Abstract
Screening for new antimicrobial agents is routinely conducted only against actively replicating bacteria. However, it is now widely accepted that a physiological state of nonreplicating persistence (NRP) is responsible for antimicrobial tolerance in many bacterial infections. In tuberculosis, the key to shortening the 6-month regimen lies in targeting this NRP subpopulation. Therefore, a high-throughput, luminescence-based low-oxygen-recovery assay (LORA) was developed to screen antimicrobial agents against NRP Mycobacterium tuberculosis. M. tuberculosis H37Rv containing a plasmid with an acetamidase promoter driving a bacterial luciferase gene was adapted to low oxygen conditions by extended culture in a fermentor with a 0.5 headspace ratio. The MICs of 31 established antimicrobial agents were determined in microplate cultures maintained under anaerobic conditions for 10 days and, for comparative purposes, under aerobic conditions for 7 days. Cultures exposed to drugs under anaerobic conditions followed by 28 h of “recovery” under ambient oxygen produced a luminescent signal that was, for most compounds, proportional to the number of CFU determined prior to the recovery phase. No agents targeting the cell wall were active against NRP M. tuberculosis, whereas drugs hitting other cellular targets had a range of activities. The calculated Z′ factor was in the range of 0.58 to 0.84, indicating the suitability of the use of LORA for high-throughput assays. This LORA is sufficiently robust for use for primary high-throughput screening of compounds against NRP M. tuberculosis.
It is now widely accepted that a physiological state of nonreplicating persistence (NRP) is responsible for antimicrobial tolerance in many bacterial infections (11). In tuberculosis, a subpopulation of Mycobacterium tuberculosis isolates in NRP is considered an important contributing factor to the long treatment duration required, and the key to shortening the currently recommended 6-month regimen (the primary goal of new anti-M. tuberculosis chemotherapy) lies in effective targeting of this phenotype (2, 6). Standard drug susceptibility assays for detection of the activities of drugs against rapidly growing bacteria may not identify such compounds (8, 11). In vitro models of M. tuberculosis isolates in NRP exist (4, 30, 37-39), and there are several reports of studies that have assessed drug activity against NRP or stationary-phase cells (20, 34-36, 42); however, these have relied upon the enumeration of CFU, thus precluding high-throughput screening (HTS) applications and requiring a minimum of 3 to 4 weeks for the completion of testing. In addition, the culture and preparation of NRP M. tuberculosis cells (5, 17, 29, 30, 38) have used batch cultures, which are not optimal for monitoring and for comparative studies. In contrast, a chemostat or a fermentor with a continuous-oxygen-monitoring culture system allows bacteria to be grown in a controlled and defined environment; there are two reports of the successful cultivation of M. tuberculosis in such systems (10, 24). We describe here a method compatible with HTS (Fig. 1) for the rapid detection of the activities of antimicrobial agents against M. tuberculosis in NRP using a low-oxygen-recovery assay (LORA) and a fermentor-grown culture of a luciferase reporter strain of M. tuberculosis adapted to low oxygen conditions.
FIG. 1.
Drug susceptibility under aerobic (replicating) and anaerobic (nonreplicating) conditions. The vertical lines and shaded arrows are proportional to the incubation times.
MATERIALS AND METHODS
Construction of reporter plasmid expressing luxAB.
The recombinant shuttle vector pFCA-luxAB was constructed by inserting the luxAB gene from pSMT1 (33) into pFPCA1 (9), which uses the acetamidase promoter to provide an enhanced signal. The luxAB genes were derived by PCR amplification with primers P1 (TAGGATCCTAAGAAAGATGAAATTTGGAAACTTCC) and P2 (TTCTTTAAATTACGAGTGGTATTTGACGATGTTGG). The amplified PCR product was cloned into the pGEM-T easy vector (Promega). After an Escherichia coli clone containing pGEMT-luxAB was obtained, the isolated plasmid was extracted and digested with BamHI and EcoRI. Following the ligation reaction with pFPCA1, the resulting plasmid, pFCA-luxAB, was used to transform E. coli One shot Top10 competent cells (Invitrogen). The plasmid was isolated, and the structure was confirmed by restriction enzyme digestion and sequencing analysis.
Electroporation of plasmid pFCA-luxAB into M. tuberculosis H37Rv ATCC 27294.
M. tuberculosis H37Rv ATCC 27294 was obtained from the American Type Culture Collection (Manassas, VA). After 5 to 7 days of culture in 200 ml Middlebrook 7H9 medium supplemented with oleic acid-albumin-dextrose-catalase, the cells were washed and transformed by mixing at least 1 μg of purified plasmid and incubating at room temperature for 30 min, followed by electroporation. The transformants were cultured on Middlebrook 7H11 agar containing 20 μg/ml kanamycin for 4 weeks. Selected colonies were transferred to 100 μl of Middlebrook 7H9 broth and sonicated at 30 W for 20 s (model S3000; Misonix Inc.) at ambient temperature prior to the measurement of the luminescence.
Antimicrobial agents.
Amikacin disulfate (Sigma), capreomycin sulfate (Sigma), ciprofloxacin (Fluka), clarithromycin (Abbott Laboratories), clindamycin hydrochloride (Sigma), clofazimine (Sigma), d-cycloserine (Sigma), ethambutol dihydrochloride (Sigma), furazolidone (Sigma), fusidic acid (sodium salt; Sigma), isoniazid (Sigma), ketoconazole (Sigma), lincomycin hydrochloride (BioChemika), linezolid (Pharmacia-Upjohn), metronidazole (Sigma), minocycline hydrochloride (Sigma), moxifloxacin (Bayer), niclosamide (Sigma), nitrofurantoin (Sigma), ofloxacin (Sigma), p-aminosalicylic acid (sodium salt; Sigma), pyrazinamide (Sigma), rifabutin (Ineti), rifampin (Fisher), RU66252 (synthesized as described elsewhere [16]), streptomycin sulfate (Sigma), tazobactam (Sigma), thiacetazone (Sigma), trimethoprim (Sigma), and vancomycin hydrochloride (Sigma) were obtained from the indicated manufacturers. The drugs were solubilized according to the manufacturers' recommendations, and stock solutions were filter sterilized (pore size, 0.22 μm) and stored at −80°C for not more than 30 days.
Growth conditions.
For the fermentor culture, recombinant H37Rv(pFCA-luxAB) was grown to NRP phase 2 (NRP-2) in 300 ml of Dubos Tween albumin broth (Becton Dickinson) in a BioStatQ fermentor (B. Braun Biotech) to mimic the Wayne oxygen-limited culture with a headspace ratio (HSR) of 0.5 and agitated at a stir rate of 120 rpm with no detectable perturbation of the surface of the medium, as described previously (10). The fermentor culture was operated and maintained within a biosafety level 3 laboratory. The dissolved oxygen concentration (DOC) was continuously monitored with an Ingold oxygen sensor probe. The optical densities of the cultures at 570 nm (A570 values), the numbers of relative light units (RLUs), and the CFU levels were determined at 3-day intervals. Bacterial samples were removed through a silicone septum with a syringe in order to preclude the introduction of oxygen. The number of CFU was estimated by plating dilutions of aliquots on Dubos oleic-albumin agar plates in triplicate and incubating the cultures at 37°C. The colonies were enumerated every week for 5 weeks. The cells were harvested at 22 days, a time when the A570 and DOC readings indicated achievement of the desired growth phase (NRP-2). Fifty-milliliter aliquots of bacterial culture samples were centrifuged (2,700 × g, 30 min, 4°C), washed once with prechilled phosphate-buffered saline (PBS; pH 7.4), suspended in 1 ml of PBS, and stored at −80°C.
In vitro LORA and conventional aerobic culture assay.
Prior to use, the cultures were thawed, diluted in Middlebrook 7H12 broth (Middlebrook 7H9 broth containing 1 mg/ml Casitone, 5.6 μg/ml palmitic acid, 5 mg/ml bovine serum albumin, and 4 μg/ml filter-sterilized catalase), and sonicated for 15s. The cultures were diluted to obtain an A570 of 0.03 to 0.05 and 3,000 to 7,000 RLUs per 100 μl. This corresponds to 5 × 105 to ∼2 × 106 CFU/ml. Twofold serial dilutions of 31 antimicrobial agents were prepared in a volume 100 μl in black 96-well microtiter plates, and 100 μl of the cell suspension was added. For LORA, the microplate cultures were placed under anaerobic conditions (oxygen concentration, less than 0.16%) by using an Anoxomat model WS-8080 (MART Microbiology) and three cycles of evacuation and filling with a mixture of 10% H2, 5% CO2, and the balance N2 (7, 32). An anaerobic indicator strip was placed inside the chamber to visually confirm the removal of oxygen. The plates were incubated at 37°C for 10 days and then transferred to an ambient gaseous condition (5% CO2-enriched air) incubator for a 28-h “recovery.” The numbers of CFU (determined by subculture onto Middlebrook 7H11 agar) during the 10-day incubation did not increase and remained essentially unchanged. On day 11 (after the 28-h aerobic recovery), 100 μl culture was transferred to white 96-well microtiter plates for determination of luminescence. For the conventional assay, the microplate cultures were placed in an incubator under ambient gaseous conditions (5% CO2-enriched air) for 7 days and 100 μl culture was transferred to white 96-well microtiter plates for determination of luminescence. A 10% solution of n-decanal aldehyde (Sigma) in ethanol was freshly diluted 10-fold in PBS, and 100 μl was added to each well with an autoinjector. Luminescence was measured in a Victor2 multilabel reader (Perkin-Elmer Life Sciences) by using a reading time of 1 s. The MIC was defined as the lowest drug concentration effecting growth inhibition of ≥90% relative to the growth for the drug-free controls. The MICs were numerically extrapolated from transformed inhibition-concentration plots, as described previously (31). Briefly, the numeric approach used commonly available spreadsheet software and was based on the calculation of averaged differences in the growth indicators (percent inhibition in the concentration intervals between the test culture and the dimethyl sulfoxide solvent control culture and linear approximation of the concentrations effecting a 90% reduction). Thus, the MICs were independent of the discrete twofold concentrations of the drug dilutions tested.
Z′ factor and statistical analysis for quality assessment of LORA.
To determine the suitability of LORA as an HTS assay, Z′-factor values (43) for 31 antimicrobial agents in LORA were calculated by using the following equation: Z′ (3 standard deviations of positive control + 3 standard deviations of negative control)/(mean of positive control − mean of negative control). All analyses were performed with Microsoft Excel software. Correlation coefficients were defined as the R2 values calculated between bacterial growth (CFU/ml) after 10 days of incubation under low-oxygen conditions and luminescence (RLUs) after 28 h of recovery.
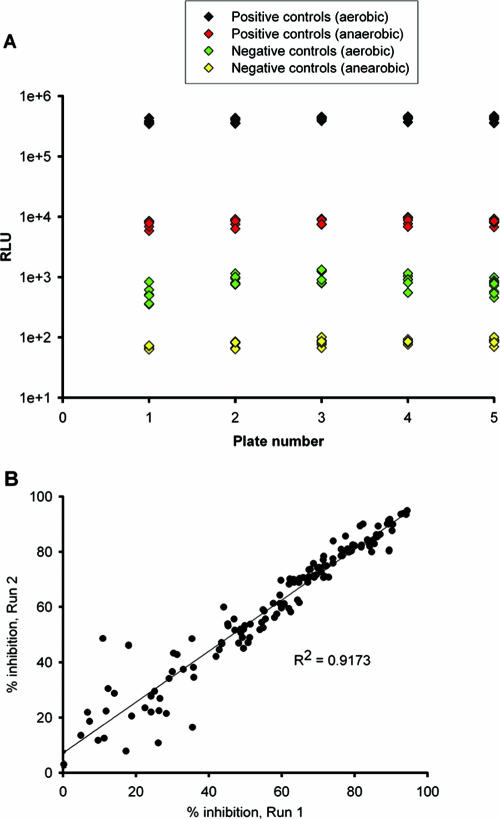
The signal-to-noise ratios determined from the controls wells of five representative plates were 453.3 and 103.1 (see Fig. 4A). The calculated Z′ factor was in the range of 0.58 to 0.84, indicating the suitability for the use of LORA for HTS. To further validate this assay, the percent inhibition by combinations of 15 compounds, including 15 positive and 15 negative controls, were determined in duplicate. A significant correlation was observed between the duplicates (Fig. 4B).
FIG. 4.
(A) Well-to-well variation for positive and negative control well signals from five plates of 31 antimicrobial agents. The calculated Z′ factor is in the range of 0.58 to 0.84. (B) Statistical analysis for reproducibility of duplicates. A significant linear correlation was observed between duplicate sets. Each of the 225 datum points represents the result of one independent experiment.
RESULTS
Inoculum preparation: growth, luminescence, and oxygen consumption of Mycobacterium tuberculosis pFCA-luxAB in a fermentor-based model.
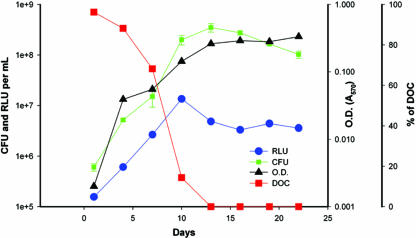
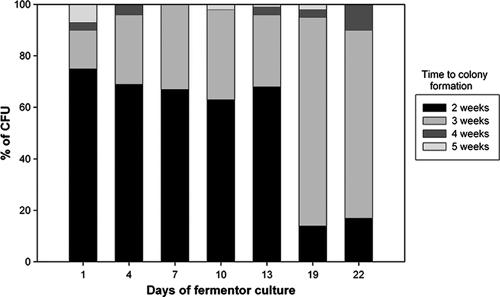
Changes in luminescence, cell growth, and DOC in actively growing cells under conditions of self-depletion of oxygen (Fig. 2) were monitored in a fermentor. The total cell number and optical density during log-phase growth peaked at day 10 and showed significant Pearson coefficients of correlation with luminescence (0.98 and 0.83, respectively). By day 13, the culture had entered into an NRP stage with no significant change in the numbers of CFU or optical density but with decreased luminescence. The proportion of the actively growing population versus that of the relatively slowly growing population was determined during the different phases of the growth curve by evaluation of the kinetics of colonial growth (Fig. 3). The proportion of bacteria able to produce visible colonies within 2 weeks of incubation on solid medium decreased dramatically at 19 days of incubation compared to the proportion at the earlier time points. The rate of appearance of colonies within 2 weeks was significantly more rapid in log phase (P = 0.0001) than in NRP-2.
FIG. 2.
Growth of an Mycobacterium tuberculosis H37Rv(pFCA-luxAB) inoculum in a 0.5 HSR fermentor culture. The numbers of CFU, RLU, optical density (OD; A570), and DOC were monitored during culture in a fermentor. The DOC is expressed as the percent saturation of the medium relative to that of the medium initially equilibrated with air.
FIG. 3.
Proportional distribution of CFU according to the time required to visually detect colonial growth on solid media. The percentage of the CFU value was determined from the proportion of the net number of CFU visible at a given time relative to the total number of CFU after 5 weeks of incubation.
Comparative MIC determination under aerobic and anaerobic culture.
For most of the 31 antimicrobial agents, the MICs for NRP M. tuberculosis (in anaerobic culture) were higher than those obtained under aerobic conditions (Table 1) (with the few exceptions belonging to the nitroaromatic compounds). These differences were most marked with isoniazid and ethambutol, two established antituberculosis agents that act on cell wall targets. The rifamycins rifampin and rifabutin were among the most active compounds under both conditions. Most of the agents acting on the 30S ribosome demonstrated either modest or potent activities against NRP M. tuberculosis; the exception was minocycline. In contrast, among those agents that act on the 50S ribosome, only RU66252, an experimental 11,12-carbazate-substituted macrolide, demonstrated relatively potent activity. Among the fluoroquinolones, moxifloxacin was highly active under both aerobic and anaerobic conditions, while the other representatives were markedly less active under the latter condition. Niclosamide and PA-824 were both active against NRP M. tuberculosis, with markedly better activities than metronidazole or the nitrofurans furazolidone and nitrofurantoin. Clofazimine demonstrated relatively potent activity against NRP M. tuberculosis.
TABLE 1.
Comparative activities of 31 antimicrobial agents against M. tuberculosis (luxAB) under anaerobic (LORA) and aerobic conditions
| Target or class | Drug | MIC (μM) derived by use ofa:
|
|||
|---|---|---|---|---|---|
| Aerobic conditions
|
Anaerobic conditions
|
||||
| T7 RLU indicator | T7 CFU indicator | T11 RLU (LORA) indicator | T10 CFU indicator | ||
| Cell wall | Vancomycin | >128 | 15.5 | >128 | 128.0 |
| Tazobactam | >128 | >128 | >128 | >128 | |
| Cycloserine | 63.7 | 60.5 | >128 | 124.8 | |
| Isoniazid | ≤0.5 | ≤0.5 | >128 | >128 | |
| Thiacetazone | 14.6 | 31.3 | >128 | 70.4 | |
| Ethambutol | 1.8 | 1.0 | >128 | >128 | |
| RNA polymerase | Rifampin | 0.5 | ≤0.5 | 1.8 | ≤0.5 |
| Rifabutin | 0.5 | ≤0.5 | ≤0.5 | ≤0.5 | |
| 30s ribosome | Amikacin | 0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| Streptomycin | 0.5 | ≤0.5 | 1.2 | 0.8 | |
| Capreomycin | 0.7 | ≤0.5 | 23.9 | 1.7 | |
| Minocycline | 7.1 | 7.2 | 53.5 | 38.4 | |
| Fusidic acid | 15.8 | 7.0 | 57.7 | 7.2 | |
| 50s ribosome | Linezolid | 6.3 | 1.9 | 65.2 | >128 |
| Lincomycin | >128 | 103.6 | >128 | >128 | |
| Clindamycin | >128 | >128 | >128 | >128 | |
| Clarithromycin | 13.4 | 1.6 | >128 | 18.7 | |
| RU66252 | 0.8 | 1.8 | 2.7 | 1.3 | |
| DNA gyrase | Moxifloxacin | 0.5 | ≤0.5 | 15.9 | ≤0.5 |
| Ciprofloxacin | 1.5 | ≤0.5 | >128 | 33.0 | |
| Ofloxacin | 2.3 | 0.7 | >128 | 24.2 | |
| Enzyme inhibitor | Ketoconazole | 13.9 | 7.6 | 53.0 | 53.3 |
| p-Aminosalicylic acid | 12.3 | 12.3 | >128 | >128 | |
| Trimethoprim | >128 | >128 | >128 | 83.2 | |
| Nitroaromatics | PA-824 | 0.6 | 0.7 | 3.1 | 12.0 |
| Furazolidone | 77.4 | 1.0 | 9.7 | 44.8 | |
| Metronidazole | >128 | >128 | >128 | 40.0 | |
| Nitrofurantoin | >128 | 0.9 | 63.9 | 32.0 | |
| Niclosamide | ≤0.5 | ≤0.5 | ≤0.5 | 0.9 | |
| Unknown | Clofazimine | ≤0.5 | 0.7 | 1.5 | 0.8 |
| Pyrazinamide (pH 6.8) | >128 | >128 | >128 | 96.0 | |
T7 RLU, RLUs determined after 7 days of aerobic incubation with test compound; T7 CFU, CFU from subcultures performed after 7 days of aerobic incubation with test compounds; T11 RLU, RLUs determined after 10 days of anaerobic incubation with test compounds and 28 h of aerobic incubation (“recovery”); T10 CFU, CFU from subcultures performed after 10 days of anaerobic incubation with test compounds.
Although in general there was a good overall correlation between the MICs determined by the use of luminescence and those determined by the use CFU for both aerobic cultures (R2 = 0.71) and hypoxic cultures (R2 = 0.78), there were some exceptions. Luminescence-derived MICs were more than eightfold higher than the CFU-derived MICs under aerobic conditions for vancomycin, clarithromycin, furazolidone, and nitrofurantoin and under anaerobic conditions for capreomycin, fusidic acid, and moxifloxacin.
Z′ factor and statistical analysis for quality assessment of LORA.
For the LORAs, all Z′-factor values derived from the RLUs determined after 7 days of aerobic incubation with test compound and the RLUs determined after 10 days of anaerobic incubation with test compound and 28 h of aerobic incubation were in the range of 0.58 to 0.84. Signal-to-noise ratios were 453.3 and 103.1 in the aerobic luminescence assay and LORA, respectively (Fig. 4A). In addition, the reproducibilities of duplicate sets of two-drug combinations in LORA yielded an R2 value of >0.9 (Fig. 4B).
DISCUSSION
A reduced luminescence signal per CFU, an increased proportion of slower-growing bacteria, and an increase in the amount of the alpha-crystallin protein (10) were all consistent with the shift to an NRP phenotype. In general, luciferase reporters function as indicators of the overall metabolic activity of a population rather than as indicators of the proportion of active cells within a population (15). This is consistent with our data, which showed a relative decrease in the proportion of the rapidly growing population (Fig. 3) but which maintained the overall luminescence level (Fig. 2). Only in actively growing cells has a good correlation between CFU and RLUs been observed. Therefore, RLUs have been used to monitor transcriptional changes and to estimate the total numbers of viable cells in such cultures. However, to date, the enumeration of nonreplicating populations in liquid culture or the organs of mice is still conducted by determination of the numbers of CFU (14, 22). In the present study, changes in the colony-forming rate were observed over time in the fermentor culture, with a progressive decrease in the population able to form colonies within 2 weeks, consistent with slow or nonreplicating populations resulting from the self-depletion of oxygen (37, 38).
Because a brief exposure of low-oxygen-adapted M. tuberculosis isolates to air was previously determined to be insufficient to reverse the NRP phenotype (39), we did not attempt to maintain a low-oxygen environment during harvesting or storage of the inoculum or during subsequent inoculation of drug-containing media. Instead, we relied upon a low temperature during harvest (4°C) and storage (−80°C) and speed during the setup of the test plate to minimize the extent of any such adaptation.
The other opportunity for the presence of oxygen to potentially confound the interpretation of the results is during the aerobic recovery phase. Therefore, the minimum time required to obtain a robust luminescence signal for the drug-free controls was used. In order to minimize the extent of the recovery phase, we attempted to optimize the signal intensity through the use of the Vibrio harveyi luciferase gene (33) (rather than the Photinus counterpart) driven by an acetamidase promoter (9) (rather than the commonly used hsp60 gene [41]) and a multicopy plasmid (rather than an integrated vector). In general, the good correlation of the response measured in CFU at the immediate termination of the anaerobic incubation period with that measured in RLUs following the aerobic recovery phase suggests that any drug activity during this relatively brief recovery phase does not constitute a significant proportion of the overall activity. Previous attempts in our laboratory to use Alamar Blue reduction (12, 18) or green (or red) fluorescent protein expression (9) as a viability endpoint following anaerobic incubation required up to 1 week of aerobic recovery to obtain an adequate signal, a time period that seriously compromised data interpretation (data not shown).
As described previously for other Wayne-type models and consistent with a lack of sterilization activity in humans, drugs that act on cell wall targets, such as isoniazid, thiacetazone, ethambutol, and cycloserine, although they are active against growing cultures, were relatively inactive in the LORA (as confirmed by determination of the numbers of CFU). Also in agreement with previously described CFU-based nonreplicating models, we observed relatively potent activities under hypoxic conditions with rifampin (40), capreomycin (20), moxifloxacin (21), niclosamide (35), and PA-824 (34).
In addition to capreomycin (20), this study identified other compounds targeting the 30S ribosome (with the exception of minocycline) that had relatively potent activities against NRP M. tuberculosis, in particular, the aminoglycosides. Streptomycin was previously found to be active in one NRP model that used a traditional Wayne-type inoculum (Y. Li and S. G. Franzblau, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 862, 1999) but not in another one (20). The lack of sterilization activity of streptomycin in vivo is likely a result of its poor activity against bacteria residing within host macrophages (28) and/or, possibly, adaptive resistance (3). Other compounds not previously identified as active against NRP M. tuberculosis included the experimental macrolide RU66252 (16) and clofazimine. The former is active against M. tuberculosis in macrophage culture and in vivo. While the latter compound is considered a third-line anti-M. tuberculosis agent, it was nonetheless capable of having nearly sterilizing activity in mice dosed orally (23) and when it was administered intravenously in liposomes at dosages up to 10 times higher than those achievable without encapsulation (1). There are no other reports of the assessment of clofazimine for its sterilization activity in animal models or in humans.
Differences in reporter gene expression versus the numbers of CFU have been noted previously, particularly with specific classes of drugs, such as fluoroquinolones and β-lactams (19, 27). The sensitivity of LORA to fluoroquinolones was enhanced by extending the anaerobic incubation time from 7 to 10 days (data not shown), but while this decreased the MIC obtained by LORA, a discrepancy between luciferase assay- and CFU-based MICs remained.
Similarly, LORA, as described here, underestimated the activities of several nitroaromatic compounds (furazolidone, metronidazole, and nitrofurantoin) compared to those determined from the CFU-based readouts. For the nitrofurans this disparity was even more pronounced in aerobic cultures. Others have reported for metronidazole a ≥1-log10 reduction in the numbers of CFU over a concentration range of ≤8 to 50 μg/ml (47 to 292 μM) (13, 20, 26, 40) in low-oxygen-based models. We obtained such a result at 6.8 μg/ml (40 μM) with the CFU endpoint, but although a steep dose-luminescence response was observed (data not shown), the level of inhibition did not exceed 80% at the maximum concentration tested (22 μg/ml [128 μM]). Our CFU-based result with metronidazole suggests that anaerobiosis was achieved in these cultures, since no other efforts were made to reduce the redox potential. In general, detection of the activities of some low-molecular-weight compounds (including nitroaromatic compounds as well as pyrazinamide) by using a luciferase readout may require primary screening at concentrations higher than 128 μM.
We have begun using LORA for the screening of compound libraries and for the testing of combinations of compounds. Screening of the Gen-Plus 960 compound library (MicroSource Discovery Systems Inc.) found that mefloquine has modest but equivalent anti-M. tuberculosis activities against both NRP M. tuberculosis and replicating cultures, and this has led to a preliminary structure-activity relationship study of mefloquine analogs for their anti-M. tuberculosis activities (25).
We propose the use of LORA for primary or secondary screening of diverse compound libraries at relatively high concentrations, after which the MICs and their correlation with the numbers of CFU can be determined. For those classes for which luminescence and CFU have a good correlation, the former assay can thereafter be used in analoging studies. Such screening could be performed in parallel with or following HTS against replicating cultures to rapidly identify those compounds with activities against both subpopulations.
Acknowledgments
We thank David Sherman for helpful discussions.
This study was supported by contract NIH/NIAID/DAIDS-01-13, subcontract 258-01-0003, from the National Hansen's Disease Programs.
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Adams, L. B., I. Sinha, S. G. Franzblau, J. L. Krahenbuhl, and R. T. Mehta. 1999. Effective treatment of acute and chronic murine tuberculosis with liposome-encapsulated clofazimine. Antimicrob. Agents Chemother. 43:1638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2001. Tuberculosis. Scientific blueprint for tuberculosis drug development. Tuberculosis (Edinburgh) 81(Suppl. 1):1-52. [DOI] [PubMed] [Google Scholar]
- 3.Barclay, M. L., and E. J. Begg. 2001. Aminoglycoside adaptive resistance: importance for effective dosage regimens. Drugs 61:713-721. [DOI] [PubMed] [Google Scholar]
- 4.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 5.Boon, C., and T. Dick. 2002. Mycobacterium bovis BCG response regulator essential for hypoxic dormancy. J. Bacteriol. 184:6760-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshoff, H. I., and C. E. Barry III. 2005. Tuberculosis—metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3:70-80. [DOI] [PubMed] [Google Scholar]
- 7.Brazier, J. S., and S. A. Smith. 1989. Evaluation of the Anoxomat: a new technique for anaerobic and microaerophilic clinical bacteriology. J. Clin. Pathol. 42:640-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burman, W. J. 1997. The value of in vitro drug activity and pharmacokinetics in predicting the effectiveness of antimycobacterial therapy: a critical review. Am. J. Med. Sci. 313:355-363. [DOI] [PubMed] [Google Scholar]
- 9.Changsen, C., S. G. Franzblau, and P. Palittapongarnpim. 2003. Improved green fluorescent protein reporter gene-based microplate screening for antituberculosis compounds by utilizing an acetamidase promoter. Antimicrob. Agents Chemother. 47:3682-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, S. H., D. Goodlett, and S. Franzblau. 2006. ICAT-based comparative proteomic analysis of non-replicating persistent Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 86:445-460. [DOI] [PubMed] [Google Scholar]
- 11.Coates, A., Y. Hu, R. Bax, and C. Page. 2002. The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Discov. 1:895-910. [DOI] [PubMed] [Google Scholar]
- 12.Collins, L., and S. G. Franzblau. 1997. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 41:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhillon, J., B. W. Allen, Y. M. Hu, A. R. Coates, and D. A. Mitchison. 1998. Metronidazole has no antibacterial effect in Cornell model murine tuberculosis. Int. J. Tuberc. Lung Dis. 2:736-742. [PubMed] [Google Scholar]
- 14.Dhillon, J., D. B. Lowrie, and D. A. Mitchison. 2004. Mycobacterium tuberculosis from chronic murine infections that grows in liquid but not on solid medium. BMC Infect. Dis. 4:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan, S., L. A. Glover, K. Killham, and J. I. Prosser. 1994. Luminescence-based detection of activity of starved and viable but nonculturable bacteria. Appl. Environ Microbiol. 60:1308-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falzari, K., Z. Zhu, D. Pan, H. Liu, P. Hongmanee, and S. G. Franzblau. 2005. In vitro and in vivo activities of macrolide derivatives against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 49:1447-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Florczyk, M. A., L. A. McCue, R. F. Stack, C. R. Hauer, and K. A. McDonough. 2001. Identification and characterization of mycobacterial proteins differentially expressed under standing and shaking culture conditions, including Rv2623 from a novel class of putative ATP-binding proteins. Infect. Immun. 69:5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzblau, S. G., R. S. Witzig, J. C. McLaughlin, P. Torres, G. Madico, A. Hernandez, M. T. Degnan, M. B. Cook, V. K. Quenzer, R. M. Ferguson, and R. H. Gilman. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 36:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galluzzi, L., and M. Karp. 2003. Amplified detection of transcriptional and translational inhibitors in bioluminescent Escherichia coli K-12. J. Biomol. Screen. 8:340-346. [DOI] [PubMed] [Google Scholar]
- 20.Heifets, L., J. Simon, and V. Pham. 2005. Capreomycin is active against non-replicating M. tuberculosis. Ann. Clin. Microbiol. Antimicrob. 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Y., A. R. Coates, and D. A. Mitchison. 2003. Sterilizing activities of fluoroquinolones against rifampin-tolerant populations of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, Y., J. A. Mangan, J. Dhillon, K. M. Sole, D. A. Mitchison, P. D. Butcher, and A. R. Coates. 2000. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J. Bacteriol. 182:6358-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jagannath, C., M. V. Reddy, S. Kailasam, J. F. O'Sullivan, and P. R. Gangadharam. 1995. Chemotherapeutic activity of clofazimine and its analogues against Mycobacterium tuberculosis. In vitro, intracellular, and in vivo studies. Am. J. Respir. Crit. Care Med. 151:1083-1086. [DOI] [PubMed] [Google Scholar]
- 24.James, B. W., A. Williams, and P. D. Marsh. 2000. The physiology and pathogenicity of Mycobacterium tuberculosis grown under controlled conditions in a defined medium. J. Appl. Microbiol. 88:669-677. [DOI] [PubMed] [Google Scholar]
- 25.Jayaprakash, S., Y. Iso, B. Wan, S. G. Franzblau, and A. P. Kozikowski. 2006. Design, synthesis, and SAR studies of mefloquine-based ligands as potential antituberculosis agents. ChemMedChem. 1:593-597. [DOI] [PubMed] [Google Scholar]
- 26.Lenaerts, A. J., V. Gruppo, K. S. Marietta, C. M. Johnson, D. K. Driscoll, N. M. Tompkins, J. D. Rose, R. C. Reynolds, and I. M. Orme. 2005. Preclinical testing of the nitroimidazopyran PA-824 for activity against Mycobacterium tuberculosis in a series of in vitro and in vivo models. Antimicrob. Agents Chemother. 49:2294-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeliger, B., I. Caldelari, A. Bizzini, P. Stutzmann Meier, P. A. Majcherczyk, and P. Moreillon. 2003. Antibiotic-dependent correlation between drug-induced killing and loss of luminescence in Streptococcus gordonii expressing luciferase. Microb. Drug Resist. 9:123-131. [DOI] [PubMed] [Google Scholar]
- 28.Maurin, M., and D. Raoult. 2001. Use of aminoglycosides in treatment of infections due to intracellular bacteria. Antimicrob. Agents Chemother. 45:2977-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muttucumaru, D. G., G. Roberts, J. Hinds, R. A. Stabler, and T. Parish. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinburgh) 84:239-246. [DOI] [PubMed] [Google Scholar]
- 30.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pauli, G. F., R. J. Case, T. Inui, Y. Wang, S. Cho, N. H. Fischer, and S. G. Franzblau. 2005. New perspectives on natural products in TB drug research. Life Sci. 78:485-494. [DOI] [PubMed] [Google Scholar]
- 32.Shahin, M., W. Jamal, T. Verghese, and V. O. Rotimi. 2003. Comparative evaluation of Anoxomat and conventional anaerobic GasPak jar systems for the isolation of anaerobic bacteria. Med. Princ. Pract. 12:81-86. [DOI] [PubMed] [Google Scholar]
- 33.Snewin, V. A., M. P. Gares, P. O. Gaora, Z. Hasan, I. N. Brown, and D. B. Young. 1999. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect. Immun. 67:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stover, C. K., P. Warrener, D. R. VanDevanter, D. R. Sherman, T. M. Arain, M. H. Langhorne, S. W. Anderson, J. A. Towell, Y. Yuan, D. N. McMurray, B. N. Kreiswirth, C. E. Barry, and W. R. Baker. 2000. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature 405:962-966. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Z., and Y. Zhang. 1999. Antituberculosis activity of certain antifungal and antihelmintic drugs. Tuber. Lung Dis. 79:319-320. [DOI] [PubMed] [Google Scholar]
- 36.Wade, M. M., and Y. Zhang. 2004. Anaerobic incubation conditions enhance pyrazinamide activity against Mycobacterium tuberculosis. J. Med. Microbiol. 53:769-773. [DOI] [PubMed] [Google Scholar]
- 37.Wayne, L. G. 1994. Dormancy of Mycobacterium tuberculosis and latency of disease. Eur. J. Clin. Microbiol. Infect. Dis. 13:908-914. [DOI] [PubMed] [Google Scholar]
- 38.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wayne, L. G., and K. Y. Lin. 1982. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect. Immun. 37:1042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wayne, L. G., and H. A. Sramek. 1994. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 38:2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams, S. L., N. B. Harris, and R. G. Barletta. 1999. Development of a firefly luciferase-based assay for determining antimicrobial susceptibility of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 37:304-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie, Z., N. Siddiqi, and E. J. Rubin. 2005. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 49:4778-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]