Abstract
Microbial amino acid biosynthesis is a proven yet underexploited target of antibiotics. The biosynthesis of methionine in particular has been shown to be susceptible to small-molecule inhibition in fungi. The first committed step in Met biosynthesis is the acylation of homoserine (Hse) by the enzyme homoserine transacetylase (HTA). We have identified the MET2 gene of Cryptococcus neoformans H99 that encodes HTA (CnHTA) by complementation of an Escherichia coli metA mutant that lacks the gene encoding homoserine transsuccinylase (HTS). We cloned, expressed, and purified CnHTA and determined its steady-state kinetic parameters for the acetylation of L-Hse by acetyl coenzyme A. We next constructed a MET2 mutant in C. neoformans H99 and tested its growth behavior in Met-deficient media, confirming the expected Met auxotrophy. Furthermore, we used this mutant in a mouse inhalation model of infection and determined that MET2 is required for virulence. This makes fungal HTA a viable target for new antibiotic discovery. We screened a 1,000-compound library of small molecules for HTA inhibitors and report the identification of the first inhibitor of fungal HTA. This work validates HTA as an attractive drug-susceptible target for new antifungal agent design.
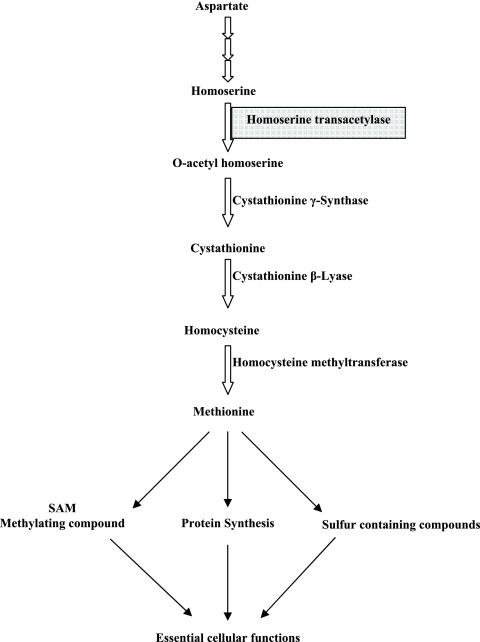
Amino acid biosynthesis is an attractive antimicrobial target because many of these biosynthetic pathways are absent in mammals. The pathway that produces Met, Thr, and Ile from the precursor Asp in bacteria and fungi is one such target (21). Met is an important amino acid due to its involvement in several microbial processes. First, it is essential for protein synthesis and is the N-terminal amino acid in most proteins. Furthermore, Met is important in the synthesis of S-adenosylmethionine, a major methylating agent, and is essential for DNA synthesis and in the synthesis of sulfur-containing compounds (Fig. 1).
FIG. 1.
Met biosynthetic branch of the Asp pathway.
This biosynthetic pathway has been studied as a potential antimicrobial target, and several component enzymes have been validated as novel targets (9, 18, 22, 25). In a search for antifungal agents, Yamaguchi et al. isolated a compound from a Streptomyces sp. that had antifungal activity. This compound, (S)-2-amino-5-hydroxy-4-oxopentanoic acid (RI-331), exhibited antifungal activity against Candida albicans but had no toxic effect when administered to mice (22). Homoserine (Hse) dehydrogenase, the third enzyme in the Asp pathway, was subsequently verified as the target of RI-331 (23, 24). Further chemical validation of the importance of Asp pathway as a potential antimicrobial target is evident in the activities of azoxybacilin (2) and rhizocticin (14). Moreover, Ejim et al. have demonstrated that cystathionine β-lyase is important for virulence of Salmonella enterica serovar Typhimurium (9). Other studies of gene disruption of different targets in Met biosynthesis such as MET3 and MET6 have demonstrated the importance of these genes in the virulence of the pathogenic fungus Cryptococcus neoformans (18, 25).
Based on this precedent, we reasoned that homoserine transacetylase (HTA), the first committed step in Met biosynthesis, would also be a good target for new antimicrobial agents. We have identified this gene in the human pathogen C. neoformans and characterized its gene product. We report its importance in virulence and a small-molecule screen that has identified the first inhibitor of a fungal HTA.
MATERIALS AND METHODS
Isolation and cloning of MET2 cDNA from C. neoformans H99.
A C. neoformans H99 cDNA library constructed with the pBluescript vector (Stratagene) was used to identify the MET2 gene. The cDNA library was transformed into an Escherichia coli metA mutant strain with a nonfunctional homoserine transsuccinylase (HTS) enzyme, which was a generous gift from Barry L. Wanner (Department of Biological Sciences, Purdue University, West Lafayette, IN) and Hirotada Mori (Institute of Science and Technology, Japan). Transformants were selected on M9 minimal medium lacking Met and induced with isopropyl β-d-thiogalactopyranoside (IPTG). The DNA insert in this isolated plasmid was completely sequenced and verified to be the MET2 gene encoding C. neoformans HTA (CnHTA).
This cDNA was amplified from the plasmid using the oligonucleotides (ML8890 and ML8891) in Table 1. The amplified gene was inserted into the TOPO 4-Blunt vector (Invitrogen) according to the manufacturer's instructions, generating a plasmid with the desired MET2 gene flanked by NheI and HindIII restriction enzyme sites. The MET2 gene was then inserted into the NheI and HindIII restriction enzyme sites of the pET28 vector (Novagen) using standard techniques. The resulting plasmid, pET28+ MET2, was used to transform E. coli BL21(DE3) cells, allowing the expression of CnHTA with an N-terminal hexa-histidine tag.
TABLE 1.
DNA oligonucleotides for PCR gene amplification
| Enzyme and oligonucleotide | Direction | Sequence |
|---|---|---|
| MET2-cDNA | ||
| ML8890 | 5′ | GTCTCTAGAGCTAGCATGTCGGACAACACACCCACACCTCA |
| ML8891 | 3′ | CGCGGATCCAAGCTT TCATGATGTGTCCTCTGTTGG |
| MET2-gDNA fragment | ||
| ML9190 | 5′ | AAAACTGCAGGAATTCCATTCTTCTTCGCTCTACATCTCC |
| ML9191 | 3′ | CCGCTCGAGAAGCTTCTCCCTGGTAGCGAAGATAGC |
| Neomycin resistance cassette | ||
| ML8803 | 5′ | GGAAGATCTCTCGAGGCTGCGAGGATGTGAGCTGGA |
| ML8804 | 3′ | GGAAGATCTCTCGAGGGTTTATCTGTATTAACACGGAAGAGATG |
Expression and purification of HTA from C. neoformans H99.
The His-tagged enzyme was expressed in E. coli BL21(DE3)/pET28+ MET2 in 1 liter of Luria-Bertani (LB) broth supplemented with 50 μg/ml kanamycin to an optical density at 600 nm of 0.75 at 37°C. Isopropyl β-d-thiogalactopyranoside was then added to a final concentration of 1 mM, and the culture was incubated for 4 h at 30°C in an orbital shaker. The cells were harvested by centrifugation at 8,000 × g for 10 min, resuspended in a final volume of 25 ml of lysis buffer (50 mM HEPES, pH 8.0, 500 mM NaCl, 20 mM imidazole, and a complete protease inhibitor cocktail tablet [Roche]), and disrupted by three passes through a French pressure cell at 10,000 lb/in2. Cell debris was removed by centrifugation at 12,000 × g for 10 min, and the supernatant was applied to a 1-ml Ni-nitrilotriacetic acid agarose column (QIAGEN) and washed with 20 ml of buffer A (50 mM HEPES, pH 8.0, 500 mM NaCl, 20 mM imidazole). A linear gradient of 20 to 250 mM imidazole in 50 mM HEPES, pH 8.0, plus 500 mM NaCl over a period of 25 min was applied, and CnHTA was eluted between 30 and 50 mM imidazole. Fractions containing the recombinant protein were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
CnHTA enzyme activity.
L-Hse acetylation reaction rates were determined by monitoring the production of free CoA through the change in absorbance at 324 nm via in situ titration with 4, 4′-dithiodipyridine (DTDP; ɛ = 19,800 M−1 cm−1). Steady-state kinetic parameters were determined from assays performed in 50 mM HEPES (pH 8.0) with 2 mM DTDP containing various concentrations of up to 10 mM L-Hse and at a fixed concentration of 1.0 mM acetyl coenzyme A (acetyl-CoA) in a final volume of 200 μl for L-Hse kinetics. The same experiment was performed with various concentrations of up to 1.0 mM acetyl-CoA at a fixed concentration of 10 mM L-Hse for acetyl-CoA kinetics. Progress curves were monitored in a Molecular Devices SpectraMAX Plus spectrophotometer using a 96-well flat-bottom polystyrene microtiter plate (VWR). Initial rates were fit to eqation 1, describing Michaelis-Menten kinetics using GraFit 4.0 software (15):
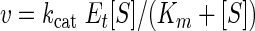 |
(1) |
Disruption of the C. neoformans MET2 gene.
C. neoformans H99 genomic DNA (gDNA) was a generous gift from J. P. Xu (McMaster University, Hamilton, Ontario, Canada). This gDNA was used as a template for the PCR to amplify an approximately 1.3-kb fragment of the MET2 gene using the oligonucleotide primers ML9190 and ML9191 described in Table 1. The amplified gene was cloned into the HindIII and PstI restriction enzyme sites of pBluescript vector (Stratagene) to generate pBluemet2. The neomycin resistance cassette was a generous gift from J. R. Perfect (Duke University, Durham, NC) (11) and was amplified using the oligonucleotides ML8803 and ML8804 outlined in Table 1. The amplicon was cloned into the BglII restriction enzyme site of pBluemet2 vector, which is approximately in the middle of the MET2 gene fragment, thus generating the pBluemet2::neoR disruption construct. This disruption construct allows the presence of 0.6 and 0.7 kb of homologous DNA to the MET2 gene flanking the neomycin cassette, which is sufficient for gene disruption in H99 (17). pBluemet2::neoR was used as a template for PCR amplification with the oligonucleotides used to amplify the MET2-gDNA fragment in Table 1 (ML9190 and ML9191). This linear amplicon was transformed into C. neoformans H99 by biolistic transformation as described by Toffaletti et al. (20). Transformants were incubated overnight at 30°C on yeast extract-peptone-dextrose (YEPD) plates supplemented with 1 M sorbitol. Then the cells were transferred to YEPD plates supplemented with the antibiotic G418 (200 μg/ml). G418-resistant transformants were further screened for Met auxotrophy on synthetic medium lacking Met. G418-resistant colonies incapable of growing without Met were further screened by PCR and Southern hybridization analysis to confirm the presence of the expected MET2 gene disruption. The strain carrying the gene disruption was termed the met2::NeoR strain.
The 1.3-kb MET2 genomic fragment from the PCR amplification described above was delivered to the met2::NeoR strain by biolistic transformation (20). Transformants were selected on synthetic medium lacking Met. Colonies that grew on this medium and that were G418 sensitive were further screened using PCR and Southern hybridization analysis to confirm the presence of the undisrupted MET2 gene. This reconstituted strain was designated the met2::MET2 strain.
H99, met2::NeoR, and met2::MET2 cells were separately grown from freshly isolated colonies from each strain in 3 ml of YEPD at 30°C overnight. Culture concentrations were adjusted to an optical density at 600 nm of unity and were monitored by serial dilutions before being applied as spots to minimal medium plates in the absence and presence of 100 μg/ml Met. Moreover, titration of Met (1,000, 500, 250, 125, 62.5, 31.25, 15.6, 7.81, 3.9, and 2.0 μM) into minimal medium with proline or ammonium sulfate as the sole nitrogen source was used to determine the minimum growth concentration required to rescue the Met auxotrophy.
Test of virulence in the murine inhalation model.
To test the role of MET2 in virulence, the murine cryptococcal inhalation model was used (7). Three groups of 10 4- to 5-week-old female A/Jcr mice were anesthetized with xylazine (5.5 mg/kg of body weight) and ketamine (80 mg/kg of body weight), and then they were suspended on a silk thread via their superior incisors. The mice were inoculated with 50 μl (5 × 104 cells) of wild-type H99, met2::NeoR, or met2::MET2 cells via intranasal instillation (dripping cell suspension into one nare). They were kept on the silk thread for at least 10 min to ensure complete inhalation into the lungs. The mice were subsequently fed ad libitum and were monitored twice daily throughout the experiment. At the first sign of morbidity, each mouse was euthanized by exposure to carbon dioxide following the UBC Animal Care Guidelines (SOP 009E4).
SpHTA ChemDiv kinase inhibitor library screen.
HTA from Schizosaccharomyces pombe (SpHTA) was produced as previously described (3). High-throughput screening was carried out by measuring the change in absorbance at 412 nm due to the production of CoA by the titration of 5, 5′-dithio-bis(2-nitrobenzoic acid) (DTNB) (ɛ = 13,600 M−1 cm−1). After selecting an optimal amount of enzyme giving linear substrate turnover for approximately 4 min, 48 high controls (no inhibitor) and 48 low controls (no enzyme) were measured to establish a Z′ factor for the assay (equation 2) (26):
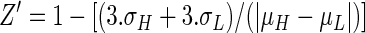 |
(2) |
The ChemDiv kinase inhibitor library (ChemDiv, San Diego, CA) contains 1,000 compounds, all resuspended in dimethyl sulfoxide at a final concentration of 1 mM. Reaction mixtures contained 100 mM HEPES, pH 8.0, 1.3 mM L-Hse, 40 μM acetyl-CoA, 1 mM DTNB, 0.001% Tween 20, and approximately 25 μM inhibitor compounds and were initiated with 54 ng of SpHTA. The final volume of the reactions was 100 μl. Assay plates were set up using a Biomek FX automated liquid handler (Beckman Coulter) at the McMaster High-Throughput Screening Facility (McMaster University, Hamilton, Ontario, Canada). Ninety-six-well flat-bottom polystyrene microtiter plates (VWR) were shaken for 5 s in a SpectraMax Plus 384 plate reader (Molecular Devices) and monitored for 4 min at a wavelength of 412 nm. Progress curve slopes for data analysis were taken from 60 to 150 s to avoid initial noise while still maintaining assay linearity. Data were visualized in a two-dimensional plot of residual enzyme activity for each replicate, calculated as per equation 3:
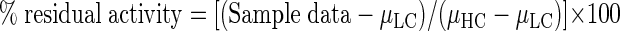 |
(3) |
where μLC is the mean of the low controls and μHC is the mean of the high controls.
FIG. 3.
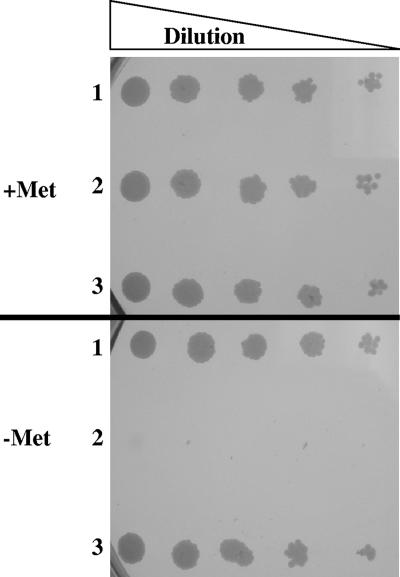
C. neoformans MET2 gene disruption mutant auxotrophy. Growth of H99 (row 1), met2::NeoR, (row 2), and met2::MET2 (row 3) cells on minimal solid medium in the presence or absence of 100 μg/ml of Met to study the effect of supplemental Met on the auxotrophy of the met2::NeoR. mutant. This mutant only grows in the presence of Met, which confirms that the amino acid is essential for rescuing the growth of C. neoformans upon MET2 gene disruption.
Hit compounds were selected for further characterization of inhibition of CnHTA activity. The concentration of inhibitor that was required for 50% inhibition of an enzyme (IC50) was determined by plotting the maximal rate versus inhibitor concentration using GraFit 4.0 software (15). Assay mixtures contained 50 mM HEPES, pH 8.0, 1.5 mM L-Hse, 200 μM acetyl-CoA, 2 mM DTDP, 0.001% Tween 20, and 20 μl of increasing inhibitor concentrations in a final volume of 200 μl. Assays were initiated by the addition of 13.5 ng of enzyme, and the reactions were monitored at a wavelength of 324 nm.
To further characterize the type of CnHTA inhibition, we monitored enzyme activity by varying one of the substrate concentrations and holding the second at a fixed concentration at different amounts of the inhibitor. Data were fit to equations 4 or 5 describing competitive or noncompetitive inhibition, respectively, using the Enzyme Kinetics Module v1.0 of Sigma Plot 2000 (6):
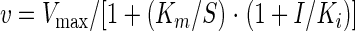 |
(4) |
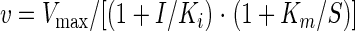 |
(5) |
RESULTS
Cloning of the C. neoformans MET2 gene.
The 19.5-Mb C. neoformans genome sequence (http://cneo.genetics.duke.edu) from Duke University was searched to identify a partial sequence of the MET2 orthologue using the S. pombe gene as a probe. The gene fragment from the C. neoformans genomic data predicted a MET2 gene with two introns. In an effort to obtain the MET2 open reading frame, we turned to a C. neoformans cDNA library constructed in the E. coli-compatible plasmid pBluescript. E. coli does not have an equivalent HTA, but rather activates L-Hse for eventual transformation into cystathionine by O-succinylation catalyzed by HTS, which is encoded by the metA gene. We reasoned, however, that HTA could complement a metA mutant if the next enzyme in the pathway, cystathionine γ-synthase, could accommodate O-acetyl homoserine in addition to O-succinyl homoserine. We therefore introduced the C. neoformans cDNA library into an E. coli metA mutant and selected for growth on media lacking Met. Plasmid DNA was isolated from the positive transformants, and the cDNA insert was sequenced to verify the presence of the C. neoformans MET2 gene.
Characterization of CnHTA.
CnHTA was overexpressed in E. coli BL21(DE3) cells and yielded 5 mg of enzyme from 1 liter of medium. The kinetic parameters of CnHTA were determined by monitoring the production of CoA, which in the presence of DTDP produces a thiolate that has a maximum absorbance at 324 nm. The steady-state kinetic parameters were determined to have a kcat of 122 s−1, kcat/Km_L-Hse of 1.30 × 105 s−1·M−1, and kcat/Km_acetyl-CoA of 8.78 × 105 s−1·M−1. The kcat of this enzyme is about 10-fold higher than that of the enzyme from S. pombe (16). Similarly, the kcat/Km_L-Hse is 100-fold higher for the CnHTA but comparable to the kcat/Km_acetyl-CoA.
Disruption and complementation of the MET2 gene in C. neoformans H99.
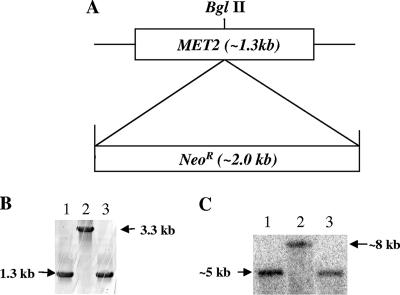
To determine whether the MET2 gene is important for C. neoformans growth in the absence of Met, we disrupted the gene by homologous recombination with a gene fragment in which MET2 was disrupted. A 1.3-kb fragment of the MET2 gene was disrupted by insertion of a neomycin resistance cassette near the center of the gene (Fig. 2A) (11). The disruption construct was introduced into the C. neoformans H99 strain by biolistic transformation (20), and the colonies that grew on YEPD supplemented with 200 μg/ml of G418 were further analyzed by their ability to grow on minimal medium lacking Met. Colonies unable to grow without supplemental Met were analyzed for the presence of the MET2 gene disruption by PCR and Southern hybridization analysis. PCR analysis of the disrupted strain showed the predicted increase in the size of the amplicon by 2.0 kb (Fig. 2B), and Southern hybridization analysis also showed an increase of 2.0 kb (Fig. 2C). The mutant with this construct (met2::NeoR) was used for the rest of the studies as the C. neoformans met2 gene disruption strain.
FIG. 2.
MET2 gene disruption strategy. (A) Diagram of the construction of the MET2 gene disruption construct with the neomycin resistance cassette (NeoR). The NeoR cassette was inserted at approximately the middle of the MET2 gene fragment. (B) PCR screen of the H99 (lane 1), met2::NeoR (lane 2), and met2::MET2 (lane 3) strains, which confirms the expected increase in the size of the MET2 gene upon the gene disruption with NeoR in the C. neoformans mutant. (C) Southern hybridization analysis of H99 (lane 1), met2::NeoR (lane 2), and met2::MET2 (lane 3) cells digested with BamHI. This also confirms the presence of the MET2 gene disruption by the increase in the fragment size upon NeoR insertion.
The met2::NeoR strain was complemented by introduction of the 1.3-kb amplicon of pBluemet2 by biolistic transformation (20). The transformants were selected on minimal medium lacking Met, and the positive transformants were further analyzed by PCR and Southern hybridization analysis to identify those with wild-type characteristics (Fig. 2B and C).
To determine the reliance of C. neoformans met2::NeoR strain growth on an exogenous Met source, we tested the growth of met2::NeoR on minimal medium plates in the presence and absence of Met. It was evident from the growth assay shown in Fig. 3 that the met2::NeoR strain could not grow in the absence of Met. This indicates the essentiality of the MET2 gene for the growth of C. neoformans in Met-deficient environments. Further experimentation revealed that when proline was the sole nitrogen source, as opposed to ammonium sulfate, then addition of 62.5 μM Met was required to rescue the met2::NeoR auxotrophy. Meanwhile, 125 μM of Met was required to rescue this auxotrophy in the presence of ammonium sulfate as the sole nitrogen source. This condition could be attributed to the low Met uptake by the cell in the presence of ammonium sulfate as the sole nitrogen source due to nitrogen repression of amino acid uptake. This effect on Met uptake had been previously studied for MET3 and MET6 gene disruptions in C. neoformans (18, 25).
MET2 is required for C. neoformans infection in the murine inhalation model.
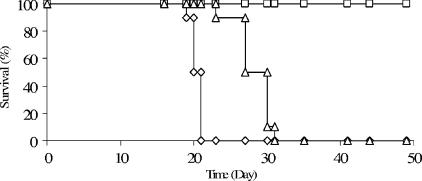
To investigate the role of MET2 in virulence, the mutant strain along with the wild-type and reconstituted strains were inoculated into mice via the nasal inhalation model (7). The survival curves shown in Fig. 4 revealed that mice infected with the wild-type strain H99 were all sacrificed upon showing signs of morbidity by the 21st day postinfection. Similarly, the met2::MET2-infected mice were all sacrificed upon showing signs of morbidity by the 31st day postinfection, indicating complementation by the wild-type allele. In contrast, mice infected with the met2::NeoR strain all survived for the entire experimental observation time postinfection. These results indicate that disruption of the MET2 gene in C. neoformans attenuates virulence in the murine inhalation model.
FIG. 4.
Murine inhalation model used to test the virulence of a MET2 gene disruption. Three groups of 10 female A/Jcr mice were inoculated with wild-type H99 (⋄), met2::NeoR (□), or met2::MET2 (Δ) cells intranasally. The time of mouse survival with an infection was used as a measure to identify the virulence of the met2::NeoR strain compared to that of the wild type. The observation period of mouse survival was a total of 50 days postinfection.
Small-molecule screen identifies inhibitors of HTA.
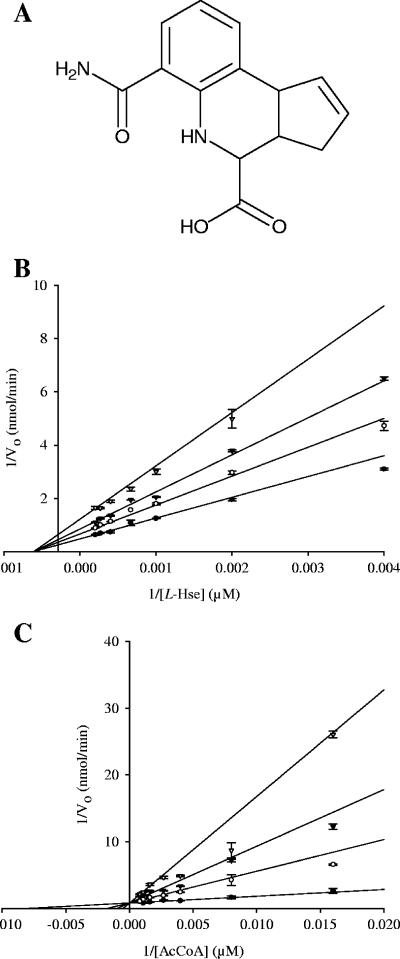
SpHTA was screened to identify inhibitors that halt its catalytic activity. Statistical analysis of high and low control assays returned a Z′ value of 0.72, indicating the assay is robust and suitable for screening. We screened a small (1,000 compound) protein kinase inhibitor library from ChemDiv, reasoning that these protein kinase inhibitors are built around a scaffold that mimics the nucleotide substrate and that these could therefore also bind to the nucleotide recognition region of the CoA binding site of HTA. Each compound was tested in duplicate and hence should render residual activities that are in agreement and fall on a line with a slope of 1. Compounds within the 50% inhibition hit zone were selected for further analysis. In total, 40 compounds were examined to identify inhibitors which were competitive for acetyl-CoA. This was accomplished by screening at two different concentrations of acetyl-CoA, 10 and 100 μM. Ideally, a competitive inhibitor for acetyl-CoA would be less effective in the presence of more substrate, yielding an increase in residual activity. Four compounds that displayed this trend were selected for further IC50 studies, but solubility problems hindered further studies on three of them. The IC50 values exhibited by 6-carbamoyl-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-4-carboxylic acid (CTCQC) (Fig. 5A) were 156 μM and 287 μM in the presence of 10 μM and 100 μM of acetyl-CoA, respectively. This compound was further used for more testing against CnHTA.
FIG. 5.
Inhibition of CnHTA by CTCQC. Panel A shows the chemical structure of CTCQC. Panel B is a Lineweaver-Burk plot of the inhibition by CTCQC versus acetyl-CoA (Ac-CoA). The intersecting lines at the y axes confirm that the compound is a competitive inhibitor of acetyl-CoA. Panel C is a Lineweaver-Burk plot of the inhibition by CTCQC versus L-Hse. This pattern of intersecting lines in the second quadrant demonstrates that the compound is a noncompetitive inhibitor of L-Hse. Inhibitor CTCQC concentrations: 0 μM (•), 50 μM (○), 100 μM (▾), and 200 μM (▿).
Characterization of CTCQC inhibition of CnHTA.
The IC50 value of CnHTA by CTCQC was determined to be approximately 4.50 μM. We further investigated the mode of CnHTA inhibition by CTCQC using a standard steady-state approach. Lineweaver-Burk plots of CnHTA activity at different concentrations of CTCQC were generated for both substrates and fit to various models of inhibition. These plots of the enzyme activity at increasing amounts of acetyl-CoA and saturating amounts of the L-Hse in the presence of increasing amounts of compound present intersecting lines at the y axes (Fig. 5B). This indicates that CTCQC is a competitive inhibitor of acetyl-CoA with a Ki value of 13.6 ± 2 μM. The same plots of the enzyme activity at increasing amounts of L-Hse and saturating amounts of acetyl-CoA in the presence of increasing amounts of CTCQC present intersecting lines in the second quadrant (Fig. 5C). This pattern of intersecting lines indicates that CTCQC acts as a noncompetitive inhibitor of L-Hse with a Ki value of 91.7 ± 11 μM (19). CTCQC had no effect on C. neoformans growth in minimal medium up to 128 μg/ml. The poor bioavailability of the compound could be due to a number of issues, including transport (influx or efflux of the compound) or metabolism to an inactive form.
DISCUSSION
Met is required for several biochemical processes, and as a result, it is essential for cell growth (25). Met is a required amino acid in mammals and must acquire it through diet, but it is synthesized in bacteria, fungi, and plants via the Asp biosynthetic pathway (21). The absence of Met biosynthesis in humans makes the associated biosynthetic enzymes attractive targets for antimicrobial discovery. This is also supported by the fact that the bioavailability of Met is too low in humans (20 μM) (10) to support C. neoformans growth.
Several research groups have studied different enzymes of the Asp pathway and revealed that they could be good candidates for antimicrobial drugs (9, 18, 22, 25). HTA is the first committed step in the biosynthesis of Met in fungi, gram-positive bacteria and some gram-negative bacteria (1). Due to the importance of Met biosynthesis in different microbes, we studied its role in virulence and inhibition by targeting HTA in the human pathogen C. neoformans.
CnHTA is the product of the MET2 gene in C. neoformans H99, which we isolated by complementing an E. coli metA mutant that is incapable of producing HTS. These two enzymes catalyze the acylation of the hydroxyl group of L-Hse, which is ultimately converted into Met. Although the two enzymes have a similar function, they share no significant primary sequence homology (4, 5). We demonstrate here for the first time that CnHTA can complement an E. coli HTS-null mutant, and we used this strategy to identify the C. neoformans MET2 open reading frame. Overexpression of MET2 in E. coli BL21(DE3) cells allowed us to confirm homoserine acetylation using steady-state kinetics after enzyme purification.
The expected role of CnHTA as an important enzyme in Met biosynthesis in C. neoformans was confirmed by disrupting the MET2 gene in this organism, which results in the predicted Met auxotrophy. As expected, the met2::NeoR mutant was not capable of growing in the absence of Met in culture media. Interestingly, when Met was added to the cultures, the mutant did not grow as well in medium supplemented with ammonium sulfate as the source of nitrogen instead of proline. Nitrogen sources have been shown to negatively affect complementation of amino acid auxotrophy (12, 13), and ammonium sulfate has been specifically shown to inhibit suppression of Met auxotrophy (18). Therefore, Met can complement the met2 mutation and allows the growth of the mutant, confirming that the MET2 gene is essential for Met production in C. neoformans. Moreover, MET2 is an essential C. neoformans gene in an environment lacking threshold concentrations of Met.
To determine the role of MET2 in virulence, the wild-type, met2::NeoR, and met2::MET2 strains were studied in a standard mouse inhalation model of infection. The wild-type H99 strain and reconstituted met2::MET2 strain were virulent, causing complete mortality to the mice in the first 21 and 31 days postinfection, respectively. The met2-null mutant strain showed attenuated virulence, with no visible signs of a pathogenic effect on the mice over the entire period (50 days) of observation postinfection. This result is encouraging with regard to drug discovery because it reveals for the first time that HTA is a viable drug target in antifungal research.
The disruption of the MET2 gene leading to a nonfunctional CnHTA supports our hypothesis that blocking the mechanism of the enzyme is detrimental to the survival of the microbe. Therefore, we followed this up with an in vitro small-molecule screen using a library of compounds directed towards protein kinase inhibition based on a purine scaffold. The reasoning was that these could interact with the purine binding site of the acetyl-CoA substrate. We identified CTCQC as a competitive inhibitor for acetyl-CoA and noncompetitive inhibitor of homoserine, consistent with our hypothesis. This is the first reported inhibitor of this enzyme and sets the stage for downstream elaboration of this hit compound to improve bioactivity.
Many studies have identified various targets in the Asp pathway as possible routes for drug discovery (8, 9, 18, 22, 25). Our work reveals for the first time that although HTA and HTS have no primary sequence homology, CnHTA can rescue an HTS-null mutant by ultimately producing Met. Moreover, CnHTA presents a novel target that is essential in Met-depleted environments. It provides a novel target for antimicrobial development in an era of increased resistance and with a desperate need for effective drugs.
Acknowledgments
The authors wish to thank Jonathan Cechetto, Jan Blanchard, and Nadine Elowe of the McMaster High Throughput Screening Laboratory for substructure searches and helpful discussions. The authors would like to thank J. R. Perfect for his generous donation of the neomycin resistance cassette and J. P. Xu for providing the C. neoformans H99 strain. Special thanks are extended to Ken Kasha and Youn-Seb Shim at the University of Guelph for providing help with the biolistic transformation machinery.
This research was supported by the Canadian Institute of Health Research, Crompton Co./Cie., and by a Canada Research Chair in Antibiotic Biochemistry to G.D.W.
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Andersen, G. L., G. A. Beattie, and S. E. Lindow. 1998. Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J. Bacteriol. 1804497-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, Y., M. Yamamoto, S. M. Hosseini-Mazinani, N. Koshikawa, K. Sugimoto, and M. Arisawa. 1996. Antifungal azoxybacilin exhibits activity by inhibiting gene expression of sulfite reductase. Antimicrob. Agents Chemother. 40127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bareich, D. C., I. Nazi, and G. D. Wright. 2003. Simultaneous in vitro assay of the first four enzymes in the fungal aspartate pathway identifies a new class of aspartate kinase inhibitor. Chem. Biol. 10967-973. [DOI] [PubMed] [Google Scholar]
- 4.Born, T. L., and J. S. Blanchard. 1999. Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded homoserine transsuccinylase. Biochemistry 3814416-14423. [DOI] [PubMed] [Google Scholar]
- 5.Born, T. L., M. Franklin, and J. S. Blanchard. 2000. Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Haemophilus influenzae met2-encoded homoserine transacetylase. Biochemistry 398556-8564. [DOI] [PubMed] [Google Scholar]
- 6.Brannan, T., B. Althoff, L. Jacobs, J. Norby, and S. Rubenstein. 2000. Sigma Plot, 6th ed. SSPS, Inc., Chicago, IL.
- 7.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ejim, L., I. A. Mirza, C. Capone, I. Nazi, S. Jenkins, G. L. Chee, A. M. Berghuis, and G. D. Wright. 2004. New phenolic inhibitors of yeast homoserine dehydrogenase. Bioorg. Med. Chem. 123825-3830. [DOI] [PubMed] [Google Scholar]
- 9.Ejim, L. J., V. M. D'Costa, N. H. Elowe, J. C. Loredo-Osti, D. Malo, and G. D. Wright. 2004. Cystathionine β-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 723310-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasman, G. D. 1976. Handbook of biochemistry and molecular biology, 3rd ed. CRC Press, Cleveland, OH.
- 11.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 21036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kingsbury, J. M., Z. Yang, T. M. Ganous, G. M. Cox, and J. H. McCusker. 2004. Cryptococcus neoformans Ilv2p confers resistance to sulfometuron methyl and is required for survival at 37 degrees C and in vivo. Microbiology 1501547-1558. [DOI] [PubMed] [Google Scholar]
- 13.Kingsbury, J. M., Z. Yang, T. M. Ganous, G. M. Cox, and J. H. McCusker. 2004. Novel chimeric spermidine synthase-saccharopine dehydrogenase gene (SPE3-LYS9) in the human pathogen Cryptococcus neoformans. Eukaryot. Cell 3752-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kugler, M., W. Loeffler, C. Rapp, A. Kern, and G. Jung. 1990. Rhizocticin A, an antifungal phosphono-oligopeptide of Bacillus subtilis ATCC 6633: biological properties. Arch. Microbiol. 153276-281. [DOI] [PubMed] [Google Scholar]
- 15.Leatherbarrow, R. J. 2001. GraFit, 4.0 ed. Erithacus Software Ltd., Staines, United Kingdom.
- 16.Nazi, I., and G. D. Wright. 2005. Catalytic mechanism of fungal homoserine transacetylase. Biochemistry 4413560-13566. [DOI] [PubMed] [Google Scholar]
- 17.Nelson, R. T., B. A. Pryor, and J. K. Lodge. 2003. Sequence length required for homologous recombination in Cryptococcus neoformans. Fungal Genet. Biol. 381-9. [DOI] [PubMed] [Google Scholar]
- 18.Pascon, R. C., T. M. Ganous, J. M. Kingsbury, G. M. Cox, and J. H. McCusker. 2004. Cryptococcus neoformans methionine synthase: expression analysis and requirement for virulence. Microbiology 1503013-3023. [DOI] [PubMed] [Google Scholar]
- 19.Segel, I. H. 1993. John Wiley & Sons, Inc., New York, NY.
- 20.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1751405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umbarger, H. E. 1978. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 47532-606. [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi, H., K. Uchida, T. Hiratani, T. Nagate, N. Watanabe, and S. Omura. 1988. RI-331, a new antifungal antibiotic. Ann. N. Y. Acad. Sci. 544188-190. [DOI] [PubMed] [Google Scholar]
- 23.Yamaki, H., M. Yamaguchi, H. Imamura, H. Suzuki, T. Nishimura, H. Saito, and H. Yamaguchi. 1990. The mechanism of antifungal action of (S)-2-amino-4-oxo-5-hydroxypentanoic acid, RI-331: the inhibition of homoserine dehydrogenase in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 168837-843. [DOI] [PubMed] [Google Scholar]
- 24.Yamaki, H., M. Yamaguchi, T. Tsuruo, and H. Yamaguchi. 1992. Mechanism of action of an antifungal antibiotic, RI-331, (S) 2-amino-4-oxo-5-hydroxypentanoic acid: kinetics of inactivation of homoserine dehydrogenase from Saccharomyces cerevisiae. J. Antibiot. (Tokyo) 45750-755. [DOI] [PubMed] [Google Scholar]
- 25.Yang, Z., R. C. Pascon, A. Alspaugh, G. M. Cox, and J. H. McCusker. 2002. Molecular and genetic analysis of the Cryptococcus neoformans MET3 gene and a met3 mutant. Microbiology 1482617-2625. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 467-73. [DOI] [PubMed] [Google Scholar]