Abstract
The frequency of tetracycline resistance in Streptococcus pneumoniae isolates in Poland is one of the highest in Europe. The aim of this study was to analyze the clonal diversity and resistance determinants of tetracycline-nonsusceptible S. pneumoniae isolates identified in Poland and to investigate the effect of tetracycline resistance on their susceptibilities to tigecycline, doxycycline, and minocycline. We have analyzed 866 pneumococcal isolates collected from 1998 to 2003 from patients with respiratory tract diseases, and 242 of these (27.9%) were found to be resistant to tetracycline. All of the resistant isolates were characterized by testing of their susceptibilities to other antimicrobials, serotyping, pulsed-field gel electrophoresis (PFGE), and identification of tetracycline resistance genes and transposons. Selected isolates representing the main PFGE types were analyzed by multilocus sequence typing. Among the isolates investigated, 27 serotypes and 146 various PFGE patterns, grouped into 90 types, were discerned. The most common PFGE type, corresponding to serotype 19F and sequence type 423, was represented by 22.3% of all of the tetracycline-resistant isolates. The tet(M) gene was the sole resistance gene in the group of isolates studied, and in over 96% of the isolates, the Tn916 family of tet(M)-containing conjugative transposons was detected. Several isolates contained specific variants of the transposons, the Tn1545-like, Tn3872-like, or Tn2009-like element. The correlation between the MICs of tetracycline, doxycycline, and minocycline was revealed, whereas no cross-resistance to tetracycline and tigecycline was observed.
Streptococcus pneumoniae is a common cause of serious and often life-threatening infections such as pneumonia, bacteremia, and meningitis, as well as upper respiratory tract infections, like otitis media or sinusitis. During the past few years, pneumococcal populations have become increasingly resistant to various antimicrobials, including tetracyclines. In Poland the rates of resistance of S. pneumoniae and other gram-positive bacteria to tetracycline are among the highest in Europe (20, 22), but despite this, the level of tetracycline consumption in Poland remains high in comparison to the levels of consumption in other European countries (http://www.esac.ua.ac.be/main.aspx?c=*ESAC2&n=4666).
Three distinct mechanisms of resistance to tetracycline in bacteria have been described so far, including active efflux, ribosome protection by chaperone proteins, and enzymatic inactivation of the compound. In S. pneumoniae, this resistance results from the acquisition of one of two genes, tet(M) or, sporadically, tet(O), both of which encode ribosome protection proteins (50). The main source of the tet(M) gene is conjugative transposons of the Tn916 family (41), the particular members of which, e.g., Tn5251, differ from Tn916 mostly in their nucleotide sequence (40). Some of these elements also contain other resistance genes, e.g., Tn1545 [erm(B), which encodes resistance to macrolides, and aph3′-III, which encodes resistance to kanamycin] (6, 7), Tn3703 [erm(B)] (27, 28), Tn3872 [erm(B) within Tn917] (32), and Tn2009 [mef(E), which encodes resistance to macrolides and which exists within the mega element] (11). On the other hand, several Tn916-like transposons with tet(M) [and, sometimes, erm(B)] got inserted into other elements, thus forming larger structures, e.g., Tn3701 with Tn3703 inside (8, 28). Some of these structures also carry the chloramphenicol resistance gene cat, e.g., Tn5253 (insertion of Tn5251 into cat-containing Tn5252) (1) and Tn3951 (21). As a result, clinical isolates of S. pneumoniae are often resistant to tetracycline and to other compounds. In the United States, more than 60% of erythromycin-resistant pneumococci are simultaneously resistant to tetracycline (13); and in some European countries, e.g., Spain and Italy, over 80% (31, 46) of erythromycin-resistant pneumococci are simultaneously resistant to tetracycline. In 95% of such isolates, the presence of the int-Tn gene, which encodes the transposase of the Tn916 family, was detected (34).
Only limited data concerning tetracycline resistance in S. pneumoniae, as well as the susceptibilities of resistant strains to a novel antibiotic, tigecycline, are available. The main objective of this analysis was to reveal the molecular epidemiology of tetracycline-resistant S. pneumoniae isolates in Poland and to investigate the relationship between tetracycline resistance and susceptibilities to tigecycline, doxycycline, minocycline, and other antimicrobials.
(Parts of this work were presented at the 15th ECCMID, 2 to 5 April 2005, Copenhagen, Denmark [R. Izdebski, E. Sadowy, and W. Hryniewicz, Abstr. 15th ECCMID, abstr. P1032, 2005], and the 5th ISPPD, 2 to 6 April 2006, Alice Springs, Australia [R. Izdebski, E. Sadowy, and W. Hryniewicz, Abstr. 5th ISPPD, abstr. PO13.03, 2006].)
MATERIALS AND METHODS
Bacterial strains.
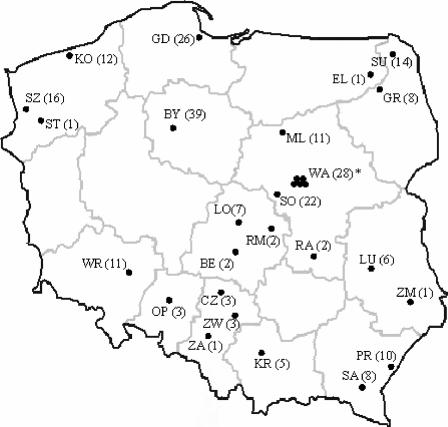
Eight hundred sixty-six clinical isolates of S. pneumoniae from respiratory tract infections were collected by the National Medicines Institute (NMI) in Warsaw, Poland, during an ongoing national surveillance program. They were recovered between 1998 and 2003 from various specimens (sputum specimens, bronchoalveolar lavage fluid, or tracheal aspirates) in 29 health care centers in 25 towns all over Poland (Fig. 1). The species identification was confirmed at NMI by conventional tests, such as susceptibility to optochin (bioMerieux, Marcy l'Etoile, France) and solubility in sodium deoxycholate (35). The collection of isolates was a part of that used in studies of the nonsusceptibility of Polish pneumococci to β-lactams and fluoroquinolones. Serotype, pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST) data, as well as the β-lactam MICs for 52 isolates among those that were nonsusceptible to penicillin and tetracycline, are published separately (44). The data for four ciprofloxacin- and tetracycline-nonsusceptible isolates are also published separately (43). In this work, the PFGE typing of these isolates was repeated for the whole group of tetracycline-resistant organisms.
FIG. 1.
Geographic distribution of tetracycline-resistant S. pneumoniae isolates in the study. The solid circles indicate the health care centers where the isolates were identified. Abbreviations of towns: BE, Be[strok]lchatów; BY, Bydgoszcz; CZ, Częstochowa; EL, E[strok]lk; GD, Gdańsk; GR, Grajewo; KO, Ko[strok]lobrzeg; KR, Kraków; LU, Lublin; LO, [strok]Lódź; ML, M[strok]lawa; OP, Opole; PR, Przemyśl; RA, Radom; RM, Rawa Mazowiecka; SA, Sanok; SO, Sochaczew; ST, Stargard; SU, Suwa[strok]lki; SZ, Szczecin; WA, Warszawa; WR, Wroc[strok]law; ZA, Zabrze; ZM, Zamość; ZW, Zawiercie. Numbers of isolates are shown in parentheses. *, 28 is the total number of isolates from five health care centers in Warsaw.
Susceptibility testing.
The initial screening of tetracycline susceptibility was performed by the disk diffusion method, as recommended by the Clinical and Laboratory Standards Institute (CLSI) (5). For all tetracycline-resistant isolates, the MICs of a wider set of antimicrobials were determined by the broth microdilution method, as recommended by the CLSI (5). The following agents were tested: tetracycline, doxycycline, penicillin G, erythromycin, and chloramphenicol (Sigma-Aldrich, Steinheim, Germany); tigecycline and minocycline (Wyeth, Pearl River, NY); clindamycin and linezolid (Pharmacia Upjohn, Kalamazoo; MI); levofloxacin and telithromycin (Aventis Pharma, Romainville, France); teicoplanin and rifampin (Gruppo Lepetit, Lainate, Italy); cefotaxime (ICN Biomedicals, Aurora, OH); meropenem (Astra Zeneca, Cheshire, United Kingdom); vancomycin (Eli Lilly, Indianapolis, IN); and trimethoprim-sulfamethoxazole (Roche, Basel, Switzerland). S. pneumoniae strain ATCC 49619 was included for quality control purposes. The appropriate CLSI criteria were used for the interpretation of the results (5). For doxycycline, minocycline, and teicoplanin, the guidelines of the Societe Francaise de Microbiologie (http://www.sfm.asso.fr/nouv/general.php?pa=2) were adopted. For tigecycline the FDA breakpoints (http://www.fda.gov/cder/foi/label/2005/021821lbl.pdf) were applied.
Analysis of resistance determinants.
Bacterial genomic DNAs were prepared with a Genomic Mini kit (A&A Biotechnology, Gdynia, Poland) and were used as templates for PCR. The isolates were tested for the presence of the tet(M) and tet(O) genes by PCR, as described elsewhere (14, 49). The PCR detection of the erm(B) and mef(E) genes was carried out with all isolates resistant to erythromycin [with primers ermBF and ermBR and primers MEF3 and MEF4 for the erm(B) and mef(E) genes, respectively], as reported previously (10, 48). The transposase gene int-Tn, specific for the Tn916 family of transposons, was detected by PCR, as described by others (38). The identification of Tn916-like and Tn1545-like transposons was performed by PCR with primers O13 and O14 as reported by Poyart et al. (37). The distinction of Tn1545-like elements was carried out by amplification of the aph3′-III gene promoter (primers aphF [5′-GGAACAGTGAATTGGAGTTC-3′] and aphR [5′-GACATTCCTTCCGTATC-3′]), as well as of the aph3′-III-erm(B) region, also with primers aphF and ermBR. For the detection of Tn5253, the region of the right junction between Tn5251 (Tn916-like) and Tn5252 was analyzed by using primers 5252F (5′-CCTCCTGATTCCAGTGTC-3′) and 5251R (5′-GATTCTTCGCTGAACGAC-3′). Tn5252 alone was detected by PCR of its transposase gene, int-Tn5252 (47). The isolates that were negative by PCR with primers O13 and O14 (37) were subjected to other analyses. Tn3872 was detected by three PCRs with primers O13 and O20, O23 and O14, and O19 and O22, respectively (37). The presence of Tn3951 was tested by PCR of the region between erm(B) and tet(M) with primers ermBF and tetMR. The isolates that were mef(E) positive were analyzed for the presence of the Tn2009 element (11) by PCR with primers MEF4 and O14 (10, 37).
In order to study the structural polymorphism of the tet(M) gene, tet(M) amplicons were digested with the SsiI restriction enzyme (an isoschizomer of AciI; Fermentas, Vilnius, Lithuania) and separated in 1% agarose gels (14).
Serotyping.
Serotyping was performed by PCR with primers specific for genes responsible for the biosynthesis of types 1, 3, 4, 6A, 6B, 14, 18C, 19A, 19F, and 23F of the capsular polysaccharide (4, 26, 36). The serotypes of the remaining isolates and of randomly selected representatives of the pneumococci serotyped by PCR were determined by the capsular swelling method at the Statens Serum Institute, Copenhagen, Denmark.
PFGE and MLST analyses.
Genomic DNA for PFGE was isolated as described by others (29), digested with the SmaI restriction enzyme (Fermentas), and separated in a CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA). Two isolates were considered indistinguishable when they shared PFGE patterns and were considered related when they showed a difference of one to three bands from each other. The dendrogram was constructed with Molecular Analyst software (version 1.12; Bio-Rad) by using the unweighted pair-group method with arithmetic averages clustering method with a Dice coefficient and a position tolerance of 1.5%. MLST of selected isolates representing the main PFGE types was performed as described previously (17). Particular allele numbers and sequence types (STs) were identified by using the MLST database (http://www.mlst.net).
Statistical analysis.
For statistical analysis, the χ2 test, the Fisher exact test, and the Kolmogorov-Smirnov test were used.
RESULTS
Tetracycline-resistant isolates.
Two hundred forty-two isolates, which represented 27.9% of all 866 isolates studied, were resistant to tetracycline. The percentages of tetracycline-resistant isolates were 25.4, 34.6, 28.5, 26.5, 26.3, and 21.6 during six consecutive years (1998 to 2003), respectively. Resistant isolates were recovered in all of the collaborating medical centers. The frequency of tetracycline resistance did not vary significantly over the 6 years studied, as shown by the χ2 and Fisher exact tests. However, a constant downward trend in the prevalence of resistant isolates was observed from 1999 to 2003 (P < 0.03; Kolmogorov-Smirnov test).
Susceptibilities to antimicrobial agents.
The tetracycline-resistant isolates showed various levels of susceptibility to the other antimicrobials studied (Table 1). A total of 80.1% and 75.6% of the isolates were nonsusceptible to doxycycline and minocycline, respectively, but all isolates were susceptible to tigecycline (MICs, ≤0.003 to 0.06 μg/ml). A total of 11.2% and 14.9% of the isolates were penicillin intermediate and penicillin resistant, respectively. The rates of resistance to erythromycin and chloramphenicol were 26.4% and 39.2%, respectively. A good correlation between the MICs of tetracycline, doxycycline, and minocycline was observed (Table 2), whereas no correlation existed between the tetracycline and the tigecycline MICs. Similarly, no link between the level of resistance to penicillin or erythromycin and the MICs of tigecycline was observed.
TABLE 1.
Distribution of MICs of 17 antimicrobial agents for tetracycline-resistant isolates
| Antimicrobial agent | No. of isolates for which the MIC (μg/ml) was:
|
MIC (μg/ml)
|
Percenta
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.003 | ≤0.0075 | 0.0075 | ≤0.015 | 0.015 | ≤0.03 | 0.03 | 0.06 | 0.12 | ≤0.25 | 0.25 | 0.5 | 1 | 2 | 4 | >4 | 8 | 16 | >16 | 32 | 64 | 128 | 50% | 90% | S | I | R | |
| Tetracycline | 14 | 48 | 109 | 69 | 2 | 32 | 64 | 100.0 | |||||||||||||||||||
| Doxycycline | 1 | 7 | 40 | 133 | 55 | 6 | 4 | 8 | 19.9 | 54.9 | 25.2 | ||||||||||||||||
| Minocycline | 3 | 23 | 33 | 89 | 71 | 23 | 4 | 8 | 24.4 | 36.8 | 38.8 | ||||||||||||||||
| Tigecycline | 2 | 37 | 123 | 73 | 7 | 0.015 | 0.03 | 100.0 | |||||||||||||||||||
| Penicillin G | 67 | 86 | 21 | 5 | 11 | 5 | 5 | 6 | 18 | 14 | 4 | 0.015 | 2 | 74.0 | 11.1 | 14.9 | |||||||||||
| Cefotaxime | 69 | 94 | 23 | 5 | 6 | 3 | 27 | 10 | 3 | 2 | 0.015 | 1 | 93.8 | 4.1 | 2.1 | ||||||||||||
| Meropenem | 173 | 1 | 17 | 5 | 14 | 30 | 2 | ≤0.015 | 0.5 | 86.8 | 12.8 | 0.4 | |||||||||||||||
| Vancomycin | 3 | 5 | 12 | 54 | 140 | 28 | 0.25 | 0.5 | 100.0 | ||||||||||||||||||
| Teicoplanin | 3 | 26 | 145 | 67 | 1 | 0.03 | 0.06 | 100.0 | |||||||||||||||||||
| Erythromycin | 175 | 3 | 1 | 3 | 3 | 1 | 56 | ≤0.25 | >16 | 72.4 | 1.2 | 26.4 | |||||||||||||||
| Clindamycin | 154 | 24 | 5 | 2 | 1 | 56 | ≤0.03 | >4 | 76.9 | 23.1 | |||||||||||||||||
| Telithromycin | 220 | 3 | 1 | 5 | 3 | 3 | 3 | 3 | 1 | ≤0.015 | ≤0.015 | 98.8 | 0.8 | 0.4 | |||||||||||||
| Chloramphenicol | 2 | 1 | 7 | 23 | 59 | 55 | 21 | 69 | 5 | 4 | 16 | 70.7 | 39.2 | ||||||||||||||
| Linezolid | 5 | 79 | 147 | 11 | 1 | 1 | 100.0 | ||||||||||||||||||||
| Rifampin | 221 | 15 | 2 | 1 | 1 | 1 | 1 | ≤0.03 | ≤0.03 | 98.8 | 0.4 | 0.8 | |||||||||||||||
| Levofloxacin | 2 | 2 | 73 | 161 | 2 | 1 | 1 | 1 | 1 | 98.8 | 0.4 | 0.4 | |||||||||||||||
| Trimethoprim-sulfamethoxazole | 1 | 3 | 2 | 39 | 27 | 39 | 37 | 32 | 41 | 16 | 5 | 2 | 8 | 30.2 | 31.4 | 38.4 | |||||||||||
S, susceptible; I, intermediate; R, resistant.
TABLE 2.
Number of isolates and correlation between MICs of tetracycline and doxycycline, tetracycline and minocycline, and tetracycline and tigecycline
| Drug and MIC (μg/ml) | No. of isolates for which the tetracycline MIC (μg/ml) was:
|
|||||
|---|---|---|---|---|---|---|
| 8 | 16 | 32 | 64 | 128 | Total | |
| Doxycycline | ||||||
| 0.5 | 1 | 1 | ||||
| 1 | 3 | 3 | 1 | 7 | ||
| 2 | 7 | 22 | 11 | 40 | ||
| 4 | 3 | 23 | 67 | 39 | 1 | 133 |
| 8 | 29 | 25 | 1 | 55 | ||
| 16 | 1 | 5 | 6 | |||
| Total | 14 | 48 | 109 | 69 | 2 | 242 |
| Minocycline | ||||||
| 0.5 | 2 | 1 | 3 | |||
| 1 | 9 | 11 | 3 | 23 | ||
| 2 | 1 | 12 | 16 | 4 | 33 | |
| 4 | 2 | 19 | 51 | 17 | 89 | |
| 8 | 5 | 27 | 37 | 2 | 71 | |
| 16 | 12 | 11 | 23 | |||
| Total | 14 | 48 | 109 | 69 | 2 | 242 |
| Tigecycline | ||||||
| ≤0.003 | 1 | 1 | 2 | |||
| 0.0075 | 4 | 8 | 13 | 12 | 37 | |
| 0.015 | 7 | 30 | 50 | 34 | 2 | 123 |
| 0.03 | 3 | 8 | 42 | 20 | 73 | |
| 0.06 | 1 | 3 | 3 | 7 | ||
| Total | 14 | 48 | 109 | 69 | 2 | 242 |
Serotyping, PFGE, and MLST analyses.
Serotypes, PFGE types, and the MLST data are shown in Table 3. Altogether, 27 serotypes were present in the group of isolates investigated. The majority of isolates (78.1%) comprised five serotypes: 19F (69 isolates [28.5%]), 23F (60 isolates [24.8%]), 6B (27 isolates [11.2%]), 6A (17 isolates [7.0%]), and 14 (16 isolates [6.6%]).
TABLE 3.
Distribution of serotypes, PFGE types and subtypes, STs, and resistance to antimicrobial agents among tetracycline-resistant isolates
| Serotype | PFGE (sub)type(s) (no. of isolates) | STa | Drug(s) to which isolate(s) was resistantb | International clone |
|---|---|---|---|---|
| 3 | 55a (2) | ND | ||
| 3 | 55a (1) | ND | DOX | |
| 3 | 1a (1) | 423 | CHL | |
| 4 | 56a (1) | ND | ||
| 6A | 31a (1) | 72 | MIN | |
| 6A | 32a (1) | 440 | MIN | |
| 6A | 24a (1) | 490 | DOX, ERY, CLI | |
| 6A | 24b (1), 29a (1) | 490 | SXT | |
| 6A | 24c (1) | 490 | MIN, SXT | |
| 6A | 25a (1), 27a (1) | 490 | ||
| 6A | 28a (1) | 490 | CHL | |
| 6A | 26a (1) | 490 | MIN, ERY, CLI, SXT | |
| 6A | 26a (1) | 1049 | MIN, DOX, ERY, CLI, SXT | |
| 6A | 30a (2) | 2185 | DOX | |
| 6A | 30a (1) | 2185 | DOX, MIN | |
| 6A | 30a (1) | 2185 | DOX, MIN, SXT | |
| 6A | 30a (1) | 2185 | DOX, MIN, ERY, CLI | |
| 6A | 30b (1) | 2184 | DOX, MIN | |
| 6B | 35a (1) | 90 | MIN, PEN, CHL, SXT | Spain6B-2 |
| 6B | 34b (1) | 273 | DOX, MIN, ERY, CLI | Greece6B-22 |
| 6B | 34c (1) | 273 | MIN, ERY, CLI | Greece6B-22 |
| 6B | 34a (1) | 273 | MIN, ERY, CLI, CHL | Greece6B-22 |
| 6B | 33a (5), 33e (1) | 315 | DOX, MIN, ERY, CLI, SXT | Poland6B-20 |
| 6B | 33f (1) | 315 | MIN, ERY, CLI, SXT | Poland6B-20 |
| 6B | 33a (1), 33i (1) | 315 | MIN, ERY, CLI | Poland6B-20 |
| 6B | 33a (2), 33f (1), 33h (1) | 315 | DOX, MIN, ERY, CLI | Poland6B-20 |
| 6B | 33d (1) | 315 | DOX, ERY, CLI | Poland6B-20 |
| 6B | 33a (1) | 315 | DOX, MIN, CLI, CHL | Poland6B-20 |
| 6B | 33g (1) | 315 | DOX, MIN, PEN, ERY, CLI, SXT | Poland6B-20 |
| 6B | 33b (1) | 1032 | DOX, MIN, MEM, ERY, CLI | SLV Poland6B-20 |
| 6B | 33b (1) | 1032 | DOX, MIN, ERY, CLI | SLV Poland6B-20 |
| 6B | 33c (1) | 1053 | DOX, ERY, CLI, SXT | SLV Poland6B-20 |
| 6B | 34a (1) | 1490 | MIN, ERY, CLI, CHL | SLV Greece6B-22 |
| 6B | 34a (1) | 1490 | ERY, CLI, CHL | SLV Greece6B-22 |
| 6B | 33a (1) | 1505 | ERY, CLI, SXT | SLV Poland6B-20 |
| 6B | 16b (1) | 2138 | ||
| 7F | 57a (1) | ND | MIN | |
| 8 | 58a (1) | ND | ||
| 9N | 59a (2) | ND | ||
| 9N | 60a (2) | ND | ||
| 10A | 61a (1) | ND | ||
| 11A | 63a (1) | 62 | DOX, MIN, ERY, SXT | |
| 11A | 62a (1) | 1010 | DOX, MIN | |
| 11A | 64a (1) | ND | DOX, MIN, CHL | |
| 11A | 65a (1) | ND | CHL | |
| 12F | 66a (1) | ND | CHL | |
| 14 | 18a (1) | 124 | CHL | |
| 14 | 19a (1) | 124 | DOX, MIN, ERY, CHL | |
| 14 | 20a (1), 21a (1) | 124 | ||
| 14 | 14a (3) | 143 | DOX, MIN, PEN, ERY, CLI, SXT | |
| 14 | 14a (1) | 143 | DOX, MIN, PEN, ERY, CLI, TEL | |
| 14 | 14a (1) | 143 | DOX, MIN, PEN, CTX, ERY, CLI | |
| 14 | 14b (1) | 143 | DOX, MIN, PEN, ERY, CLI | |
| 14 | 15a (1) | 143 | PEN, ERY, CLI | |
| 14 | 22a(1) | 156 | PEN, ERY, CLI, SXT | Spain9V-3 |
| 14 | 23a (1) | 378 | ||
| 14 | 14a (1) | 1477 | DOX, MIN, PEN, ERY, CLI | |
| 14 | 16a (1) | 2138 | ||
| 14 | 17a (1) | 2139 | ||
| 15A | 67a (1) | 63 | DOX, MIN, ERY, CLI | Sweden15A-25 |
| 15A | 70a (1) | 124 | DOX, MIN, SXT | |
| 15A | 68a (1) | 410 | DOX, CHL | |
| 15A | 69a (1) | 410 | CHL, SXT | |
| 15A | 16b (1) | 2138 | ||
| 15B/C | 71a (1) | ND | ERY, CLI, SXT | |
| 15B/C | 72a (3) | ND | ||
| 15B/C | 73a (1) | ND | MIN, CHL | |
| 15F | 74a (1) | ND | MIN | |
| 18B | 76a (1) | ND | ||
| 18C | 1a (1) | 423 | DOX, CHL | |
| 18C | 75a (1) | 1016 | CHL, SXT | |
| 18C | 75b (1), 75c (1) | 1016 | ||
| 19A | 77a (1) | 276 | ERY, SXT, RIF | |
| 19A | 77b (1) | 276 | ERY, CLI | |
| 19A | 77b (1) | ND | MIN, PEN, ERY, CLI, SXT | |
| 19A | 77b (1) | ND | PEN, ERY, CLI, SXT | |
| 19A | 78a (2) | 1625 | SXT | |
| 19F | 13a (1) | 81 | PEN, CHL, SXT | Spain23F-1 |
| 19F | 7a (1) | 179 | DOX, ERY, CLI | SLV Portugal19F-21 |
| 19F | 6a (1) | 236 | DOX, MIN, PEN, CTX, ERY, SXT | Taiwan19F-14 |
| 19F | 9a (1) | 236 | SXT | Taiwan19F-14 |
| 19F | 11a (1) | 236 | MIN | Taiwan19F-14 |
| 19F | 8a (1) | 257 | ERY, CLI, SXT | |
| 19F | 5a (1) | 410 | DOX, CHL | |
| 19F | 1a (20) | 423c | CHL | |
| 19F | 10a (1) | 423 | CHL | |
| 19F | 1a (2) | ND | DOX, MIN, CHL | |
| 19F | 1a (1) | ND | DOX, CHL | |
| 19F | 1a (1) | ND | MIN, CHL | |
| 19F | 2b(1) | 423 | MIN, CHL | |
| 19F | 1a (7) | ND | CHL, SXT | |
| 19F | 12a (1) | 423 | CHL, SXT | |
| 19F | 1a (1) | ND | DOX, MIN, CHL, SXT | |
| 19F | 1a (3) | 423c | MIN, CHL, SXT | |
| 19F | 2a (1) | 423 | MIN, CHL, SXT | |
| 19F | 1a (2) | 423c | SXT | |
| 19F | 1a (2) | ND | MIN, SXT | |
| 19F | 1a (1) | 423 | ERY | |
| 19F | 1a (1) | ND | CHL, RIF | |
| 19F | 1a (3) | ND | ||
| 19F | 1e (1), 2c (1) | 423 | ||
| 19F | 1b (1) | 423 | DOX, MIN, SXT | |
| 19F | 1c (2) | 423c | SXT | |
| 19F | 1c (1) | ND | CHL, SXT | |
| 19F | 1g (1) | 423 | CHL, SXT | |
| 19F | 1f (1), 1g (1), 1i (1) | 423 | CHL | |
| 19F | 1h (1) | 423 | ||
| 19F | 3a (1) | 1489 | DOX, ERY, CLI | |
| 19F | 1d (1) | 1815 | ||
| 19F | 4a (1) | 2136 | DOX, MIN, SXT | |
| 20 | 79a (1) | ND | ||
| 22F | 80a (1) | ND | CHL | |
| 22F | 81a (1) | ND | ||
| 23F | 52a (1) | 72 | DOX, MIN, ERY, CLI | |
| 23F | 37a (3), 37d (1), 37e (2), 37g (1) | 81 | PEN, CHL, SXT | Spain23F-1 |
| 23F | 37d (1) | 81 | DOX, PEN, CHL, SXT | Spain23F-1 |
| 23F | 37a (2), 37h (1) | 81 | CHL, SXT | Spain23F-1 |
| 23F | 37a (1), 37f (1) | 81 | PEN, ERY, CHL, SXT | Spain23F-1 |
| 23F | 37b (1) | 81 | PEN, SXT | Spain23F-1 |
| 23F | 37c (1) | 81 | PEN, CHL, SXT, LVX | Spain23F-1 |
| 23F | 38a (1) | 81 | PEN, ERY, CLI, CHL, SXT | Spain23F-1 |
| 23F | 54a (1) | 242 | DOX, MIN, PEN, ERY, CLI | Taiwan23F-15 |
| 23F | 47b (2) | 272 | DOX, MIN, PEN, ERY, CLI, CHL, SXT | SLV Poland23F-16 |
| 23F | 47c (2) | 272 | DOX, MIN, PEN, CTX, ERY, CLI, CHL, SXT | SLV Poland23F-16 |
| 23F | 47c (1) | 272 | MIN, PEN, CTX, ERY, CLI, CHL, SXT | SLV Poland23F-16 |
| 23F | 48a (1) | 410 | CHL, SXT | |
| 23F | 1a (1) | 423 | SXT | |
| 23F | 39a (3), 39f (2), 42a (1), 43a (1), 45a (1) | 440 | MIN | |
| 23F | 39a (2), 42b (1), 44a (1) | 440 | ||
| 23F | 39a (1) | 440 | DOX, MIN, CHL | |
| 23F | 39a (1) | 440 | MIN, SXT | |
| 23F | 46a (1) | 440 | MIN, ERY, CLI | |
| 23F | 39a (1), 39c (1), 39e (1), 41a (1) | 602 | ||
| 23F | 39a (1), 39b (1), 40a (1) | 602 | MIN | |
| 23F | 39a (3) | 602 | MIN, SXT | |
| 23F | 37d (1) | 1051 | PEN, CHL, SXT | SLV Spain23F-1 |
| 23F | 51a (1) | 1116 | ||
| 23F | 53a (1) | 1364 | MIN, CHL | |
| 23F | 37e (1) | 1476 | CHL, SXT | SLV Spain23F-1 |
| 23F | 39d (1) | 1479 | MIN, ERY, SXT | |
| 23F | 39d (1) | 1479 | MIN, SXT | |
| 23F | 50a (1) | 2137 | CHL, SXT | |
| 23F | 47a (1) | 2140 | DOX, MIN, PEN, ERY, CLI, CHL, SXT | SLV Poland23F-16 |
| 23F | 49a (1) | 2141 | DOX | |
| 23F | 39a (1) | ND | ||
| 28 | 82a (1), 83a (1) | ND | ||
| 28F | 84a (1) | ND | MIN | |
| 28F | 85a (1) | ND | ||
| 33F | 66b (1) | ND | CHL | |
| 35C | 86a (1) | ND | ||
| 35C | 87a (1) | ND | MIN | |
| 37 | 88a (1) | ND | MIN | |
| 37 | 89a (1) | ND | CHL, SXT | |
| Rough | 90a (1) | 317 | MIN, ERY, CLI, SXT | |
| Rough | 36a (1) | 410 | DOX, CHL |
New STs are shown in boldface; allelic profiles are as follows: ST2136, 13-16-19-5-17-20-26; ST2137, 15-17-4-12-6-1-6; ST2138, 2-12-1-18-17-4-14; ST2139, 16-44-1-16-9-11-8; ST2140, 7-13-8-1-10-6-171; ST2141, 10-25-12-1-6-20-28; ST2184, 1-10-9-18-5-1-6; ST2185, 1-10-9-43-5-1-6. ND, not determined.
CTX, cefotaxime; CHL, chloramphenicol; CLI, clindamycin; DOX, doxycycline; ERY, erythromycin; LVX, levofloxacin; MEM, meropenem; MIN, minocycline; PEN, penicillin; RIF, rifampin; TEL, telithromycin; SXT, trimethoprim-sulfamethoxazole.
ST was determined for selective representative only.
The PFGE analysis revealed 146 various DNA patterns and classified these into 90 types. Among the most prevalent isolates of serotype 19F, 13 PFGE types were present, with type 1 (55 isolates) being highly predominant. Together with PFGE types 2, 10, and 12, type 1 corresponded to ST423 and its single-locus variant (SLV), ST1815. Consideration of three non-type 19F isolates of PFGE type 1 and ST423 as well shows that the ST423 clonal group consisted of 63 tetracycline-resistant isolates (26.0%) altogether. Isolates of serotype 23F showed higher levels of diversity, comprising 19 PFGE types. Almost half of these isolates (26 isolates) were represented by ST602 and its two SLVs, ST440 and ST1479. The isolates of serotype 6B represented four PFGE types. This group was dominated by ST315, which corresponds to international clone Poland6B-20 (16 isolates).
Tetracycline resistance determinants.
All of the tetracycline-resistant isolates possessed the tet(M) gene and were negative by the tet(O) PCR. For the further analysis of tet(M)-carrying transposons, the isolates were split into four phenotypic groups with respect to resistance to tetracycline (TETr), erythromycin (ERYr), and chloramphenicol (CHLr), namely, TETr; TETr and ERYr; TETr and CHLr; and TETr, ERYr, and CHLr. Among the 96 TETr isolates, the int-Tn gene, which codes for the transposase of the Tn916 family of elements, was detected in 91 isolates; and the O13-O14 PCR, positive results of which indicate an intact structure of the Tn916-like transposons behind the tet(M) gene (37), was positive for 94 isolates (PCR product size, ∼600 bp). Forty-eight of the 51 isolates in the TETr and ERYr group were positive by PCR of the int-Tn gene. Four of these most probably carried the Tn1545-like elements, as suggested by the positive O13-O14 PCR result (37) and amplification of the aph3′-III gene promoter (product size, ∼100 bp) and the aph3′-III-erm(B) region (∼3.5 kb). Five further isolates were found to contain the Tn3872-like transposons, being negative by the O13-O14 PCR but yielding products of the expected size in PCRs with primers O13 and O20 (∼1.4 kb), O23 and O14 (∼2.7 kb), and O19 and O22 (∼2.4 kb) (37). The vast majority of the remaining isolates of the TETr and ERYr group were positive by the O13-O14 PCR and possessed the erm(B) gene; however, PCR of the erm(B)-tet(M) region excluded the possibility of the presence of a transposon associated with Tn3951 (28). Two isolates were erm(B) negative but carried the mef(E) gene, which is responsible for erythromycin resistance; and these two isolates were found to contain the Tn2009-like elements, as suggested by the positive PCR with primers MEF4 and O14 (product size, ∼3.3 kb). All of the 82 isolates in the TETr and CHLr group carried the int-Tn gene and were positive by the O13-O14 PCR (37). Only five of these isolates produced the amplicon of the transposase gene specific for the cat gene-containing Tn5252 (product size, ∼900 bp); however, the Tn916-like and Tn5252-like elements did not form a Tn5253-like structure in any of the isolates, as revealed by the lack of a product by PCR mapping of the junction region (1). All of the 13 isolates in the TETr, ERYr, and CHLr group carried the int-Tn gene. Three of these contained the Tn3872-like element and one contained the Tn1545-like element. The presence of an isolate with Tn3951 carrying cat, erm(B), and tet(M) (28) was excluded. One other isolate possessed mef(E) instead of erm(B); however, tet(M) and mef(E) were not located in the Tn2009-like structure (11). The cat gene of this isolate was present in the Tn5252-like transposon but, again, not in the Tn5253-like configuration (1).
The tet(M) PCR-RFLP analysis showed seven polymorphs of the gene with three predominant profiles: profiles D (38.8%), A (27.7%), and G (19.0%). Three of these corresponded to restriction maps of tet(M) alleles previously identified in Tn916 (GenBank accession number U09422; profile G), Tn5251 (GenBank accession number X90939; profile A), and Tn1545 (GenBank accession number X04388; profile B). There was no strict correlation between tet(M) polymorphs and particular transposon variants, as illustrated by the fact that profile B (2.5%) was found in only the Tn916 family of elements, which had no Tn1545-like structure, whereas profile A was observed not only in Tn916-like transposons but also in Tn1545-like and Tn3872-like transposons. The majority of the tet(M) polymorphs were present in multiple S. pneumoniae clones; the only exception was polymorph B, which fully correlated with ST272 and ST2140 (SLVs of Poland23F-16).
Characterization of tetracycline-susceptible isolates.
Since the group of tetracycline-resistant S. pneumoniae isolates contained a highly prevalent fraction of closely related 19F/ST423 isolates, we decided to compare them with isolates of the same serotype from among the remaining 624 tetracycline-susceptible isolates. Among these, only three isolates (0.5%) of serotype 19F were found, and these represented ST423 (two isolates) and unrelated ST2142 (one isolate). One of the ST423 isolates, for which the tetracycline MIC was 2 μg/ml, possessed the tet(M) gene; and its PFGE profile was identical to that of PFGE subtype 1a, which was the most prevalent in the tetracycline-resistant serotype 19F group. The other ST423 and ST2142 isolates had tetracycline MICs of 0.12 μg/ml and lacked the tet(M) and tet(O) genes. The ST423 isolate had a unique PFGE pattern, although its PFGE pattern was related to that of the most frequent subtype, subtype 1a (pattern 1k).
DISCUSSION
Tetracyclines are low-cost antibiotics, and because of that they are attractive for use in countries with limited health care budgets. This could be one of explanations for their high level of consumption in Poland (http://www.esac.ua.ac.be/main.aspx?c=*ESAC2&n=4666), despite the significant prevalence of resistance to these drugs among clinically important pathogens (20, 22). Although tetracycline-resistant S. pneumoniae isolates have been reported in all parts of the world, only a limited number of studies have addressed the question of the genotypic and phenotypic diversities of such isolates. In particular, only fragmentary data of this kind have been available in Poland and other countries of Central and Eastern Europe. In accordance with the findings of previous studies (15, 20, 22), we found a relatively high frequency of resistance to this compound (27.9%; standard deviation, 4.2%), with a slight downward trend over the period from 1999 to 2003. This tendency may be associated with the fact that tetracycline consumption has been kept at a relatively constant level in recent years (4.6 defined daily doses per 1,000 inhabitants per day [DID] in 1997 and 4.5 DID in 2001), whereas the total level of antibiotic use in Poland has been systematically increasing (19.3 DID in 1997 to 27.2 DID in 2001) (http://www.esac.ua.ac.be/main.aspx?c=*ESAC2&n=4666).
A positive correlation between tetracycline MICs and those of doxycycline and minocycline was observed in the analysis, which was in congruence with the findings presented in earlier reports (12, 25). All of the tetracycline-resistant isolates retained clear susceptibility to tigecycline, similar to the findings of previous reports from Europe, North America, and Asia (3, 12, 19, 52; T. S. Bouchillon, T. S., B. Johnson, J. Johnson, D. Hoban, M. Hackel, M. Person, M. Dowzicky, 15th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P805, 2005; T. S. Bouchillon, T. S., B. Johnson, J. Johnson, D. Hoban, M. Hackel, M. Person, M. Dowzicky, 15th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P806, 2005; S. B. Johnson, T. Stevens, J. Johnson, D. Hoban, A. Hsiung, and M. Dowzicky, 15th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P807, 2005). So far no cross-resistance to tetracycline and tigecycline has been observed in pneumococci, and it is possible that the risk of selection of tigecycline-resistant organisms with tetracyclines may be low by definition. These findings, together with those presented in reports on the excellent activity of tigecycline against other agents that cause community-acquired respiratory tract infections (2, 16, 24, 42), position it to be among the promising therapeutic options under certain circumstances for these types of infections (18).
The tet(M) gene was the only determinant of resistance identified in the isolates studied, which was in accordance with the seldom occurrence of the tet(O) gene (30, 51). To our knowledge, in Europe only the tet(M) gene has been detected so far in pneumococci (15, 45). The analysis of the tet(M)-carrying mobile elements revealed their remarkable diversity in the population studied. Two hundred thirty-four isolates (96.7%) carried the int-Tn gene, which codes for the transposase of the Tn916 family of transposons, which has confirmed the role of these conjugative elements as major determinants of tetracycline resistance in S. pneumoniae (34, 39). The sporadic isolates that were negative by the int-Tn PCR could either possess specific structural variants of the Tn916-like elements or, eventually, other transposon types. Interestingly, among the isolates that, apart from their resistance to the tetracyclines, were resistant to erythromycin and/or chloramphenicol, the presence of elements that determine such coresistance that have been described so far seems to be rare, as exemplified by the fact that only eight isolates had a Tn3872-like transposon; five isolates had a Tn1545-like transposon; two isolates had a Tn2009-like transposon; and no isolates had a Tn5253, Tn3951, or Tn2010 transposon (9). It is possible that in most of these isolates, tet(M) resided in Tn916-like elements, whereas the erm(B) and cat genes were located in separate transposons. This could, for example, be the case for several chloramphenicol-resistant isolates, in which cat-containing transposon Tn5252 was detected, but not in association with tet(M), as is the case in Tn5253. However, the possibility could not be excluded that a significant fraction of isolates resistant to tetracycline and erythromycin possessed tet(M) and erm(B) located in other than Tn1545-like or Tn3872-like structures of the Tn916 family, such as Tn3701 (28).
A single S. pneumoniae serotype may include genetically diverse clones that can be distinguished by PFGE and MLST. In this study, a remarkable diversity of PFGE patterns and STs was observed in all major serotypes (serotypes 6A, 6B, 14, 19F, and 23F). Each of these serotypes was dominated by specific clones, namely, ST423 in serotype 19F; ST81, ST440, and ST602 in serotype 23F; ST315 in serotype 6B; ST490 in serotype 6A; and ST143 in serotype 14. This observation suggests that the high prevalence of resistance to tetracycline in S. pneumoniae in Poland is mainly the result of the spread of a relatively few clones, especially ST423 of serotype 19F. The particular role of serotype 19F is underlined by the fact that only 3 of 624 tetracycline-susceptible isolates in the study represented this common serotype, which means that almost each Polish pneumococcus of serotype 19F is resistant to tetracycline. Of the 26 international multiresistant clones of pneumococci recognized so far (33), 17 clones are resistant to tetracycline. In our analysis, representatives of eight such clones (with two SLVs) were detected, and these constituted 24.0% of the isolates. These results demonstrated that, apart from ST423, the international S. pneumoniae clones contribute significantly to the spread of tetracycline resistance as well. The currently available pneumococcal vaccines, PCV7 and PPV23, cover 75.2% and 86.8% of the serotypes of the isolates studied, respectively. However, clones of vaccine serotypes may be gradually replaced by those of nonvaccine serotypes because of, among other reasons, capsule switching (23). It probably occurred among the study isolates, although mostly to serotypes covered by the vaccines; e.g., three ST423 isolates represented serotype 3, 18C, or 23F instead of serotype 19F.
In summary, this work has provided useful comparative data for future studies, presenting the findings of a complex molecular analysis of Polish tetracycline-resistant S. pneumoniae isolates. The observed high frequency of resistance to tetracycline among the S. pneumoniae organisms isolated in Poland appears to arise from the spread of a relatively few epidemic clones that harbor the tet(M) gene, located in transposons of the Tn916 family, as the sole resistance determinant. The data obtained confirmed the observations on the cross-resistance between tetracycline, doxycycline, and minocycline and showed that tigecycline has very good activity against the tetracycline-resistant isolates. Moreover, the results of the study create a strong experimental basis for elaboration of therapeutic recommendations aimed at the prevention of the further dissemination of tetracycline resistance among S. pneumoniae isolates in Poland.
Acknowledgments
This study was supported by the grant from the Polish Ministry of Education and Science (grant 2P05D 095 29).
We acknowledge the use of the S. pneumoniae MLST database, which is located at Imperial College, London, United Kingdom, and which is funded by the Wellcome Trust. We thank Wyeth Research for the tigecycline standard.
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Ayoubi, P., A. O. Kilic, and M. N. Vijayakumar. 1991. Tn5253, the pneumococcal omega (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J. Bacteriol. 173:1617-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betriu, C., M. Gomez, I. Rodriguez-Avial, E. Culebras, and J. J. Picazo. 2005. In vitro activity of tigecycline against ampicillin-resistant Haemophilus influenzae isolates. J. Antimicrob. Chemother. 55:809-810. [DOI] [PubMed] [Google Scholar]
- 3.Betriu, C., I. Rodriguez-Avial, B. A. Sanchez, M. Gomez, J. Alvarez, and J. J. Picazo. 2002. In vitro activities of tigecycline (GAR-936) against recently isolated clinical bacteria in Spain. Antimicrob. Agents Chemother. 46:892-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brito, D. A., M. Ramirez, and H. de Lencastre. 2003. Serotyping Streptococcus pneumoniae by multiplex PCR. J. Clin. Microbiol. 41:2378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; sixteenth informatonal supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Courvalin, P., and C. Carlier. 1987. Tn1545: a conjugative shuttle transposon. Mol. Gen. Genet. 206:259-264. [DOI] [PubMed] [Google Scholar]
- 7.Courvalin, P., and C. Carlier. 1986. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol. Gen. Genet. 205:291-297. [DOI] [PubMed] [Google Scholar]
- 8.David, F., G. de Cespedes, F. Delbos, and T. Horaud. 1993. Diversity of chromosomal genetic elements and gene identification in antibiotic-resistant strains of Streptococcus pneumoniae and Streptococcus bovis. Plasmid 29:147-153. [DOI] [PubMed] [Google Scholar]
- 9.Del Grosso, M., R. Camilli, F. Iannelli, G. Pozzi, and A. Pantosti. 2006. The mef(E)-carrying genetic element (mega) of Streptococcus pneumoniae: insertion sites and association with other genetic elements. Antimicrob. Agents Chemother. 50:3361-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Grosso, M., A. Scotto d'Abusco, F. Iannelli, G. Pozzi, and A. Pantosti. 2004. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshpande, L. M., A. C. Gales, and R. N. Jones. 2001. GAR-936 (9-t-butylglycylamido-minocycline) susceptibility test development for streptococci, Haemophilus influenzae and Neisseria gonorrhoeae: preliminary guidelines and interpretive criteria. Int. J. Antimicrob. Agents 18:29-35. [DOI] [PubMed] [Google Scholar]
- 13.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzierzanowska-Fangrat, K., K. Semczuk, P. Gorska, S. Giedrys-Kalemba, M. Kochman, A. Samet, S. Tyski, D. Dzierzanowska, and K. Trzcinski. 2006. Evidence for tetracycline resistance determinant tet(M) allele replacement in a Streptococcus pneumoniae population of limited geographical origin. Int. J. Antimicrob. Agents 27:159-164. [DOI] [PubMed] [Google Scholar]
- 16.Edelstein, P. H., W. J. Weiss, and M. A. Edelstein. 2003. Activities of tigecycline (GAR-936) against Legionella pneumophila in vitro and in guinea pigs with L. pneumophila pneumonia. Antimicrob. Agents Chemother. 47:533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144(Pt 11):3049-3060. [DOI] [PubMed] [Google Scholar]
- 18.Fritsche, T. R., J. T. Kirby, and R. N. Jones. 2004. In vitro activity of tigecycline (GAR-936) tested against 11,859 recent clinical isolates associated with community-acquired respiratory tract and gram-positive cutaneous infections. Diagn. Microbiol. Infect. Dis. 49:201-209. [DOI] [PubMed] [Google Scholar]
- 19.Hoellman, D. B., G. A. Pankuch, M. R. Jacobs, and P. C. Appelbaum. 2000. Antipneumococcal activities of GAR-936 (a new glycylcycline) compared to those of nine other agents against penicillin-susceptible and-resistant pneumococci. Antimicrob. Agents Chemother. 44:1085-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hryniewicz, W. 2003. Alexander Project—5 years in Poland. Pol. Merkuriusz Lek. 14:5-8. (In Polish.) [PubMed] [Google Scholar]
- 21.Inamine, J. M., and V. Burdett. 1985. Structural organization of a 67-kilobase streptococcal conjugative element mediating multiple antibiotic resistance. J. Bacteriol. 161:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, and R. N. Gruneberg. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 23.Jefferies, J. M., A. Smith, S. C. Clarke, C. Dowson, and T. J. Mitchell. 2004. Genetic analysis of diverse disease-causing pneumococci indicates high levels of diversity within serotypes and capsule switching. J. Clin. Microbiol. 42:5681-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenny, G. E., and F. D. Cartwright. 2001. Susceptibilities of Mycoplasma hominis, M. pneumoniae, and Ureaplasma urealyticum to GAR-936, dalfopristin, dirithromycin, evernimicin, gatifloxacin, linezolid, moxifloxacin, quinupristin-dalfopristin, and telithromycin compared to their susceptibilities to reference macrolides, tetracyclines, and quinolones. Antimicrob. Agents Chemother. 45:2604-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitzis, M. D., A. Ly, and F. W. Goldstein. 2004. In vitro activities of tigecycline (GAR-936) against multidrug-resistant Staphylococcus aureus and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:366-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence, E. R., D. B. Griffiths, S. A. Martin, R. C. George, and L. M. Hall. 2003. Evaluation of semiautomated multiplex PCR assay for determination of Streptococcus pneumoniae serotypes and serogroups. J. Clin. Microbiol. 41:601-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Bouguenec, C., G. de Cespedes, and T. Horaud. 1988. Molecular analysis of a composite chromosomal conjugative element (Tn3701) of Streptococcus pyogenes. J. Bacteriol. 170:3930-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Bouguenec, C., G. de Cespedes, and T. Horaud. 1990. Presence of chromosomal elements resembling the composite structure Tn3701 in streptococci. J. Bacteriol. 172:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luna, V. A., and M. C. Roberts. 1998. The presence of the tetO gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. J. Antimicrob. Chemother. 42:613-619. [DOI] [PubMed] [Google Scholar]
- 31.Marchese, A., E. Tonoli, G. Balistreri, E. Debbia, and G. C. Schito. 2000. Antibiotic susceptibility patterns and serotypes of antibiotic resistant and/or invasive Streptococcus pneumoniae strains circulating in Italy. Microb. Drug Resist. 6:163-170. [DOI] [PubMed] [Google Scholar]
- 32.McDougal, L. K., F. C. Tenover, L. N. Lee, J. K. Rasheed, J. E. Patterson, J. H. Jorgensen, and D. J. LeBlanc. 1998. Detection of Tn917-like sequences within a Tn916-like conjugative transposon (Tn3872) in erythromycin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2312-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montanari, M. P., I. Cochetti, M. Mingoia, and P. E. Varaldo. 2003. Phenotypic and molecular characterization of tetracycline- and erythromycin-resistant strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2236-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. A. Pfaller, R. H. Yolken (ed.). 2003. Manual of clinical microbiology, 8th ed. Washington, DC.
- 36.Pai, R., J. Limor, and B. Beall. 2005. Use of pyrosequencing to differentiate Streptococcus pneumoniae serotypes 6A and 6B. J. Clin. Microbiol. 43:4820-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poyart, C., G. Quesne, P. Acar, P. Berche, and P. Trieu-Cuot. 2000. Characterization of the Tn916-like transposon Tn3872 in a strain of Abiotrophia defectiva (Streptococcus defectivus) causing sequential episodes of endocarditis in a child. Antimicrob. Agents Chemother. 44:790-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1989. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 8:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1991. Nucleotide sequences specific for Tn1545-like conjugative transposons in pneumococci and staphylococci resistant to tetracycline. Antimicrob. Agents Chemother. 35:1657-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Provvedi, R., R. Manganelli, and G. Pozzi. 1996. Characterization of conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol. Lett. 135:231-236. [DOI] [PubMed] [Google Scholar]
- 41.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roblin, P. M., and M. R. Hammerschlag. 2000. In vitro activity of GAR-936 against Chlamydia pneumoniae and Chlamydia trachomatis. Int. J. Antimicrob. Agents 16:61-63. [DOI] [PubMed] [Google Scholar]
- 43.Sadowy, E., R. Izdebski, A. Skoczynska, M. Gniadkowski, and W. Hryniewicz. 2005. High genetic diversity of ciprofloxacin-nonsusceptible isolates of Streptococcus pneumoniae in Poland. Antimicrob. Agents Chemother. 49:2126-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadowy, E., R. Izdebski, A. Skoczynska, P. Grzesiowski, M. Gniadkowski, and W. Hryniewicz. 2007. Phenotypic and molecular analysis of penicillin-nonsusceptible Streptococcus pneumoniae isolates in Poland. Antimicrob. Agents Chemother. 51:40-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz, F. J., M. Perdikouli, A. Beeck, J. Verhoef, and A. C. Fluit. 2001. Molecular surveillance of macrolide, tetracycline and quinolone resistance mechanisms in 1191 clinical European Streptococcus pneumoniae isolates. Int. J. Antimicrob. Agents 18:433-436. [DOI] [PubMed] [Google Scholar]
- 46.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garcia, and R. Gomez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 47.Shiojima, T., Y. Fujiki, H. Sagai, S. Iyobe, and A. Morikawa. 2005. Prevalence of Streptococcus pneumoniae isolates bearing macrolide resistance genes in association with integrase genes of conjugative transposons in Japan. Clin. Microbiol. Infect. 11:808-813. [DOI] [PubMed] [Google Scholar]
- 48.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trzcinski, K., B. S. Cooper, W. Hryniewicz, and C. G. Dowson. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45:763-770. [DOI] [PubMed] [Google Scholar]
- 50.Widdowson, C. A., and K. P. Klugman. 1998. The molecular mechanisms of tetracycline resistance in the pneumococcus. Microb. Drug Resist. 4:79-84. [DOI] [PubMed] [Google Scholar]
- 51.Widdowson, C. A., K. P. Klugman, and D. Hanslo. 1996. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2891-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhanel, G. G., L. Palatnick, K. A. Nichol, T. Bellyou, D. E. Low, and D. J. Hoban. 2003. Antimicrobial resistance in respiratory tract Streptococcus pneumoniae isolates: results of the Canadian Respiratory Organism Susceptibility Study, 1997 to 2002. Antimicrob. Agents Chemother. 47:1867-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]