Abstract
We showed that a Candida albicans petite mutant in which oxidative phosphorylation is uncoupled was eightfold more resistant to fluconazole and voriconazole than SC5314 but equally susceptible to ketoconazole, itraconazole, and amphotericin B. Strain P5 significantly overexpressed MDR1, which likely accounts for the decreased drug susceptibility.
The azole antifungal agents are widely used to treat diverse Candida albicans infections. Resistance to these drugs can contribute to treatment failures (28, 29). The molecular bases of resistance are best characterized for fluconazole (12, 23, 42). Mechanisms include drug efflux due to overexpression of ATP-binding cassette (ABC) transporters encoded by CDR1 and CDR2 (10, 30, 32) and major facilitator transporters encoded by MDR1 and FLU1 (5, 15, 30); increased expression of ERG11, which encodes the target enzyme 14-α-demethylase; mutations in ERG11 (21, 38); and mutations in ERG3 that inactivate Δ5,6 desaturase and cause accumulation of growth-supporting 14α-methylfecosterol (19, 20, 27). More recently, aneuploidy, particularly of chromosome 5, has been implicated and considered likely due to increased copy numbers of ERG11 and other genes (36).
Petite mutant strains of Candida glabrata and Saccharomyces cerevisiae are resistant to fluconazole and other azoles through mechanisms that are not fully defined (1-4, 34, 42). Although a C. albicans petite mutant was tolerant to amphotericin B (14), susceptibility of such strains to azoles is unknown. In a previous study, we serially passed C. albicans strain SC5314 through the spleens of mice and recovered a petite mutant (called P5) in which oxidative phosphorylation was uncoupled (7). The primary objectives of this study were to determine the susceptibility of SC5314 and strain P5 to azoles and, in the event of differences, to study previously characterized mechanisms of resistance.
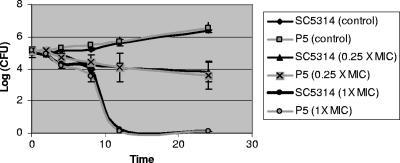
Using a standard broth macrodilution method (24), we found the MICs of fluconazole and voriconazole to be eightfold higher against strain P5 than SC5314 but still within susceptible ranges (Table 1). The MICs of ketoconazole and itraconazole did not differ between the strains, nor did MICs of amphotericin B (Table 1). Moreover, amphotericin B time-kill curves (8) were similar at drug concentrations 0.25× and 1× MIC (Fig. 1).
TABLE 1.
In vitro susceptibilities of strains SC5314 and P5 to antifungal agents and other drugs
| Drug | MIC (μg/ml)a
|
|||
|---|---|---|---|---|
| 24 h
|
48 h
|
|||
| SC5314 | P5 | SC5314 | P5 | |
| Fluconazole | 0.125 | 1.0 | 0.125 | 1.0 |
| Ketoconazole | 0.064 | 0.064 | 0.064 | 0.064 |
| Itraconazole | 0.016 | 0.016 | 0.016 | 0.016 |
| Voriconazole | 0.004 | 0.032 | 0.004 | 0.032 |
| Amphotericin B | 0.06 | 0.06 | 0.125 | 0.125 |
| Brefeldin A | 4 | 16 | 8 | 32 |
| Cerulenin | 2 | 4 | 2 | 8 |
| Rhodamine 6G | 8 | 8 | 8 | 8 |
| Cycloheximide | 400 | 400 | 400 | 800 |
| Nitroquinoline-N-oxide | 0.05 | 0.05 | 0.05 | 0.05 |
| Benomyl | 20 | 20 | 20 | 20 |
| Crystal violet | 0.2 | 0.2 | 0.4 | 0.4 |
| Sulfometuron | 10 | 10 | 10 | 10 |
Values in bold indicate MICs that differ between strains SC5314 and P5.
FIG. 1.
Amphotericin B time-kill curves against strains SC5314 and P5. Data presented are means ± standard deviations from three experiments.
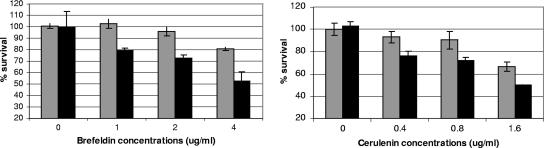
We used a broth microdilution method to measure the MICs of several drugs that are structurally unrelated to fluconazole but subjected to efflux by ABC and/or Mdr1p transporters (Table 1). MICs were identical against both strains for all drugs except brefeldin A and cerulenin. MICs of these agents were consistently 4- and ≥2-fold higher, respectively, against strain P5. As a further measurement of susceptibility, we diluted overnight cultures of the strains (yeast extract-peptone-dextrose at 30°C) to an optical density at 599 nm (OD599) of 0.1 in fresh yeast extract-peptone-dextrose and coincubated them at sub-MIC concentrations of brefeldin A and cerulenin (Fig. 2). As shown, survival of strain P5 was greater at all concentrations.
FIG. 2.
Susceptibility of strains SC5314 (black bars) and P5 (gray bars) to brefeldin A and cerulenin at concentrations below the MIC. Data presented are means ± standard deviations from three experiments.
Using semiquantitative reverse transcription-PCR (RT-PCR) (Table 2), we demonstrated that MDR1 expression was up-regulated in strain P5 and CDR1 was up-regulated in SC5314 (data not shown). Levels of CDR2 expression were very low in both strains. The expression of ERG11, ERG3, and FLU1 did not differ between the strains. To corroborate and better quantify our findings for MDR1, CDR1, and CDR2, we performed real-time RT-PCR (Table 2). We demonstrated that expression of MDR1 by strain P5 was 8.3- to 12.5-fold greater than by SC5314, whereas expression of CDR1 was 3.1- to 5.7-fold lower (Table 3). The expression of CDR2 by strain P5 was slightly higher than by SC5314 (Table 3).
TABLE 2.
Primers and probes used for RT-PCR and real-time PCR
| Gene | Expt | Probe or primers | Sequence(s)a | Reference |
|---|---|---|---|---|
| EFB1 | RT-PCR | Primers | 5′-ATTGAACGAATTCTTGGCTGAC 5′-CATCTTCTTCAACAGCAGCTTG | 35 |
| ERG3 | RT-PCR | Primers | 5′-GGAAGAACCCATCAACTGGATGG 5′-GTGCCACTACTGCCATTCCA | 22 |
| ERG11 | RT-PCR | Primers | 5′-ATTGGTATTCTTATGGGTGGTCAACATAC 5′-CCCAATACATCTATGTCTACCACCACC | 16 |
| CDR1 | RT-PCR | Primers | 5′-TTTCTGGTGCCATGACTCCTGCTAC 5′-CAATATAAATGGCCAAAAAGAATACG | 25 |
| CDR2 | RT-PCR | Primers | 5′-GGGTATTGGCTGGTCCTAATGTGATTC 5′-CTAGCCAACCAGTAAAAGAAAATAGTAA | 25 |
| MDR1 | RT-PCR | Primers | 5′-AGAGCCATCACCGGTAACGACAG 5′-CCAACCAAAAATGAAAAGACCTGAAG | 25 |
| FLUI1 | RT-PCR | Primers | 5′-CACTGCCTTGGCTGGTAAC 5′-ACATCGTGCAAAAGGAAGAAC | 25 |
| CDR1 | Real-time | Probe | 6-FAM-TTAACCCATATGTCAGAAGTGCCCGGG-TAMRA | 6 |
| PCR | Primers | TTTAGCCAGAACTTTCACTCATGATT TATTTATTTCTTCATGTTCATATGGATTGA | 6 | |
| CDR2 | Real-time | Probe | 6-FAM-TCCCGGGTTTTGGATTTTCATGTACAGA-TAMRA | 6 |
| PCR | Primers | GGTATTGGCTGGTCCTAATGTGA GCTTGAATCAAATAAGTGAATGGATTAC | 6 | |
| MDR1 | Real-time | Probe | 6-FAM-TCGCAAGGCTAAAAGATTGAGAGCCATCA-TAMRA | 6 |
| PCR | Primers | TTACCTGAAACTTTTGGCAAAACA ACTTGTGATTCTGTCGTTACCG | 6 |
6-FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
TABLE 3.
Differences in expression of CDR1, CDR2, and MDR1 by strain P5 compared to SC5314, as measured by real-time RT-PCR
| Expt no. | Fold difference in expression
|
||
|---|---|---|---|
| CDR1 | CDR2 | MDR1 | |
| 1 | ↓ 5.74 (3.41-9.65) | ↑ 1.59 (0.96-2.63) | ↑ 12.5 (7.14-20.0) |
| 2 | ↓ 3.10 (1.74-5.50) | ↑ 1.54 (1.01-2.38) | ↑ 8.33 (5.55-12.50) |
The sequence of ERG11 in strain P5 differed at two residues from that of our SC5314 strain and the published sequence (http://www.candidagenome.org/): F14S and A409T. ERG3 sequences did not differ from the published sequence. Using whole-genome single-nucleotide polymorphism (SNP) microarrays to analyze the strains' genotypes (11), we found that the SNP profiles were identical. These results effectively excluded aneuploidy (A. Forche, personal communication).
To our knowledge, this is the first study of azole susceptibility in a C. albicans petite mutant strain. Similar to strain P5, previously described C. glabrata petite mutants have shown diminished susceptibility to fluconazole. Our results differed in several ways from earlier reports, however. Most notably, the C. glabrata petite mutants were fully resistant to both fluconazole and itraconazole, which was attributed to up-regulation of CDR1 with a lesser contribution from CDR2 (2, 3, 34). In strain P5, the diminished susceptibilities to fluconazole and voriconazole were most likely mediated by increased drug efflux due to overexpression of MDR1 and, to a lesser extent, CDR2.
Several pieces of evidence are consistent with a role for MDR1 overexpression. First, MDR1 was found by real-time RT-PCR to be significantly up-regulated in strain P5. Previous studies have shown that activation of MDR1 is associated with diminished fluconazole susceptibility but not necessarily full resistance (40). Second, strain P5 exhibited decreased susceptibility to brefeldin A and cerulenin, drugs that are also effluxed from cells by Mdr1p and by Mdr1p and Cdr2p, respectively (18, 41). Strain P5 and SC5314 were equally susceptible to ketoconazole, itraconazole, benomyl, sulfoneturon, and crystal violet, drugs that are not substrates for Mdr1p (17, 26, 30, 31, 37, 41), cycloheximide, which is pumped by both Mdr1p and ABC transporters, and rhodamine 6G, which is pumped by ABC transporters only. Finally, we excluded other molecular causes of resistance, including overexpression of ERG11, mutations in ERG3, or aneuploidy. The ERG11 mutations in strain P5 have not been previously associated with diminished azole susceptibility (33). Nevertheless, we cannot definitively conclude that they did not contribute to our results, particularly since the levels of drug resistance were low. Indeed, we must acknowledge that C. albicans is likely to possess as-yet-uncharacterized resistance mechanisms (39) which might also have influenced our findings.
Whereas petite mutant yeasts have generally demonstrated diminished azole susceptibility, amphotericin B results have been less consistent. In addition to our findings, amphotericin B-tolerant C. albicans and hypersusceptible C. glabrata strains have been previously reported (2, 3, 9, 13, 14). The diverse types of mitochondrial damage that cause petite mutant phenotypes are known to alter sterol synthesis in different ways (13, 14). Results to date suggest that the alterations to synthetic pathways have more consistent effects on susceptibility to azoles than amphotericin B. Along these lines, strain P5 is potentially a unique tool with which to study relationships between mitochondrial function, oxidative phosphorylation, sterol synthesis, and mechanisms of azole resistance.
Acknowledgments
Experiments were performed in C. Clancy and K. Nguyen's labs at the North Florida/South Georgia Veterans Health System.
This study was supported by the Medical Research Service of the Department of Veterans Affairs and the National Institute of Allergy and Infectious Diseases (NIH PO1 AI061537-01).
We thank Anja Forche and Judith Berman of the University of Minnesota for performing SNP microarray experiments.
Footnotes
Published ahead of print on 26 February 2007.
REFERENCES
- 1.Bouchara, J.-P., R. Zouhair, S. L. Boudouil, G. F. R. Renier, D. Chabasse, J.-N. Hallet, and A. Defonatine. 2000. In-vivo selection of an azole-resistant petite mutant of Candida glabrata. J. Med. Microbiol. 49977-984. [DOI] [PubMed] [Google Scholar]
- 2.Brun, S., C. Aubry, O. Lima, R. Filmon, T. Berges, D. Chabasse, and J. P. Bouchara. 2003. Relationships between respiration and susceptability to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 47847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun, S., T. Berges, P. Poupard, C. Vauzelle-Moreau, D. Chabasse, and J.-P. Bouchara. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 481788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun, S., F. Dalle, P. Saulnier, G. Renier, A. Bonnin, D. Chabasse, and J. P. Bouchara. 2005. Biological consequences of petite mutations in Candida glabrata. J. Antimicrob. Chemother. 56307-314. [DOI] [PubMed] [Google Scholar]
- 5.Calabrese, D., J. Bille, and D. Sanglard. 2000. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 1462743-2754. [DOI] [PubMed] [Google Scholar]
- 6.Chau, A. S., C. A. Mendrick, F. J. Sabatelli, D. Loebenberg, and P. M. McNicholas. 2004. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob. Agents Chemother. 482124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, S., C. J. Clancy, Z. Zhang, B. Hao, W. Wang, K. A. Iczkowski, M. A. Pfaller, and M. H. Nguyen. 2007. Uncoupling of oxidative phosphorylation enables Candida albicans to resist killing by phagocytes and persist in tissue. Cell. Microbiol. 9492-501. [DOI] [PubMed] [Google Scholar]
- 8.Clancy, C. J., H. Huang, S. Cheng, H. Derendorf, and M. H. Nguyen. 2006. Characterizing the effects of caspofungin on Candida albicans, Candida parapsilosis, and Candida glabrata isolates by simultaneous time-kill and postantifungal-effect experiments. Antimicrob. Agents Chemother. 502569-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Defontaine, A., J. P. Bouchara, P. Declerk, C. Planchenault, D. Chabasse, and J. N. Hallet. 1999. In-vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J. Med. Microbiol. 48663-670. [DOI] [PubMed] [Google Scholar]
- 10.Dominique, I. F. M., and M. B. Jacques. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 134405-416. [DOI] [PubMed] [Google Scholar]
- 11.Forche, A., G. May, and P. T. Magee. 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot. Cell 4156-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morshhauser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 423065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geraghty, P., and K. Kavanagh. 2003. Erythromycin, an inhibitor of mitoribosomal protein biosynthesis, alters the amphotericin B susceptibility of Candida albicans. J. Pharm. Pharmocol. 55179-184. [DOI] [PubMed] [Google Scholar]
- 14.Geraghty, P., and K. Kavanagh. 2003. Disruption of mitochondrial function in Candida albicans leads to reduced cellular ergosterol levels and elevated growth in the presence of amphotericin B. Arch. Microbiol. 179295-300. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, V., K., K. Avmeet, P. Shankarling, A. Neeti, S. Panwar, and R. Prasad. 1998. Identification of polymorphic mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance, and its in vitro transcriptional activation. Curr. Genet. 34192-199. [DOI] [PubMed] [Google Scholar]
- 16.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 442693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiller, D., D. Sanglard, and J. Morschhauser. 2006. Overexpression of the MDR1 gene is sufficient to confer increased resistance to toxic compounds in Candida alibicans. Antimicrob. Agents Chemother. 501365-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes, A. R., S. Tsao, S. W. Ong, E. Lampling, K. Niimi, M. C. Monk, M. Niimi, A. Kaneko, B. R. Holland, J. Schmid, and R. D. Cannon. 2006. Heterozygosity and functional allelic variation in the Candida albicans efflux pump genes CDR1 and CDR2. Mol. Microbiol. 62170-186. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, S. L., D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele. 1996. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet 3481523-1524. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, and H. Hebart. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 40080-82. [DOI] [PubMed] [Google Scholar]
- 21.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. S. Ramaekers, F. C. Odds, and H. V. Bossche. 1999. Contribution of mutations in the cytochrome P450 14 α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 1452701-2713. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki, T., Y. Miyazaki, K. Izumikawa, H. Kakeya, S. Miyakoshi, J. E. Bennett, and S. Kohno. 2006. Fluconazole treatment is effective against a Candida albicans erg3/erg3 mutant in vivo despite in vitro resistance. Antimicrob. Agents Chemother. 50580-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morschhauser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta 1587240-248. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. NCCLS document M27-A. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 25.Niewerth, M., D. Kunze, M. Seibold, M. Schaller, H. C. Korting, and B. Hube. 2003. Ciclopirox olamine treatment affects the expression pattern of Candida albicans genes encoding virulence factors, iron metabolism proteins, and drug resistance factors. Antimicrob. Agents Chemother. 471805-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niimi, K., K. Maki, F. Ikeda, A. R. Holmes, E. Lamping, M. Niimi, B. C. Monk, and R. D. Cannon. 2006. Overexpression of Candida alibicans CDR1, CDR2, or MDR1 does not produce significant changes in echinocandin susceptibilty. Antimicrob. Agents Chemother. 501148-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolte, F. S., T. Parkinson, D. J. Falconer, S. Dix, J. Williams, C. Gilmore, R. Geller, and J. R. Wingard. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 41196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaller, M. A., D. J. Diekema, and D. J. Sheehan. 2006. Interpretive breakpoints for fluconazole and Candida revisited: a blueprint for the future of antifungal susceptibility testing. Clin. Microbiol. Rev. 19435-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rex, J. H., M. A. Pfaller, J. N. Galgiani, M. S. Bartlett, A. Espinel-Ingroff, M. A. Ghannoum, M. Lancaster, F. C. Odds, M. G. Rinaldi, T. J. Walsh, A. L. Barry, et al. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24235-247. [DOI] [PubMed] [Google Scholar]
- 30.Sanglard, D., K. Kuchler, F. Ischer, J.-L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 392378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilites of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 402300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143405-416. [DOI] [PubMed] [Google Scholar]
- 33.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1997. Amino acid substitutions in the cytochrome P-450 lanosterol 14 α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to antifungal agents. Antimicrob. Agents Chemother. 42241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 451174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaller, M., W. Schafer, H. C Korting, and B. Hube. 1998. Differential expression of secreted aspartyl proteinases in a model of human oral candidiasis and in patient samples from the oral cavity. Mol. Microbiol. 29605-615. [DOI] [PubMed] [Google Scholar]
- 36.Selmecki, A., A. Forcher, and J. Berman. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313367-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 411482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, T. C., S. Holleman, F. Dy, L. F. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 461704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirsching, S., S. Michel, and J. Morschhauser. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36856-865. [DOI] [PubMed] [Google Scholar]
- 41.Wirsching, S., S. Michel, G. Kohler, and J. Morschhauser. 2000. Activation of the multiple drug resistance gene MDR1 in fluconazole-resistant, clinical Candida albicans strains is caused by mutations in a trans-regulatory factor. J. Bacteriol. 182400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, Y.-L., and J.-L. Lo. 2001. Mechanisms of antifungal agent resistance. J. Microbiol. Immunol. Infect. 3479-86. [PubMed] [Google Scholar]