Abstract
Isolates (3,845) obtained from German adults with invasive pneumococcal disease between 1992 and 2004 were investigated. Of these, 430 isolates (11.2%) were erythromycin A nonsusceptible. Macrolide resistance genotypes and multilocus sequence types were determined. Among the isolates, 35.6% were erm(B) positive and 63.5% were mef positive. Over the study period, the frequency of resistance rose significantly from 2.2 to 17.0% (P < 0.001). A serotype 14, sequence type 9 clone was the most widespread.
Streptococcus pneumoniae continues to be a significant cause of morbidity and mortality in humans. The worldwide increase in antibiotic resistance in pneumococci has become a serious infectious-disease problem within the last 20 years. Macrolide resistance in S. pneumoniae is usually caused by the presence of the erm(B) or the mef(E) or mef(A) resistance determinant. The erm(B) gene encodes a 23S rRNA methylase that confers resistance to 14-, 15-, and 16-member-ring macrolides, lincosamides, and streptogramin B. The mef(E) and mef(A) genes, which are carried by different genetic elements, encode an efflux pump that leads to resistance to 14- and 15-member-ring macrolides (the M phenotype) (14, 20). Other rare mechanisms of macrolide resistance are changes in a highly conserved region of domain V of 23S rRNA, which plays a key role in macrolide binding, and in ribosomal proteins L4 and L22 (3, 5, 18, 21).
Multilocus sequence typing (MLST) is a recently developed technique that produces unambiguous molecular typing data by using the sequences of seven loci to obtain an allelic profile of each strain (6, 11; http://www.mlst.net). The present study used this technique to analyze the genetic relatedness of clinical erythromycin A-resistant strains of S. pneumoniae isolated from adults with invasive pneumococcal disease in Germany.
The German National Reference Center for Streptococci received consecutive isolates from 126 clinical microbiological laboratories throughout Germany. Inclusion criteria were isolation from an individual of >16 years and isolation from a normally sterile body site.
MIC testing, serotyping, the determination of resistance genotypes and phenotypes, and MLST of 62 randomly selected macrolide-resistant strains were performed as described previously (11, 13).
Multilocus sequence types were analyzed using the program eBURST, which displays relationships between closely related isolates of a bacterial species or population. eBURST, unlike cluster diagrams, trees, or dendrograms, uses a simple but appropriate model of bacterial evolution in which an ancestral (or founding) genotype increases in frequency in the population and, while doing so, begins to diversify to produce a cluster of closely related genotypes that are all descended from the founding genotype. This cluster of related genotypes is referred to as a clonal complex (8; http://eburst.mlst.net). A phylogenetic tree using maximum likelihood was created with the program Puzzle (19; http://genius.dkfz-heidelberg.de).
A total of 3,845 isolates were consecutively collected at 126 centers from 1992 to 2004. Of these, 3,134 strains (81.5%) were isolated from blood, 426 (11.1%) were from cerebrospinal fluid, 119 (3.1%) were from pleural fluid, and 166 (4.3%) were from other normally sterile body sites.
Macrolide-resistant S. pneumoniae strains showed cross-resistance to other 14- and 15-member-ring macrolides. All strains were telithromycin susceptible and were inhibited by 1 μg of telithromycin/ml (MIC at which 90% of the tested isolates were inhibited, 0.12 μg/ml); 4% of the isolates were clindamycin resistant. Over the study period (1992 to 2004), a statistically significant increase in the frequency of erythromycin A resistance (2.2% to 17%) was observed (P < 0.001) (Table 1). In total, 153 of the 430 nonsusceptible isolates (35.6%) were erm(B) positive, and 273 (63.5%) were mef positive. Two isolates were both erm(B) and mef positive, and two isolates contained neither erm(B) nor mef.
TABLE 1.
Development of resistance among S. pneumoniae isolates from adults with invasive pneumococcal disease in Germany from 1992 to 2004
| Yr of isolation | % of erythromycin A-resistant isolates | % of mef-positive isolates (% of macrolide-resistant isolates)a | % of erm(B)-positive isolates (% of macrolide-resistant isolates)a |
|---|---|---|---|
| 1992 | 2.2 | 0.7 (31) | 1.5 (69) |
| 1993 | 3.9 | 2.2 (58) | 1.7 (42) |
| 1994 | 3.8 | 1.1 (29) | 2.7 (71) |
| 1995 | 6.9 | 3.1 (45) | 3.9 (55) |
| 1996 | 7.5 | 4.5 (60) | 3.0 (40) |
| 1997 | 12.5 | 7.7 (62) | 4.8 (38) |
| 1998 | 12.1 | 6.7 (55) | 5.5 (45) |
| 1999 | 17.3 | 10.1 (58) | 6.1 (42) |
| 2000 | 15.3 | 8.5 (56) | 6.8 (44) |
| 2001 | 15.2 | 8.2 (54) | 6.7 (46) |
| 2002 | 13.3 | 8.9 (67) | 4.2 (32) |
| 2003 | 15.9 | 11.7 (74) | 4.2 (26) |
| 2004 | 17.0 | 13.3 (78) | 3.9 (22) |
| Total | 11.0 | 6.7 (61) | 4.2 (39) |
Percentage of macrolide-resistant isolates positive for the indicated gene.
The serotyping of resistant isolates resulted in the following distribution: serotype 14, 48.8%; 6B, 9.5%; 23F, 7.7%; 9V, 4.4%; 19A, 4.0%; and 19F, 3.5%. Fewer than 10 isolates of other serotypes were encountered (data not shown).
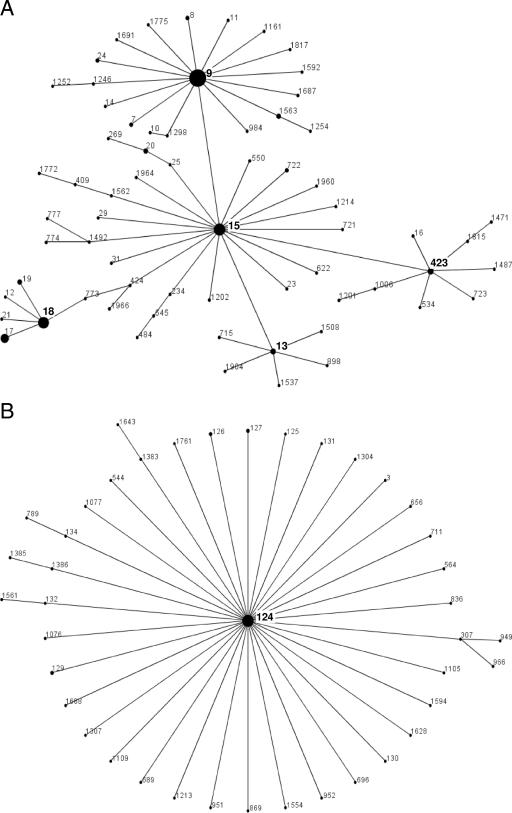
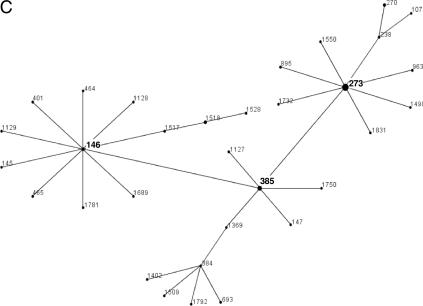
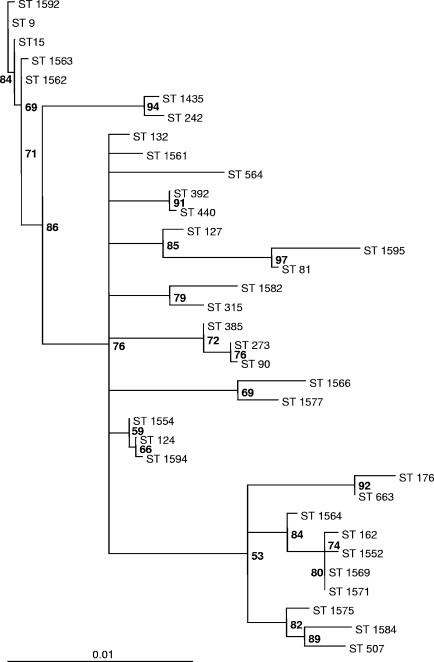
Macrolide resistance was caused by the oligoclonal spread of some multilocus sequence types. Among those, sequence type 9 (ST 9; serotype 14; United Kingdom and Germany) was by far the most important, followed by another serotype 14 clone (ST 124) and a serotype 6B clone (ST 273). In addition, 16 macrolide-resistant clones were described for the first time in this investigation (Table 2). eBURST analysis showed ST 9 to be the cofounder of a large clonal complex also containing ST 15 (Fig. 1A). ST 124 is the founder of a very large clonal complex (Fig. 1B). ST 273 forms a complex with ST 146 and ST 385 (Fig. 1C). All isolates of ST 9 carried the mef(A) gene, and both isolates of ST 242 carried the mef(E) gene. A maximum-likelihood tree of the concatenated alleles of the 35 multilocus sequence types found in this study, developed using the program Puzzle, showed ST 9 and ST 124 to be relatively separated from all other multilocus sequence types (Fig. 2).
TABLE 2.
Distribution of 62 STs of macrolide-resistant pneumococcal isolates from adults with invasive pneumococcal disease in Germany
| STa | Macrolide resistance phenotype,b genotype | Clone designation or descriptionc | No. of isolates | % of total isolates | Predominant country(ies) of origin |
|---|---|---|---|---|---|
| 9 | M, mef(A) | England14-9 | 15 | 24.2 | United Kingdom, Germany |
| 124 | cMLSB, erm(B) | PMEN global clone “clone 35” | 6 | 9.7 | Germany, The Netherlands |
| 273 | cMLSB, erm(B) | PMEN global clone Greece6B-22 | 5 | 8.1 | Portugal |
| 15 | cMLSB, erm(B) | Single-locus variant of England14-9 | 3 | 4.8 | Brazil |
| 81 | cMLSB, erm(B) | Spain23F-1 | 2 | 3.2 | Spain |
| 242 | M, mef(E) | Taiwan23F clone | 2 | 3.2 | Taiwan |
| 90 | cMLSB, erm(B) | Spain6B-2 | 1 | 1.6 | Spain, Australia |
| 127 | cMLSB, erm(B) | 1 | 1.6 | United Kingdom, Germany | |
| 132 | cMLSB, erm(B) | 1 | 1.6 | The Netherlands | |
| 162 | M, mef | 1 | 1.6 | United Kingdom | |
| 176 | M, mef | 1 | 1.6 | Poland | |
| 315 | cMLSB, erm(B) | Poland6B-20 | 1 | 1.6 | Poland |
| 385 | iMLSB, mef | 1 | 1.6 | United States | |
| 392 | cMLSB, erm(B) | 1 | 1.6 | United Kingdom | |
| 440 | cMLSB, erm(B) | 1 | 1.6 | United Kingdom | |
| 507 | M, mef | 1 | 1.6 | Finland | |
| 564 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 663 | M, no erm(B), no mef | 1 | 1.6 | United States | |
| 1435 | M, mef | Single-locus variant of Taiwan23F-15 | 1 | 1.6 | Japan, Germany |
| 1552 | M, mef | 1 | 1.6 | Germany | |
| 1554 | M, mef | 1 | 1.6 | Germany | |
| 1561 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1562 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1563 | M, mef | Single-locus variant of England14-9 | 1 | 1.6 | Germany |
| 1564 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1566 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1569 | cMLSB, erm(B) | Single-locus variant of Spain9V-3 | 1 | 1.6 | Germany |
| 1571 | M, mef | 1 | 1.6 | Germany | |
| 1575 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1577 | cMLSB, erm(B), mef | 1 | 1.6 | Germany | |
| 1582 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1584 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1592 | M, mef | 1 | 1.6 | Germany | |
| 1594 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| 1595 | cMLSB, erm(B) | 1 | 1.6 | Germany | |
| Total | 62 | 100.0 |
New STs first described in the present study are highlighted in bold.
cMLSB, constitutive resistance to 14-, 15-, and 16-member-ring macrolides, lincosamides, and streptogramin B; iMLSB, inducible resistance to14-, 15-, and 16-member-ring macrolides, lincosamides, and streptogramin B.
PMEN, Pneumococcal Molecular Epidemiology Network.
FIG. 1.
(A) ST 9 is part of a clonal complex of 66 STs. The predicted founder is ST 15, but ST 9 is the founder of a large subgroup of 20 different STs. (B) ST 124 is the predicted founder of a group of 41 STs. Isolates of ST 124 from Germany, The Netherlands, Scandinavia, the United Kingdom, Australia, and Canada have been reported. All isolates in the database are reported to be penicillin G and macrolide sensitive, except for the German isolates, which are macrolide resistant. Other members of this group are mostly penicillin G and macrolide sensitive. (C) ST 273 is part of a clonal complex of 32 STs with the predicted founder ST 146. ST 273 is the cofounder of a subgroup of 11 STs.
FIG. 2.
Maximum-likelihood tree of 35 STs found in this study constructed by using the program Puzzle. Numbers at branching points represent the percentages of agreement in 1,000 puzzle steps. Model of substitution, HKY (10). Transition/transversion parameter (estimated from the data set), 7.17 (standard error, 1.34). Nucleotide frequencies (estimated from the data set): A, 27.8%; C, 21.5%; G, 22.6%; T, 28.1%. Expected transition/transversion ratio, 6.10. Expected pyrimidine transition/purine transition ratio, 0.96.
Within the last 10 years, macrolide resistance in S. pneumoniae has emerged on a dramatic scale throughout Europe (4, 16). This study shows that resistance to macrolides in Germany has drastically increased over the last 12 years. In a previous study, our group showed a clear correlation between macrolide consumption and erythromycin resistance in Germany (15). Both the mef and erm resistance determinants could be identified. In 1992, 1994, and 1995, the erm(B) gene was the most predominant. From 1996 until 2004, the mef gene was the main resistance determinant. Of note, only two of the strains were found to be mef(A) and erm(B) negative, suggesting the presence of one of the recently described novel macrolide resistance mechanisms (23S rRNA mutations and alterations in L4 and L22 ribosomal proteins).
eBURST analysis showed that only three clones represented 58% of all macrolide-resistant isolates (ST 9 complex, ST 124 complex, and ST 273 complex). Phylogenetic analysis showed these three complexes as relatively separated subgroups in a maximum-likelihood tree. The prevalence of macrolide resistance genotypes varies substantially among countries. In a recent study, most isolates from France, Spain, Switzerland, and Poland were found to be erm(B) positive, whereas high levels of mef-positive strains from Greece and Germany were reported (17). Clones England14-9 and Taiwan19F-14 are the major contributors to the worldwide dissemination of M phenotype strains (7, 12; http://www.mlst.net). The England14-9 clone harboring mef(A) has been described as being predominant among M phenotype pneumococci isolated in England, Italy, and Greece (1, 9). In contrast, the strains of the England14-9 clone described in the United States carried the mef(E) gene (12). The England14-9 clone described in this paper carried mef(A) and was of ST 9 exclusively, suggesting the clonal spread of this strain in Germany. In a recent report from Spain on isolates obtained from 1998 to 2003, the rate of erythromycin resistance among pneumococci was 34.4%. Interestingly, although the macrolide-lincosamide-streptogramin B resistance phenotype was the most prevalent (94.7%), the frequency of the M phenotype increased from 3.3% to 8.9%. The clonal dissemination of mef(E)-carrying strains of the serotype 14 variant of the Spain9V-3 clone was the major contributor to this increase (2). In August 2006, a general recommendation for the 7-valent pneumococcal conjugate vaccine was issued in Germany; however, it is too early to see any effects.
In summary, the present investigation demonstrates the clonal spread of macrolide-resistant strains in Germany and underscores the high value of MLST in analyzing the genetic relatedness of antibiotic-resistant pneumococcal strains. The results are in accordance with findings of investigators from other countries and indicate that the horizontal spread of the mef gene among clinical pneumococcal isolates may be a major contributor to the emergence of macrolide-resistant pneumococci and is, therefore, a worrying infectious-disease problem with international significance.
Acknowledgments
This study was supported in part by grant RKI-415/1369235 from the German Ministry of Health (Bundesminister für Gesundheit) and the German Ministry for Science and Technology (BMFT; CAP net).
We thank the various microbiological laboratories in Germany for cooperation and for providing the isolates. We thank Nelli Neuberger for excellent technical assistance. We thank Irene Seegmüller and Florian Burckhardt for help with statistical analysis.
Footnotes
Published ahead of print on 26 February 2007.
REFERENCES
- 1.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 403313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardanuy, C., A. Fenoll, S. Berron, L. Calatayud, and J. Linares. 2006. Increase of the M phenotype among erythromycin-resistant Streptococcus pneumoniae isolates from Spain related to the serotype 14 variant of the Spain9V-3 clone. Antimicrob. Agents Chemother. 503162-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decousser, J. W., P. Pina, F. Viguier, F. Picot, P. Courvalin, and P. Allouch. 2004. Invasive Streptococcus pneumoniae in France: antimicrobial resistance, serotype, and molecular epidemiology findings from a monthly national study in 2000 to 2002. Antimicrob. Agents Chemother. 483636-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae identification of clones associated with serious invasive disease. Microbiology 1443049-3060. [DOI] [PubMed] [Google Scholar]
- 7.Farrell, D. J., S. G. Jenkins, S. D. Brown, M. Patel, B. S. Lavin, and K. P. Klugman. 2005. Emergence and spread of Streptococcus pneumoniae with erm(B) and mef(A) resistance. Emerg. Infect. Dis. 11851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fotopoulou, N., P. T. Tassios, D. V. Beste, S. Ioannidou, A. Efstratiou, E. R. Lawrence, J. Papaparaskevas, R. C. George, and N. J. Legakis. 2003. A common clone of erythromycin-resistant Streptococcus pneumoniae in Greece and the UK. Clin. Microbiol. Infect. 9924-929. [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22160-174. [DOI] [PubMed] [Google Scholar]
- 11.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 953140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEllistrem, M. C., J. M. Adams, K. Shutt, L. T. Sanza, R. R. Facklam, C. G. Whitney, J. H. Jorgensen, and L. H. Harrison. 2005. Erythromycin-nonsusceptible Streptococcus pneumoniae in children, 1999-2001. Emerg. Infect. Dis. 11969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montanari, M. P., M. Mingoia, E. Giovanetti, and P. E. Varaldo. 2001. Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J. Clin. Microbiol. 391311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinert, R. R. 2004. Clinical efficacy of ketolides in the treatment of respiratory tract infections. J. Antimicrob. Chemother. 53918-927. [DOI] [PubMed] [Google Scholar]
- 15.Reinert, R. R., A. Al-Lahham, M. Lemperle, C. Tenholte, C. Briefs, S. Haupts, H. H. Gerards, and R. Lütticken. 2002. Emergence of macrolide and penicillin resistance among invasive pneumococcal isolates in Germany. J. Antimicrob. Chemother. 4961-68. [DOI] [PubMed] [Google Scholar]
- 16.Reinert, R. R., S. Reinert, M. van der Linden, M. Y. Cil, A. Al-Lahham, and P. Appelbaum. 2005. Antimicrobial susceptibility of Streptococcus pneumoniae in eight European countries from 2001 to 2003. Antimicrob. Agents Chemother. 492903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinert, R. R., A. Ringelstein, M. van der Linden, M. Y. Cil, A. Al-Lahham, and F. J. Schmitz. 2005. Molecular epidemiology of macrolide-resistant Streptococcus pneumoniae isolates in Europe. J. Clin. Microbiol. 431294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinert, R. R., A. Wild, P. Appelbaum, R. Lütticken, M. Y. Cil, and A. Al-Lahham. 2003. Ribosomal mutations conferring resistance to macrolides in Streptococcus pneumoniae clinical strains isolated in Germany. Antimicrob. Agents Chemother. 472319-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18502-504. [DOI] [PubMed] [Google Scholar]
- 20.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 401817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tait-Kamradt, A., T. Davies, M. Cronan, M. R. Jacobs, P. C. Appelbaum, and J. Sutcliffe. 2000. Mutations in 23S rRNA and ribosomal protein L4 account for resistance in pneumococcal strains selected in vitro by macrolide passage. Antimicrob. Agents Chemother. 442118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]