were introduced into regional acute-care hospitals in large numbers.
(This work was presented at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy, 29 September 2006, San Francisco, CA [J. D. D. Pitout, N. Hamilton, D. Boyd, M. R. Mulvey, L. Campbell, B. L. Chow, D. B. Gregson, and D. L. Church, Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1664, 2006].)
MATERIALS AND METHODS
Patient population and definitions.
The Calgary Health Region provides all publicly funded health care services to the 1.2 million people residing in the Canadian cities of Calgary and Airdrie and numerous adjacent communities covering an area of 37,000 km2. Acute care is provided principally through one pediatric hospital and three large adult hospitals. Calgary Laboratory Services provides clinical microbiology services for both the hospital and community care sites within the Calgary Health Region through a regional centralized laboratory system (8). Community-onset infections were recognized in individuals who were either (i) outpatients or (ii) hospitalized patients whose first positive cultures were obtained within 48 h of hospital admission. Other hospitalized patients and patients from nursing homes were deemed to have hospital-onset infections.
Bacterial isolates.
Consecutive nonduplicate isolates of E. coli collected at Calgary Laboratory Services from January 2000 to December 2005 were included in this study. These organisms were screened for ESBL production and were then investigated for the presence of CTX-M β-lactamases. Strains were identified to the species level with the Vitek system (Vitek AMS; bioMérieux Vitek Systems Inc., Hazelwood, MO).
Antimicrobial susceptibility testing.
The MICs of the following drugs were determined with the Vitek system (Vitek AMS; bioMérieux Vitek Systems Inc.): piperacillin-tazobactam, imipenem, gentamicin, tobramycin, trimethoprim-sulfamethoxazole, and ciprofloxacin. Throughout this study, the results were interpreted by using the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) criteria for broth dilution (18). The quality control strains used for this part of the study were E. coli ATCC 25922, E. coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853.
Screening for and confirmation of ESBLs.
Clinical isolates of E. coli were evaluated for the presence of ESBLs by using the CLSI criteria for ESBL screening and disk confirmation tests (18). BD BBL Sensi-Discs consisting of ceftazidime/clavunanic acid (30/10 μg) and cefotaxime/clavulanic acid (30/10 μg) for ESBL confirmation tests were obtained from Becton Dickson and Company (Sparks, MD), while ceftazidime (30 μg) and cefotaxime (30 μg) disks were obtained from Oxoid Inc. (Nepean, Ontario, Canada). K. pneumoniae ATCC 700603 and E. coli ATCC 25922 were used as positive and negative controls, respectively.
β-Lactamase gene and insertion sequence identification.
Isoelectric focusing (IEF), which included cefotaxime hydrolysis and inhibitor profiles in polyacrylamide gels, was performed with all the isolates that tested positive by the CLSI ESBL confirmation test by using freeze-thaw extracts, as described previously (26). The β-lactamases were assessed for isoelectric points (pIs) and substrate and inhibitor profiles in polyacrylamide gels with overlays of 0.75 μg of cefotaxime per ml, 1,000 μM clavulanic acid, and 1,000 μM cloxacillin prior to overlay with nitrocefin agar. PCR amplification for blaCTX-M, blaOXA, blaSHV, and blaTEM β-lactamases was carried out with all the isolates with a GeneAmp 9700 ThermoCycler instrument (Applied Biosystems, Norwalk, CT) by using the PCR conditions and primers described previously (10, 22, 26). The isolates that tested positive for blaCTX-M β-lactamases underwent additional amplification with primers specific for CTX-M groups 1, 2, 8, 14, and 25 (24). By correlating the results obtained by IEF, PCR, and pulsed-field gel electrophoresis (PFGE), isolates were selected for the identification of the blaCTX-M genes and insertion sequences ISEcp1, IS10, and IS903 by using the primers and conditions described previously (13, 17). The following isolates (n = 241) were sequenced: all the 14NR (where NR indicates not related to 14A, 14AR [where AR indicates related to A], or 14B), 14NT (where NT indicates not typeable), 15NT, and 15NR (where NR indicates not related to 15A, 15AR, 15B, or 15BR) strains (n = 116); 110 isolates representing clusters 14A, 14AR, 14B, 15A, 15AR, 15B, and 15BR (where BR indicates related to B); and strains producing CTX-M-2 (n = 5), CTX-M-3 (n = 4), CTX-M-24 (n = 4), and CTX-M-27 (n = 2). Strains (n = 113) within the same cluster and with identical IEF patterns and identical results with group-specific primers were considered to produce the same CTX-M (e.g., 24 of the strains belonging to cluster 15A were not sequenced but had IEF and PCR results identical to those for the other strains of 15A that were sequenced). Automated sequencing was performed with the PCR products with an ABI Prism 3100 genetic analyzer (Applied Biosystems) and Sequence Analysis software. The sequences of the different amplicons were compared to each other and to homologous sequences by using the Sequence Navigator software. The nucleotide and the deduced protein sequences were analyzed by using the software available over the Internet at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Plasmid analysis.
The plasmid fingerprints of the CTX-M-15-producing E. coli isolates were determined with HpaI by using the protocols and conditions described previously (6).
PFGE.
All CTX-M-producing E. coli were typed by PFGE following the extraction of genomic DNA and digestion with XbaI by using the standardized E. coli (O157:H7) protocol established by the Centers for Disease and Prevention, Atlanta, GA (27), and standardized in 2005 (12). The subsequent PFGE analyses were performed on a CHEF-MAPPER XA apparatus (Bio-Rad Laboratories, Hercules, CA). Gel images in tiff format were exported to BioNumerics software (version 3.0; Applied Maths, Sint-Martens-Latem, Belgium) for analysis. Comparisons for E. coli were made by using the band-based Dice coefficient, which is a binary coefficient that measures similarity based upon common and different bands. Dendrograms were generated by using the unweighted pair group method with arithmetic averages method with 1.5% position tolerance. DNA relatedness was calculated on the basis of the Dice coefficient, and isolates were considered genetically related if the Dice coefficient correlation was 80% or greater, which corresponds to the “possibly related (four- to six-band difference)” criteria of Tenover et al. (28).
Statistical methods.
Fisher's exact test was used to compare group categorical data by using the Stata 9.0 program (Stata Corp., College Station, TX).
RESULTS
Bacterial isolates and susceptibilities.
During the 6-year study period, 94,545 E. coli were isolated from patients at Calgary Laboratory Services and 557 (0.6%) tested positive for ESBL production (5 isolates were unavailable for further analysis). The majority of ESBL-producing isolates (490 [88%]) were recovered from urine samples. The remaining isolates were recovered from the following sites: 33 (6%) from blood, 17 (3%) simultaneously from blood and urine, and 17 (3%) from various other specimens (e.g., respiratory, abdominal, and wound specimens). Of the 552 viable ESBL-producing clinical isolates included in this study, 28 (5%) were resistant to piperacillin-tazobactam, 321 (58%) were resistant to trimethoprim-sulfamethoxazole, 346 (63%) were resistant to ciprofloxacin, 191 (35%) were resistant to gentamicin, and 211 (38%) were resistant to tobramycin (Table 1). No imipenem resistance was detected. A greater proportion of CTX-M-positive isolates than CTX-M-negative ESBL isolates was resistant to ciprofloxacin (308/354 [87%] and 38/198 [19%], respectively [P < 0.0001]). Escherichia coli isolates producing CTX-M-15 were more resistant to trimethoprim-sulfamethoxazole, gentamicin, and tobramycin than E. coli isolates producing CTX-M-14 (Table 1). CTX-M-2-producing isolates were multiresistant, being sensitive only to imipenem.
TABLE 1.
Antimicrobial susceptibilities of ESBL-producing Escherichia coli isolated in the Calgary Health Region from January 2000 to December 2005
| β-Lactamase | No. of isolates (column %) resistant toa:
|
||||
|---|---|---|---|---|---|
| TZP | SXT | CIP | GEN | TOB | |
| CTX-M negative (n = 198) | 7 (4) | 160 (81) | 38 (19) | 55 (28) | 53 (27) |
| CTX-M-14 (n = 211) | 5 (2) | 64 (30) | 177 (84) | 47 (22) | 46 (22) |
| CTX-M-15 (n = 128) | 10 (8) | 86 (67) | 118 (92) | 79 (62) | 102 (80) |
| CTX-M-2 (n = 5) | 5 (100) | 5 (100) | 5 (100) | 5 (100) | 5 (100) |
| CTX-M-3 (n = 4) | 1 (25) | 4 (100) | 4 (100) | 3 (75) | 3 (75) |
| CTX-M-24 (n = 4) | 0 | 1 (25) | 3 (75) | 1 (25) | 1 (25) |
| CTX-M-27 (n = 2) | 0 | 1 (50) | 1 (50) | 1 (50) | 1 (50) |
| Total (n = 552) | 28 (5) | 321 (58) | 346 (63) | 191 (35) | 211 (38) |
TZP, piperacillin-tazobactam; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; GEN, gentamicin; TOB, tobramycin.
CTX-M-producing isolates.
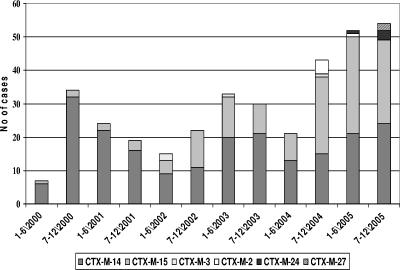
Of the 552 ESBL-producing E. coli isolated, 354 (64%) were positive for blaCTX-M β-lactamases; 211/354 (60%) produced CTX-M-14, 128/354 (36%) produced CTX-M-15, 5/354 (1%) produced CTX-M-2, 4/354 (1%) produced CTX-M-3, 4/354 (1%) produced CTX-M-24, and 2/354 (1%) produced CTX-M-27 (Table 1). The patients with CTX-M-producing E. coli isolates were scattered throughout the Calgary Health Region, with no apparent clustering of patients in a certain area or center. All the CTX-M-producing isolates were positive for insertion sequence ISEcp1, which was specifically linked with blaCTX-M, while some CTX-M-14- producing isolates that belonged to clusters 14A and 14AR were also positive for IS10, which was also specifically linked with blaCTX-M. CTX-M-producing isolates also produced the following additional β-lactamases: CTX-M-14 (except for clusters 14A and 14AR), CTX-M-2, CTX-M-3, CTX-M-24, and CTX-M-27. The CTX-M-27-producing isolates also produced TEM-1. The CTX-M-15-producing isolates also produced OXA-1 and TEM-1 (Table 2). No SHV enzymes were detected. The distribution of the different CTX-M-producing E. coli isolates over the study period is illustrated in Fig. 1. E. coli isolates producing CTX-M-14 and -15 were first isolated during March and June 2000, respectively; CTX-M-3-producing isolates were first encountered in 2002; and CTX-M-2-producing isolates were first encountered in 2004, while CTX-M-24 and CTX-M-27 appeared in 2005 (Fig. 1). CTX-M-15 producers constituted 3/41 (7%) of CTX-M-producing E. coli isolates in 2000, and this figure increased to 54/106 (51%) during 2005 (P < 0.0001) (Fig. 1).
TABLE 2.
Characteristics of CTX-M-producing Escherichia coli isolated in the Calgary Health Region from January 2000 to December 2005
| Type of CTX-M | PFGE groupa | β-Lactamase(s) | No. (%) of patients with the following location of onset of infectionb:
|
||
|---|---|---|---|---|---|
| Community | Hospital | Nursing home | |||
| CTX-M-14 (n = 211) | 14A (n = 98) | CTX-M-14 | 71 (72) | 25 (26) | 2 (2) |
| 14AR (n = 22) | CTX-M-14 | 11 (50) | 8 (36) | 3 (14) | |
| 14B (n = 12) | CTX-M-14, TEM-1 | 9 (75) | 3 (25) | 0 | |
| 14NR (n = 78) | CTX-M-14, TEM-1 | 55 (71) | 21 (27) | 2 (2) | |
| NT (n = 1) | CTX-M-14, TEM-1 | 1 (100) | 0 | 0 | |
| CTX-M-15 (n = 128) | 15A (n = 49) | CTX-M-15, OXA-1, TEM-1 | 25 (51) | 18 (37) | 6 (12) |
| 15AR (n = 21) | CTX-M-15, OXA-1, TEM-1 | 12 (57) | 9 (43) | 0 | |
| 15B (n = 10) | CTX-M-15, OXA-1, TEM-1 | 5 (50) | 5 (50) | 0 | |
| 15BR (n = 8) | CTX-M-15, OXA-1, TEM-1 | 4 (50) | 4 (50) | 0 | |
| 15NR (n = 38) | CTX-M-15, OXA-1, TEM-1 | 22 (58) | 15 (39) | 1 (3) | |
| NT (n = 2) | CTX-M-15, OXA-1, TEM-1 | 1 (50) | 1 (50) | 0 | |
| CTX-M-2 (n = 5) | NT (n = 5) | CTX-M-2, TEM-1 | 3 (60) | 1 (20) | 1 (20) |
| CTX-M-3 (n = 4) | 15B (n = 4) | CTX-M-3, TEM-1 | 2 (50) | 2 (50) | 0 |
| CTX-M-24 (n = 4) | NRB (n = 4) | CTX-M-24, TEM-1 | 1 (25) | 2 (50) | 1 (25) |
| CTX-M-27 (n = 2) | NRC (n = 2) | CTX-M-27, TEM-1 | 1 (50) | 1 (50) | 0 |
| Total (n = 354) | |||||
The 14A, 14AR, and 14B isolates formed separate clusters with >80% similar PFGE profiles. The 14AR isolates exhibited >60% similarity of profiles to 14A. The 14NR isolates were more distantly related. The 15A, 15AR, 15B, and 15BR isolates formed separate clusters with >80% similar PFGE profiles. The 15AR and 15BR isolates exhibited >60% similarity to profiles t15A and 15B, respectively, which indicates that 15AR is related to 15A, while 15BR is related to 15B. The 15NR isolates were more distantly related. The CTX-M-2-, 1 CTX-M-14-, and 2 CTX-M-15-producing isolates did not generate DNA bands by the PFGE method used in this study and were named NT and CTX-M-3-producing isolates were related to cluster 15B, while isolates producing CTX-M-24 and CTX-M-27 (PFGE groups NRB and NRC) were not clonally related; i.e., they exhibited PFGE profiles that were <80% similar.
Community-onset infections were diagnosed in individuals who were either outpatients or patients admitted to a hospital whose first positive cultures were obtained within 48 h of hospital admission. Other hospitalized patients and all residents of nursing homes were deemed to have nosocomial infections.
FIG. 1.
Distribution of E. coli isolates producing different CTX-M β-lactamases divided into 6-month periods from January 2000 to December 2005 in the Calgary Health Region.
PFGE.
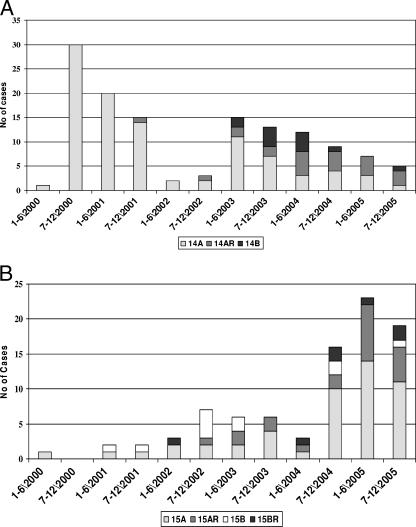
PFGE identified three closely related groups of E. coli producing CTX-M-14, designated clusters 14A (n = 98) and 14AR (n = 22), which were closely related, and a separate cluster named 14B (n = 12) (Table 2). The 14A, 14AR, and 14B isolates formed separate clusters with >80% similar PFGE profiles (data not shown). The profiles of the 14AR isolates exhibited >60% similarity to the profiles of the 14A isolates, which indicates that 14AR is related to 14A, although the significance is unknown. The remaining 78 CTX-M-14-producing isolates were not related to the isolates in cluster 14A, 14AR, or 14B or to each other and were designated 14NR. The 14NR isolates were more distantly related to these three groups (data not shown). One CTX-M-14-producing isolate did not generate DNA bands by the PFGE method used in this study and was named NT. The distribution of the different CTX-M-14 genotypes over the 6-year period showed that 14A caused an outbreak during 2000 and 2001 and again in 2003, while 14B was first encountered in 2003 and practically disappeared in 2005 (Fig. 2A). The 14NR isolates replaced clusters 14A, 14AR, and 14B in 2005.
FIG. 2.
(A) Distributions of different genotypes of E. coli producing CTX-M-14-β-lactamases divided into 6-month periods from January 2000 to December 2005 in the Calgary Health Region. The 14A, 14AR, and 14B isolates formed separate clusters with PFGE profiles that were >80% similar. The profiles of the 14AR isolates exhibited >60% similarity to the profiles of the 14A isolates. Data for the 14NR isolates are not shown here. (B) Distributions of different genotypes of E. coli producing CTX-M-15-β-lactamases divided into 6-month periods from January 2000 to December 2005 in the Calgary Health Region. The 15A, 15AR, 15B, and 15BR isolates formed separate clusters with PFGE profiles that were >80% similar. The profiles of the 15AR and 15BR isolates exhibited >60% similarity to the profiles of the 15A and 15B isolates, respectively, which indicates that 15AR is related to 15A and that 15BR is related to 15B. Data for the 15NR isolates are not shown here.
PFGE identified four closely related groups of E. coli producing CTX-M-15, designated clusters 15A (n = 49) and 15AR (n = 21), which were closely related, as well as separate clusters designated 15B (n = 10) and 15BR (n = 8) (Table 2). The 15A, 15AR, 15B, and 15BR isolates formed separate clusters with >80% similar PFGE profiles (data not shown). The profiles of the 15AR and 15BR isolates exhibited >60% similarity to the profiles of 15A and 15B, which indicates that 15AR is related to 15A, while 15BR is related to 15B, although the significance is unknown. The remaining 38 CTX-M-15-producing isolates were not related to isolates in cluster 15A, 15AR, 15B, or 15BR or to each other and were designated 15NR. The 15NR isolates were more distantly related to these four groups. Two CTX-M-15-producing isolates did not generate DNA bands by the PFGE method used in this study and were designated NT. The distribution of the different CTX-M-15-genotypes over the 6-year period illustrated that the increase in CTX-M-15 isolates during 2004 and 2005 was largely due to clusters 15A and 15AR (Fig. 2B).
The remaining CTX-M-producing isolates demonstrated the following PFGE patterns: CTX-M-2-producing isolates (n = 5) did not generate DNA bands by the PFGE method used in this study and were named NT, and CTX-M-3 (n = 4)-producing isolates were related to cluster 15B, while isolates producing CTX-M-24 (n = 4) and CTX-M-27 (n = 2) (PFGE groups NRB and NRC, respectively) were not clonally related; i.e., they exhibited PFGE profiles with <80% similar (data not shown).
The majority of CTX-M-producing isolates (223/354 [63%]) originated from the community (Table 2), and CTX-M-14 isolates (147/211 [70%]) were more likely than CTX-M-15 isolates (69/128 [52%]) (P = 0.005) to be isolated from the community. E. coli isolates producing CTX-M-15 tended to become more prevalent in the hospital and nursing home sectors during the last 18 months of the study period. Overall 40/77 (52%) of CTX-15-producing E. coli isolates were recovered during the last 18 months from hospital and nursing home sites, whereas only 19/51 (37%) were isolated from hospital and nursing home sites during the first 54 months (P = 0.11). Of particular interest was the fact that the majority of CTX-M-15 producers isolated from the hospital and nursing home sites during the last 18 months of the study belonged to clusters 15A and 15AR (26/40 [65%]), whereas 5/19 (26%) were isolated from hospital and nursing home sites during the first 54 months (P = 0.11).
DISCUSSION
The molecular epidemiology of CTX-M-producing E. coli strains on a countrywide scale has been previously described in United Kingdom (29), Spain (19), Italy (16), and Canada (17).
Our study differs from the studies mentioned above because it describes the molecular epidemiology of CTX-M-producing E. coli isolates in a large well-defined geographical region with a single centralized laboratory system over a 6-year period. Routine testing for ESBL-producing E. coli isolates and Klebsiella spp. was initiated at Calgary Laboratory Services during 1999, and CTX-M-producing E. coli isolates were first isolated in the Calgary Health Region during March 2000. All isolates recovered from patients admitted to acute-care hospitals or nursing homes or patients accessing community care sites since 2000 were included in this study. Overall the Calgary Health Region has relatively low rates of resistance to antimicrobial agents, particularly among isolates of the family Enterobacteriaceae, and the major resistant bacteria isolated at Calgary Laboratory Services are community-associated methicillin-resistant Staphylococcus aureus (9) and metallo-β-lactamase-producing P. aeruginosa (14). Since the Calgary Health Region currently has low rates of resistance to antimicrobial agents, it was surprising to find such a degree of diversity among CTX-M enzyme types, with CTX-M-14 and CTX-M-15 being the most abundant, while isolates producing CTX-M-2, -3, -24, and -27 were relatively rare (Fig. 1). This study describes the unique epidemiology of both CTX-M-14- and CTX-M-15-producing E. coli isolates in our region; the prevalence of CTX-M-14- and CTX-M-15-producing E. coli isolates changed during the study period, as did the clustering of these strains within different health care locations. In addition, CTX-M-15-producing E. coli isolates were more likely than CTX-M-14-producing strains to be multidrug resistant (Table 1). We had a predominance of CTX-M-14 during the first 4 years of the study period, with community outbreaks associated with cluster 14A during 2000, 2001, and 2003 (Fig. 2A). The numbers of the CTX-M-15-producing isolates progressively increased during the first 4 years, with a substantial increase during the last 18 months of the study that was mostly due to an increase in isolates of clusters 15A and 15AR in the hospital and the nursing home sectors. This increase was not due to the clustering of patients in a specific acute-care center or nursing home but, rather, was due to the introduction of these strains from the community.
This study also showed that the horizontal and the vertical transmission of CTX-M-15 had occurred in the Calgary Health Region. We wanted to compare the plasmids from our CTX-M-15-producing E. coli isolates with pC15-1a, a plasmid from a CTX-M-15-producing E. coli isolate responsible for an outbreak in a long-term-care center in Toronto, Ontario, Canada, during 2000 that was sequenced previously (6). The restriction profiles of plasmids isolated from the strains in clusters 15A, 15B, and 15NR were not related to the profile of pC15-1a (data not shown).
It is interesting to note that organisms producing specific CTX-M β-lactamases have been isolated from different countries, such as CTX-M-9 and CTX-M-14, which are mostly present in Spain; CTX-M-14 in Canada and China; CTX-M-1 in Italy; CTX-M-3 in Poland; and CTX-M-2 in several South American countries, Japan, and Israel, while CTX-M-15-producing strains have been described from all continents except Antarctica (7). Organisms producing CTX-M-24 and CTX-M-27 are relatively rare on a worldwide basis and have previously been reported from Southeast Asia, Africa, and France (2, 4, 5, 11, 30). This is the first study to describe the isolation of CTX-M-2-, CTX-M-3-, CTX-M-24-, and CTX-M-27-producing organisms from the North American continent.
In summary our study demonstrates that the molecular epidemiology of CTX-M-producing E. coli strains in the Calgary Health Region is dynamic, in that there is a continuous change of proportions between different CTX-M-producing strains over a period of time. CTX-M-14-producing E. coli isolates were more likely to be isolated from the community in our region, while certain clusters of CTX-M-15-producing isolates (cluster 15A and 15AR) established themselves during 2004 and 2005 in the hospital and the nursing home sectors after being introduced from the community.
In conclusion, this study highlights the important role of molecular surveillance for the tracking of CTX-M-producing E. coli isolates in the community and investigating their influx into hospitals. Our study illustrated the emergence of clonally related CTX-M-15-producing E. coli strains over a period of 6 years in our region and supports the findings of the pandemic nature of these CTX-M-producing bacteria (7, 16, 19, 29). We recommend that a simple, standardized, and cost-effective typing protocol be established for monitoring the spread of different clusters of CTX-M-15-producing E. coli isolates throughout the world. This protocol should be distributed to different laboratories across the world. Typing images could then be forwarded to a center for comparison purposes, which will ensure the tracking of these important CTX-M-producing bacteria. We also recommend that future investigations be undertaken to study the microbiological and ecological factors that make CTX-M-15 producers such successful pathogens to help prevent future infections caused by these medically important pathogens.
Acknowledgments
We thank Lorraine Campbell; Nina Hamilton of Calgary Laboratory Services, Calgary, Alberta, Canada; and David Boyd from National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Manitoba, Canada, for their excellent technical support of this study and Terry Ross for database management.
This study was funded jointly by the University of Calgary Dean's Starter Grant (grant 75-4777), the Antibiotic Resistant Organism (ARO) Research fund (a partnership between Calgary Laboratory Services, the Calgary Health Trust, the University of Calgary, and the Calgary Health Region) and AMMI, Canada.
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Babic, M., A. M. Hujer, and R. A. Bonomo. 2006. What's new in antibiotic resistance? Focus on β-lactamases. Drug Resist. Update 9:142-156. [DOI] [PubMed] [Google Scholar]
- 2.Bin, C., W. Hui, Z. Renyuan, N. Yongzhong, X. Xiuli, X. Yingchun, Z. Yuanjue, and C. Minjun. 2006. Outcome of cephalosporin treatment of bacteremia due to CTX-M-type extended-spectrum β-lactamase-producing Escherichia coli. Diagn. Microbiol. Infect. Dis. [E-pub ahead of print.] [DOI] [PubMed]
- 3.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, R., C. Recule, R. Baraduc, C. Chanal, D. Sirot, C. De Champs, and J. Sirot. 2003. Effect of D240G substitution in a novel ESBL CTX-M-27. J. Antimicrob. Chemother. 52:29-35. [DOI] [PubMed] [Google Scholar]
- 5.Bouallegue-Godet, O., Y. Ben Salem, L. Fabre, M. Demartin, P. A. Grimont, R. Mzoughi, and F. X. Weill. 2005. Nosocomial outbreak caused by Salmonella enterica serotype Livingstone producing CTX-M-27 extended-spectrum β-lactamase in a neonatal unit in Sousse, Tunisia. J. Clin. Microbiol. 43:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canton, R., and T. M. Coque. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 8.Church, D. L., C. Don-Joe, and B. Unger. 2000. Effects of restructuring on the performance of microbiology laboratories in Alberta. Arch. Pathol. Lab. Med. 124:357-361. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert, M., J. MacDonald, D. Gregson, J. Siushansian, K. Zhang, S. Elsayed, K. Laupland, T. Louie, K. Hope, M. Mulvey, J. Gillespie, D. Nielsen, V. Wheeler, M. Louie, A. Honish, G. Keays, and J. Conly. 2006. Outbreak in Alberta of community-acquired (USA300) methicillin-resistant Staphylococcus aureus in people with a history of drug use, homelessness or incarceration. Can. Med. Assoc. J. 175:149-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson, N. D., E. S. Moland, A. Hossain, S. A. Neville, I. B. Gosbell, and K. S. Thomson. 2002. Unusual Salmonella enterica serotype Typhimurium isolate producing CMY-7, SHV-9 and OXA-30 β-lactamases. J. Antimicrob. Chemother. 49:1011-1014. [DOI] [PubMed] [Google Scholar]
- 11.Ho, P. L., R. H. Shek, K. H. Chow, R. S. Duan, G. C. Mak, E. L. Lai, W. C. Yam, K. W. Tsang, and W. M. Lai. 2005. Detection and characterization of extended-spectrum β-lactamases among bloodstream isolates of Enterobacter spp. in Hong Kong, 2000-2002. J. Antimicrob. Chemother. 55:326-332. [DOI] [PubMed] [Google Scholar]
- 12.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lartigue, M. F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 234:201-207. [DOI] [PubMed] [Google Scholar]
- 14.Laupland, K. B., M. D. Parkins, D. L. Church, D. B. Gregson, T. J. Louie, J. M. Conly, S. Elsayed, and J. D. Pitout. 2005. Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-β-lactamase (MBL)-producing strains. J. Infect. Dis. 192:1606-1612. [DOI] [PubMed] [Google Scholar]
- 15.Livermore, D. M. 2003. Bacterial resistance: origins, epidemiology, and impact. Clin. Infect. Dis. 36:S11-S23. [DOI] [PubMed] [Google Scholar]
- 16.Mugnaioli, C., F. Luzzaro, F. De Luca, G. Brigante, M. Perilli, G. Amicosante, S. Stefani, A. Toniolo, and G. M. Rossolini. 2006. CTX-M-type extended-spectrum β-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents Chemother. 50:2700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulvey, M. R., E. Bryce, D. Boyd, M. Ofner-Agostini, S. Christianson, A. E. Simor, and S. Paton. 2004. Ambler class A extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella spp. in Canadian hospitals. Antimicrob. Agents Chemother. 48:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing; 14th informational supplement M100-S14. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 19.Oteo, J., C. Navarro, E. Cercenado, A. Delgado-Iribarren, I. Wilhelmi, B. Orden, C. Garcia, S. Miguelanez, M. Perez-Vazquez, S. Garcia-Cobos, B. Aracil, V. Bautista, and J. Campos. 2006. Spread of Escherichia coli strains with high-level cefotaxime and ceftazidime resistance between the community, long-term care facilities, and hospital institutions. J. Clin. Microbiol. 44:2359-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitout, J. D., D. B. Gregson, D. L. Church, S. Elsayed, and K. B. Laupland. 2005. Community-wide outbreaks of clonally related CTX-M-14 β-lactamase-producing Escherichia coli strains in the Calgary Health Region. J. Clin. Microbiol. 43:2844-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitout, J. D. D., N. Hamilton, D. L. Church, P. Nordmann, and L. Poirel. 2007. Development and validation of a molecular diagnostic assay to detect CTX-M-type β-lactamases in Enterobacteriaceae. Clin. Microbiol. Infect. 13:291-297. [DOI] [PubMed] [Google Scholar]
- 23.Pitout, J. D., N. D. Hanson, D. L. Church, and K. B. Laupland. 2004. Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum β-lactamases: importance of community isolates with blaCTX-M genes. Clin. Infect. Dis. 38:1736-1741. [DOI] [PubMed] [Google Scholar]
- 24.Pitout, J. D., A. Hossain, and N. D. Hanson. 2004. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 42:5715-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitout, J. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56:52-59. [DOI] [PubMed] [Google Scholar]
- 26.Pitout, J. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, E. S. Moland, and C. C. Sanders. 1998. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob. Agents Chemother. 42:1350-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]
- 30.Yu, Y., S. Ji, Y. Chen, W. Zhou, Z. Wei, L. Li, and Y. Ma. 2006. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J. Infect. [E-pub ahead of print.] [DOI] [PubMed]