Abstract
A sul3 domain (IS440-sul3-orf1-IS26) was found linked to an unusual 3′ conserved sequence region (qacH) of class 1 integrons and detected among nontyphoid Salmonella isolates (n = 47) from different sources. Three types of integrons differing in the gene cassette array (dfrA12-orfF-aadA2-cmlA1-aadA1, dfrA12-orfF-aadA2/1, and estX-psp-aadA2-cmlA1-aadA1) were found associated with this sul3 domain. They were associated with particular clones and specific high-molecular-weight plasmids.
Antimicrobial agents of the sulfonamide group have been widely used in the treatment of human infections and also administrated to food animals (14, 17), a practice which has been argued to contribute to the maintenance or emergence of resistance (4). Sulfonamide resistance in gram-negative bacilli generally arises from the acquisition of one of the three genes, sul1, sul2, and sul3, encoding forms of dihydropteroate synthase that are not inhibited by the drug (14, 17). The sul3 gene is the most recently described gene conferring resistance to sulfonamides (14) and has been increasingly identified in humans and principally among food animal isolates in the family Enterobacteriaceae. Several studies have demonstrated the dissemination of the sul3 gene among Escherichia coli isolates from different origins and countries (5, 6, 7, 10, 14) and among Salmonella isolates from different sources and of different serotypes and clones (1, 8). The association of the genetic determinants for sulfonamide resistance with horizontally transferable capture genetic units may facilitate their dissemination. The spread of the sul1 gene seems to be related to class 1 integrons, and the sul2 gene is incorporated in plasmids (17). The dissemination and genetic background of the sul3 gene have not been completely characterized. The sul3 gene found in a Swiss E. coli isolate was flanked by transposable elements (sul3 domain) inserted into a conjugative plasmid (14). Other studies have reported that sul3 could be found on different large plasmids (6, 8). Only one recent report described the linkage of sul3 to other resistance gene cassettes as part of an integron in swine E. coli isolates from the United States (5). Also in a previous study, we observed that all of the Salmonella isolates carrying the sul3 gene contained a class 1 integrase, and in several of these isolates, sul3 was the only sulfonamide resistance gene detected (1). The objective of this study was to characterize the genetic background of the sul3 gene, including its association with integron structures, in nontyphoid Salmonella isolates in order to understand the dissemination of this sulfonamide resistance gene.
This study included 45 sul3-carrying Salmonella isolates from different sources (humans, food products, and the environment) and of different serotypes detected among the sulfonamide-resistant isolates (n = 331) of a collection of 1,511 Portuguese nontyphoid Salmonella isolates recovered between 2002 and 2004. These 331 isolates were recovered from human clinical sources (n = 204), food products (n = 114), the environment (n = 7), and unknown sources (n = 6). Two isolates previously collected in a central hospital in 2000 were also included in the present study. Characterization of the sulfonamide-resistant isolates was performed as previously described (1, 2). The characteristics of the sul3-producing Salmonella isolates are presented in the Table 1. Sixteen of the sul3-positive isolates were from food products, principally pork products (n = 10), collected from distinct locations in Portugal. The 27 human Salmonella isolates were recovered from diverse sources (predominantly feces) and 14 hospitals in geographically dispersed regions. Three isolates were obtained from environmental sources (bathing water). The isolates carrying sul3 belonged to four serotypes and eight PFGE clones, with most of them identified as S. enterica serotype Typhimurium (five clones) and S. enterica serotype Rissen (one clone). It is of note that isolates from two clones of S. enterica serotype Typhimurium were the predominant ones carrying the sul3 gene. All of the sul3-positive isolates showed resistance to several antimicrobials; coresistance to streptomycin (aadA), chloramphenicol (cmlA, catA), tetracycline (tetA tetB), and trimethoprim (dfrA12) (Table 1) was observed in 60% of the isolates. The cmlA gene was observed in all of the chloramphenicol-resistant isolates and in one of them with catA.
TABLE 1.
Characteristics of the sul3-producing Salmonella isolates used in this study
| Integron type, serotype (no. of isolates) | PFGEb | Sourcec (no. of isolates) | Isolation: yr/location (no. of isolates) | Resistance phenotyped and gene profile | Integron size (bp) and/or gene cassette (5′CS-3′CS)f | Size(s) (kb)e of plasmid(s) carrying-sul3-associated integrons (no. of isolates) |
|---|---|---|---|---|---|---|
| I | ||||||
| Rissen (2) | N | S (1), U (1) | 2002 (1), 2004 (1)/north (1), south (1) | StrrChlrTetrSulrTmpraadA2-aadA1cmlA1tetAsul3dfrA12 | int1 | 100 (2) |
| Typhimurium (7) | Q | H (6), S (1) | 2000 (1), 2003 (2), 2004 (4)/north (7) | ChlrSulrTmpr (StrrKanr Tetr) cmlA1sul3dfrA12 (aadA2-aadA1aphA1tetB | int1 | 100 (1), 160 (1), 165 (1) |
| IIIb:65:lv:enxz15 (1) | S | H (1) | 2003 (1)/south (1) | StrrChlrSulrTmpraadA2-aadA1cmlA1sul3dfrA12 | int1 | 100 (1) |
| Haifa (1) | Z | H (1) | 2004 (1)/north (1) | StrrAmprChlr TetrSulrTmpraadA2-aadA1blaTEMcmlA1tetA sul2-sul3dfrA12 | int1 | 165 (1) |
| Typhimurium DT104 (1)a | A | U (1) | 2004 (1)/north (1) | Strr Ampr Chlr Tetr Sulr TmpraadA2-aadA1 blaTEMcmlA1 tetA sul1 sul3 dfrA12 | 1,000; aadA2 | 135 (1) |
| II | ||||||
| Rissen (3) | N | S (3) | 2002 (2), 2003 (1)/ north (3) | StrrAmprTetrSulrTmpraadA2-aadA2/1blaTEMtetAsul1-sul3dfrA12 | 2,000; dfrA12orfFaadA2 | 70 (2) |
| III | ||||||
| Typhimurium (1) | J | S (1) | 2003 (1)/south (1) | StrrKanrAmprChlrTetrSulrTmpraadA2-aadA1aphA1blaTEMcmlA1-catAtetBsul1-sul3dfrA1 | 1,700; dfrA1aadA1 | 240 (1) |
| Typhimurium DT104 (2)a | X | H (2) | 2004 (2)/south (2) | Strr Genr Ampr Chlr Tetr Sulr TmpraadA2-aadA1 aac(3)-IV blaTEMcmlA1 tetA sul1-sul2-sul3 dfrA12 | 2,000; dfrA12 orfF aadA2 | 220 (1) |
| Typhimurium DT104 (29)a | O | H (17), S (4), P (1), C (2), U (1), E (3), UN (1) | 2000 (1), 2002 (3), 2003 (12), 2004 (13)/north (19), center (1), south (8), UN (1) | Strr Chlr Sulr (Genr Kanr Ampr Tetr Tmpr Nalr) aadA2-aadA1 cmlA1 sul1-sul2-sul3 [floR aac(3)-IV blaTEMtetA tetB dfrA12] | 2,000; dfrA12 orfF aadA2 | 150 (1), 170 (5), 220 (3), 150 + 170 (1) |
PCR assay for the identification of S. enterica serotype Typhimurium DT104 and U302 phage types (15) positive.
Clones are designated by capital letters as previously described (2).
H, humans; S, pork products; P, poultry products; C, beef products; U, unknown food product; E, environment; UN, unknown source.
Antibiotic resistances and resistance genes transferred to transconjugants are underlined. Variable antibiotic resistance and resistance gene among isolates of the same PFGE are in parentheses. The following genes implicated in antimicrobial resistance were detected by PCR (1, 2, 9): blaPSE-1, blaOXA-30, and blaTEM encoding β-lactam resistance; floR, cmlA1, and catA encoding chloramphenicol resistance; tetA, tetB and tetG encoding tetracycline resistance; sul1, sul2 and sul3 encoding sulfonamide resistance; aac(3)-IV and aphA1 encoding, respectively, gentamicin and kanamycin resistance. The characterization of genes associated with streptomycin (aadA) and trimethoprim (dfrA) resistance was done by PCR amplification and DNA sequencing in isolates carrying those genes inserted into integrons, as described previously (2). Antimicrobial agents were tested as previously described (2).Abbreviations: Strr, streptomycin resistance; Genr, gentamicin resistance; Kanr, kanamycin resistance; Ampr, ampicillin resistance; Nalr, nalidixic acid resistance; Chlr, chloramphenicol resistance; Tetr, tetracycline resistance; Sulr, sulfamethoxazole resistance; Tmpr, trimethoprim resistance.
Plasmid profiles obtained by S1 PFGE in the isolates selected for testing; the plasmids transferred to transconjugants are underlined.
int1 gene was detected, as previously described (2) in isolates without a positive 5′CS-3′CS PCR assay.
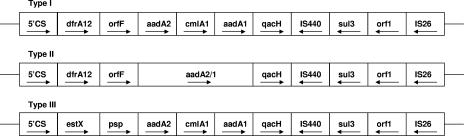
Structure of the sul3-associated integrons.
Several methodologies were used to determine the structure of the sul3-genetic elements: (i) PCR was used to screen isolates for the sul3 gene, with plasmid pVP440 (14) used as a positive control. (ii) PCR amplification was then used to determine the organizational structure of sul3-associated integrons as described previously (5), with TripleMaster enzyme mix (Eppendorf, Hamburg, Germany) and the primer combinations 5′CS (13) and cmlA-B (9), cmlA-F (9) and sul3F (14), and INT/5CS (16) and sul3F (14). (iii) Typing of sul3-carrying integrons was performed by a PCR-restriction fragment length polymorphism analysis in which PCR products corresponding to the amplification of the 5′ conserved sequence (5′CS)-cmlA region and of the cmlA-sul3 region were purified and further digested with TaqI endonuclease (New England BioLabs, Ipswich, MA). PCR products representing the two different amplicons or the complete integron containing the sul3 gene were sequenced through a primer-walking strategy with specific designed primers. Sequence comparisons were made with the BLAST program available at the National Center for Biotechnology Information website, and the sequence data for each integron type were deposited in the GenBank database.
The following three sul3-associated integrons presenting distinct gene cassette organizations were observed (Fig. 1): type I, 5′CS-dfrA12-orfF-aadA2-cmlA1-aadA1-qacH-IS440-sul3 (7,085 bp); type II, 5′CS-dfrA12-orfF-aadA2/1-qacH-IS440-sul3 (4,525 bp); type III, 5′CS-estX-psp-aadA2-cmlA1-aadA1-qacH-IS440-sul3 (7,304 bp). All of the types presented genes coding for streptomycin resistance (aadA), including one new hybrid aadA gene, and two (types I and III) of them contained the cmlA1 gene coding for chloramphenicol resistance. In all of the class 1 integron types described in this report, it seems that the sul3 gene had replaced sul1 in addition to the replacement of qacEΔ1 by the qacH gene in the 3′CS region.
FIG. 1.
Organization of the three types of sul3-carrying integrons in Salmonella (not to scale). Gene orientations are indicated by arrows.
Two sul3-carrying class 1 integrons (types I and II), presenting an original gene cassette organization, were observed in multidrug-resistant (MDR) isolates of four serotypes (Rissen, Typhimurium, IIIb:65:lv:enxz15, and Haifa) corresponding to five clones. Interestingly, of the S. enterica serotype Rissen clone isolates, two contained the type I integron and three contained the type II integron. The structure of 7,085 bp (type I) contained the first three gene cassettes (dfrA12-orfF-aadA2) typical of a class 1 integron widely disseminated among members of the family Enterobacteriaceae, including Salmonella (2) and downstream an organization identical to part of the type III structure. In 10 of 12 isolates carrying the type I genetic structure, sul3 was the only sul gene detected. The other new integron structure (type II), only described in the MDR S. enterica serotype Rissen clone, contained the first two gene cassettes (dfrA12-orfF) typical of class 1 integrons and also present in the type I integron described here, but with a hybrid aadA gene cassette as the third gene, suggesting recombination between the dfrA12-orfF-aadA2 cassette array and an aadA1 gene cassette (the crossover in the hybrid aadA2/1 cassette was located between positions 289 and 299 in the cassette). This genetic event and the absence of the cmlA gene, before the qacH-tnp-sul3 sequence, observed in isolates from the same clone, suggest that the type II integron may be a result of evolution (e.g., by an incorrect cassette excision event) from the sul3-carrying class 1 type I integron.
Class 1 integron structure type III, identical to that recently described by Bischoff et al. (5) in swine E. coli isolates, was observed in the majority (n = 32) of the sul3-producing Salmonella isolates (including the oldest strain from 2000). This genetic structure was located only in MDR S. enterica serotype Typhimurium isolates corresponding to three clones. Most of those isolates (24/32) also carried another class 1 integron (dfrA12-orfF-aadA2 in 23 and dfrA1-aadA1 in 1) with the qacEΔ1-sul1 genes as part of the 3′CS. With the exception of one isolate, all presented the three types of sul gene simultaneously.
Finally, characterization of the sul3 genetic context was also performed by PCR with several specific primer combinations (TNP-F-SUL3F and SUL3FR-TNP-R) and sequencing (PCR products and plasmids extracted from transconjugants). Characterization of the sul3 genetic vicinity showed a gene cluster comprising sul3 and transposase-like sequences (IS440-sul3-orf1-IS26) identical in all of the isolates, with the exception of one (deletion of ca. 500 bp in the orf1-IS26 region). The tnp gene of IS440 was identical to the one described in an S. choleraesuis plasmid (GenBank accession no. AY509004), to the E. coli plasmids characterized by Bischoff et al. (5), and also to the partial sequence of the truncated IS440 sequence described by Perreten and Boerlin (14). The insertion sequence observed downstream of the sul3 gene was identical to IS26, already observed flanking resistance genes, including in several MDR S. enterica plasmids (e.g., GenBank accession no. AY333434 and AJ628353). It is of note that the orientation of the IS440-sul3-orf1-IS26 element is the opposite of that previously described by Perreten and Boerlin (14), suggesting a different sul3 gene acquisition event.
Genetic locations of sul3-associated integrons.
Several methods were used to determine the locations of sul3-containing genetic elements. (i) Conjugation assays with E. coli K802N (Nalr Rifr) as a recipient strain were attempted by mating in agar plates. Transconjugants were selected on Mueller-Hinton agar 2 (bioMérieux, Marcy l'Étoile, France) containing sulfamethoxazole (256 μg/ml) plus nalidixic acid (64 μg/ml) (or 100 μg/ml rifampin if the donor was nalidixic acid resistant). (ii) Plasmid DNA was isolated from donors and transconjugants by several methods (11, 12). The relatedness of plasmids harbored by the transconjugants was determined by single-restriction analysis with EcoRI. For better resolution and sizing of high-molecular-weight plasmids, extraction from selected isolates was also performed by nuclease S1 (Amersham Biosciences, Uppsala, Sweden) digestion prior to PFGE (3). (iii) Southern blot hybridizations of S1-PFGE patterns were performed by standard methods, by using a nonradioactive technique (Amersham Biosciences) with probes for sul3 from pVP440 (14) and probes for class 1 integron- and gene cassette-specific sequences (5′CS-dfrA12 and estX-psp).
The sul3 gene was originally identified on a 54-kb conjugative plasmid (14), but it also appeared to be located on large plasmids of different sizes (6, 8, 14). Sulfonamide resistance and the sul3 gene were transferred, by conjugation assays, in 5 of 12 isolates carrying genetic structure type I and in all of the isolates carrying genetic structure type II, with cotransference of resistance to trimethoprim and streptomycin. Plasmid characterization by S1-PFGE, followed by Southern hybridizations, demonstrated that plasmids carrying the sul3 gene cohybridized with specific probes for class 1 integrons and gene cassette-specific sequences. The sul3 gene and sul3-associated type I and II integrons were located on large plasmids of different sizes (≥100 kb) and on identical conjugative plasmids of ca. 70 kb, respectively (Table 1). The conjugative plasmid of ca. 100 kb was the most disseminated among the different clones; although the RFLP with EcoRI showed different profiles between isolates from different clones, several bands were shared between them, suggesting that they are highly related (data not shown). Conjugation experiments failed to demonstrate the occurrence of conjugative transfer of resistance determinants, including sul3, from all of the isolates of S. enterica serovar Typhimurium DT104 carrying sul3-associated integron type III. However, Southern hybridizations after S1-PFGE experiments demonstrated that the sul3 gene and sul3-associated integron type III were located on large plasmids of different sizes (between 150 and 220 kb) (Table 1). One isolate from the environment presented two copies of the sul3 gene on plasmids of different sizes (150 and 170 kb), both also detected in human and food isolates of the same clone. The conjugative plasmid of ca. 240 kb carrying sul3 integron type III was only detected on a non-DT104 S. enterica serotype Typhimurium isolate (Table 1).
We describe the dissemination of sul3 associated with plasmid-borne class 1 integrons containing an unusual 3′CS site. The presence of similar sul3-integron platforms containing different gene cassette arrays or hybrid genes suggests evolution of the genetic background by different recombinatorial events. The association with epidemic plasmids and particular MDR clones of Salmonella might contribute to the maintenance and further spread of modular antibiotic resistance elements from food animals to hospitalized humans, as reported for other Salmonella genetic elements (2).
Nucleotide sequence accession numbers.
The nucleotide sequences of the sul3-carrying integrons reported in this study have been submitted to the EMBL/GenBank sequence databases and assigned accession numbers EF051037 (type I), EF051038 (type II), and EF051039 (type III).
Acknowledgments
We are very grateful to Teresa Coque (Hospital Ramon et Cajal, Madrid, Spain) for critical reading of the manuscript, V. Perreten (Institute of Veterinary Bacteriology, University of Bern, Bern, Switzerland) for the control strain with plasmid pVP440, Centro Nacional de Salmonella (Lisboa, Portugal) for serotyping of the strains, and CDC for the PFGE protocols and control strain S. enterica serotype Braenderup H9812.
The present work was partially supported by FSE/FEDER (POCI/AMB/61814/2004).
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Antunes, P., J. Machado, J. C. Sousa, and L. Peixe. 2005. Dissemination of sulfonamide resistance genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica strains and relation with integrons. Antimicrob. Agents Chemother. 49:836-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antunes, P., J. Machado, and L. Peixe. 2006. Characterization of antimicrobial resistance and class 1 and 2 integrons in Salmonella enterica isolates from different sources in Portugal. J. Antimicrob. Chemother. 58:297-304. [DOI] [PubMed] [Google Scholar]
- 3.Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 4.Bean, D. C., D. M. Livermore, I. Papa, and L. M. C. Hall. 2005. Resistance among Escherichia coli to sulphonamides and other antimicrobials now little used in man. J. Antimicrob. Chemother. 56:962-964. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, K. M., D. G. White, M. E. Hume, T. L. Poole, and D. J. Nisbet. 2005. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 243:285-291. [DOI] [PubMed] [Google Scholar]
- 6.Grape, M., L. Sundström, and G. Kronvall. 2003. Sulfonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J. Antimicrob. Chemother. 52:1022-1024. [DOI] [PubMed] [Google Scholar]
- 7.Guerra, B., E. Junker, A. Schroeter, B. Malorny, S. Lehmann, and R. Helmuth. 2003. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J. Antimicrob. Chemother. 52:489-492. [DOI] [PubMed] [Google Scholar]
- 8.Guerra, B., E. Junker, and R. Helmuth. 2004. Incidence of the recently described sulfonamide resistance gene sul3 among German Salmonella enterica strains isolated from livestock and food. Antimicrob. Agents Chemother. 48:2712-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerra, B., E. Junker, A. Miko, R. Helmuth, and M. C. Mendoza. 2004. Characterization and localization of drug resistance determinants in multidrug-resistant, integron-carrying Salmonella enterica serotype Typhimurium strains. Microb. Drug Resist. 10:83-91. [DOI] [PubMed] [Google Scholar]
- 10.Hammerum, A. M., D. Sandvang, S. R. Andersen, A. M. Seyfarth, L. J. Poesbo, N. Frimodt-Moller, and O. E. Heuer. 2006. Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 106:235-237. [DOI] [PubMed] [Google Scholar]
- 11.Handwerger, S., J. Skoble, L. F. Discotto, and M. J. Pucci. 1995. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob. Agents Chemother. 39:362-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kado, C. L., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perreten, V., and P. Boerlin. 2003. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47:1169-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchett, L. C., M. E. Konkel, J. M. Gay, and T. E. Besser. 2000. Identification of DT104 and U302 phage types among Salmonella enterica serotype Typhimurium isolates by PCR. J. Clin. Microbiol. 38:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sköld, O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist. Updates 3:155-160. [DOI] [PubMed] [Google Scholar]